Abstract
To determine the relative fitness of oseltamivir-resistant strains compared to susceptible wild-type viruses, we combined mathematical modeling and statistical techniques with a novel in vivo “competitive-mixtures” experimental model. Ferrets were coinfected with either pure populations (100% susceptible wild-type or 100% oseltamivir-resistant mutant virus) or mixed populations of wild-type and oseltamivir-resistant influenza viruses (80%:20%, 50%:50%, and 20%:80%) at equivalent infectivity titers, and the changes in the relative proportions of those two viruses were monitored over the course of the infection during within-host and over host-to-host transmission events in a ferret contact model. Coinfection of ferrets with mixtures of an oseltamivir-resistant R292K mutant A(H3N2) virus and a R292 oseltamivir-susceptible wild-type virus demonstrated that the R292K mutant virus was rapidly outgrown by the R292 wild-type virus in artificially infected donor ferrets and did not transmit to any of the recipient ferrets. The competitive-mixtures model was also used to investigate the fitness of the seasonal A(H1N1) oseltamivir-resistant H274Y mutant and showed that within infected ferrets the H274Y mutant virus was marginally outgrown by the wild-type strain but demonstrated equivalent transmissibility between ferrets. This novel in vivo experimental method and accompanying mathematical analysis provide greater insight into the relative fitness, both within the host and between hosts, of two different influenza virus strains compared to more traditional methods that infect ferrets with only pure populations of viruses. Our statistical inferences are essential for the development of the next generation of mathematical models of the emergence and spread of oseltamivir-resistant influenza in human populations.
The neuraminidase (NA) inhibitors are a class of influenza antiviral drugs that are specifically designed to inhibit the enzymatic function of the NA, thereby preventing normal viral replication. Since 1999, two NA inhibitors (NAIs), oseltamivir (Tamiflu) and zanamivir (Relenza), have been shown to be effective for the treatment and prophylaxis of patients infected with not only seasonal influenza, but also highly pathogenic A(H5N1) and the newly emerged A(H1N1) pandemic virus. Prior to 2007, resistance to this class of drugs was considered relatively uncommon, particularly in comparison with the other class of influenza antivirals, the adamantanes, which readily select for viral resistance in treated patients. During early clinical trials, oseltamivir resistance was detected in only 1 to 2% of adults (14) and 5 to 6% of children (33) under treatment, although later studies detected resistance in up to 18% of oseltamivir-treated children (16). In contrast, resistance following zanamivir treatment is rare, with only one reported case observed in an immunocompromised patient (6). Influenza viruses that develop resistance to these drugs typically contain mutations in the NA which, either directly or indirectly, alter the shape of the NA enzymatic site, thereby reducing the ability of the drugs to bind to this specific pocket. One of the most commonly observed mutations in oseltamivir-resistant A(H3N2) viruses is an arginine-to-lysine mutation at residue 292 (R292K) of the NA, while the predominant NA mutation in oseltamivir-resistant A(H1N1) viruses is a histidine-to-tyrosine mutation at residue 274 (H274Y) (N2 NA amino acid numbering, equivalent to residue 275 based on N1 numbering). Both of these mutations have an indirect impact on drug binding, as they affect the ability of the glutamic acid residue at position 276 to reorientate, as required for slow binding by oseltamivir (3). Many mutations that cause NAI resistance also cause reduced NA enzyme activity and, consequently, can compromise viral fitness.
Previous studies have demonstrated that viruses with an R292K NA mutation demonstrated compromised growth in vitro (36) and in ferrets were significantly less infectious and did not transmit (9). The replication and transmission fitness of the H274Y mutation has also been studied previously. An H274Y mutant A(H1N1) strain isolated from a patient under oseltamivir treatment demonstrated compromised growth in cell culture compared to a wild-type (WT) virus (13), although a strain carrying the same mutation selected in vitro was found to replicate as well as the wild type (32). The infectivity and transmissibility of an H274Y mutant were found to be restricted in ferrets (13), although a second study demonstrated that transmission of the mutant virus between ferrets was possible, but required a greater viral dose of the mutant compared to the wild type (10). These results suggest that resistant virus variants with the same NA mutation may differ in replication or transmission fitness depending on other viral components. Nevertheless, based on these data and the viral fitness of other resistant mutants, it was believed that NAI-resistant viruses were unlikely to spread throughout the community due to their compromised viral fitness in the absence of drug selective pressure. This was proven incorrect during the Northern Hemisphere 2007-2008 influenza season, when large numbers of oseltamivir-resistant seasonal A(H1N1) viruses with an H274Y mutation were detected in patients who had not been treated with oseltamivir (4, 24). The mutant strain continued to spread to the Southern Hemisphere, such that by late 2008 virtually all circulating seasonal A(H1N1) viruses were oseltamivir resistant (11). The rapid global spread of this strain clearly suggested that the oseltamivir-resistant seasonal A(H1N1) virus had fitness equivalent to or greater than that of the previous oseltamivir-sensitive A(H1N1) strain. The reasons for enhanced viral fitness in this strain, when previous studies demonstrated that the acquisition of an H274Y mutation led to reduced viral fitness, remain unclear but probably involve compensatory mutations or reassortment events which may have improved the hemagglutinin (HA)/NA balance, allowing efficient transmission (5, 26).
Experimental methods have been developed to assess the relative fitness of NAI-resistant strains compared with respective wild-type viruses, both in vitro and in vivo. Ferrets have been considered the most appropriate model animal for influenza research, and fitness studies have assessed variables such as minimum dose required to achieve infection, duration of viral shedding, and levels of viral load to allow comparisons between viruses. The guinea pig model has also been previously used to assess the viral fitness of influenza viruses, particularly in comparing the transmissibility of strains via either the contact or aerosol route (2). As an alternative to these traditional approaches, we have investigated a methodology that involves coinfection of ferrets with a mixture of two influenza viruses. Daily monitoring of changes in the relative proportion of those viruses over the course of the infection allows determination of the relative replication fitness of the viruses. Monitoring of recipient ferrets exposed to the infected ferrets enables the relative transmissibility of the viruses (henceforth, the relative transmission fitness) to be determined. In this study, the “competitive-mixtures” methodology was used to assess the relative replication and transmission fitness of an oseltamivir-resistant R292K mutant A(H3N2) virus compared with an oseltamivir-sensitive A(H3N2) wild-type strain and also to asses the relative replication and transmission fitness of an oseltamivir-resistant H274Y seasonal A(H1N1) mutant compared with an oseltamivir-sensitive A(H1N1) wild-type strain. Quantitative estimates for the replication fitness of mutant viruses were determined using a simple mathematical model of within-host viral replication and mixed-effects statistical tests. Transmission fitness was evaluated by application of a graphical technique that demonstrated the relationship between the proportion of mutant virus in the infectee ferrets as a function of the proportion of mutant virus in the infector ferrets.
Inferences drawn from the statistical analyses presented here are essential for the refinement of existing mathematical models that simulate the spread of influenza in the human population and model the deployment of antiviral agents. These models are designed to assess the likely impact of different antiviral agent deployment strategies to control pandemic influenza (18, 21, 35). At present, data on the probability of emergence of NAI-resistant strains, the relative transmission fitness of these strains, and the probability of an individual's infection reverting to an NAI-sensitive strain in the absence of ongoing selective pressure are severely limited. In consequence, human population-level models of influenza spread must make gross assumptions on the likely characteristics of NAI-resistant strains. Data such as those presented here will be used to inform new models of drug deployment and result in improved pandemic policy advice (20, 23).
MATERIALS AND METHODS
Viruses.
Two oseltamivir-resistant viruses were paired with respective wild-type viruses. The first pair of viruses was composed of a wild-type (WT) NAI-sensitive strain, A/Fukui/20/2004 A(H3N2) (referred to as “R292 WT”), and an oseltamivir-resistant mutant, A/Okayama/23/2004 A(H3N2), which contained an R292K NA mutation (referred to as “R292K MUT”). The second pair of viruses was composed of a wild-type NAI-susceptible seasonal A(H1N1) strain, A/Brisbane/59/2007 (referred to as “H274 WT”), and an oseltamivir-resistant seasonal A(H1N1) strain, A/Sydney/142/2007, which contained an H274Y NA mutation (N2 numbering) (referred to as “H274Y MUT”). Each of the viruses was isolated in Madin-Darby canine kidney (MDCK) cells (obtained from ATCC; #CCL-34) and had undergone at least two further passages in MDCK cells prior to two rounds of plaque purification in MDCK cells to achieve a homogeneous population. The viruses were then expanded once to achieve sufficient stock material for in vitro and in vivo experiments. The infectivity titers of the R292 WT, R292K MUT, H274 WT, and H274Y MUT stock viruses were 2.69 × 106, 1.58 × 106, 3.16 × 108, and 3.16 × 108 50% tissue culture infectious doses (TCID50)/ml, respectively.
Full-genome sequence analysis of the viruses demonstrated a high genetic homology between each pair. The H274 WT and H274Y MUT viruses shared 99.8% amino acid similarity across the entire genome. Comparison of each segment for the H274 WT and H274Y MUT viruses revealed no amino acid differences in the HA and M segments (100% similarity) and two changes in the NA: the H274Y mutation (that confers oseltamivir resistance) and a G354D mutation (which has previously been shown to be associated with H274Y mutant viruses [26]). A high genetic similarity in the PB1, PB2, PA, NP, and NS segments was also observed between the H274Y WT and H274Y MUT strains, with amino acid homology ranging from 99.7% to 99.9%. The R292 WT and R292K MUT viruses also showed a high degree of similarity, although slightly lower than that seen with the H274 WT and H274Y MUT viruses, with 98.8% amino acid homology across the entire genome and segment-specific homology ranging from 96.6% to 99.6%. Of the few amino acid differences detected between each pair, none was in a location known to impact on growth, replication, or virulence.
Plaque purification and cell culture.
For plaque purification, confluent MDCK cells in six-well tissue culture plates were prepared a day prior to infection in MDCK cell culture growth medium containing Dulbecco's modified Eagle's medium (DMEM)-Coons basal medium (SAFC Biosciences) supplemented with 2 mM l-glutamine (SAFC Biosciences), 1% nonessential amino acids (NE) (SAFC Biosciences), 0.05% sodium bicarbonate (SAFC Biosciences), 1 M HEPES (SAFC Biosciences), 2% penicillin-streptomycin (Sigma-Aldrich), and inactivated fetal bovine serum (JRH Biosciences). MDCK cells were infected with 0.4 ml of virus per well (from a dilution range of 10−1 to 10−8). The plates were incubated at 35°C for 30 min in a 5% CO2-gassed incubator. After 30 min of incubation, the inocula were removed using a plastic Pasteur pipette, and 4 ml of overlay containing equal proportions of liquefied 1% agarose and 2× minimal essential medium (MEM) plus NE (MEM/NE) supplemented with 8 μg/ml trypsin (SAFC Bioscience) was added to each well. The plates were kept in the biohazard cabinet until the overlay solidified and then were placed back in the 5% CO2-gassed incubator. The appearance of foci was examined after 4 days of incubation at 35°C. The three most-separated and smallest plaques were harvested by removing a plug with a sterile 100-μl micropipette tip. Each of the plugs was placed into 0.5 ml of CaMg-free phosphate-buffered saline (PBS) and stored at −70°C prior to further plaque purification or viral expansion. Viral expansion in MDCK cells was done in maintenance medium containing DMEM-Coons basal medium (SAFC Bioscience), 2 mM l-glutamine (SAFC Bioscience), 1% nonessential amino acids (SAFC Bioscience), 0.05% NaHCO3 (SAFC Bioscience), 1 M HEPES (SAFC Bioscience), 2% penicillin-streptomycin (Sigma-Aldrich), 2 μg/ml amphotericin (Gibco), and 4 μg/ml trypsin (SAFC Bioscience).
RNA extraction, RT-PCR, and sequencing.
Total RNA was extracted from the cell culture supernatant using the robotic MagNA Pure LC instrument and the MagNA Pure LC total nucleic acid isolation kit (Roche Applied Science). The RNA extraction procedure was performed according to manufacturer's instructions, with a total elution volume of 50 μl. RNA extracts were stored at −70°C. A 5-μl aliquot of RNA was used to amplify the selected influenza virus gene using specific primers (sequences available by request) and SuperScript III Platinum one-step reverse transcription (RT)-PCR system (Invitrogen) reagents. Amplicons were visualized on a 2% agarose gel. PCR products were purified for use in a sequencing reaction using the QIAquick PCR purification kit (Qiagen). DNA sequencing was carried out using the ABI Prism dye terminator III cycle sequencing kit (Applied Biosystems) followed by the removal of excess dye terminators using a DyeEx spin kit (Qiagen). The sequence was determined using an automated capillary DNA sequencer (ABI Prism 377 located at the Institute of Medical and Veterinary Science, Adelaide, Australia). Sequences were assembled and aligned using the DNAStar Lasergene 8 software.
Determination of viral infectivity.
To determine the infectivity titer of each virus, confluent MDCK cells were infected with 10-fold serially diluted influenza virus supernatant to determine the TCID50. Cells (2 × 105 cells/well) were prepared in 96-well flat-bottom plates in MDCK growth medium 24 h before use. The MDCK growth medium was removed, and the monolayer washed twice with CaMg-free PBS. One hundred microliters of each virus diluted in MDCK maintenance medium was inoculated into the wells in triplicate. The inoculated viruses were incubated at 35°C in 5% CO2 for 2 h, after which virus inocula were removed and 200 μl of maintenance medium supplemented with 4 μg/ml of trypsin was added. The plates were incubated at 35°C in 5% CO2 for 4 days. Cell viability in virus-infected wells was determined using a neutral red staining method (31). The log10 TCID50/ml was calculated as described by Reed and Muench (27).
In vitro replication kinetics.
In vitro replication kinetics experiments were performed in conventional MDCK cells and also in MDCK-SIAT1 cells (kindly provided by Hans-Dieter Klenk, University of Marburg, Marburg, Germany) as this modified cell line has an increased surface expression of human-like α-2,6-linked sialic acid receptors and may be better suited to analysis of NAI-resistant viruses (19, 36). The replication efficiency of each virus was investigated using both a single-step replication cycle, which involved the infection of MDCK or MDCK-SIAT1 cells with a high virus titer and sampling of the supernatant over a short time period, and multiple-step replication cycles, which involved the infection of MDCK or MDCK-SIAT1 cells with a lower virus titer and sampling of the supernatant over a longer period of time. Both growth studies were performed in triplicate. For the single-step replication cycle, confluent MDCK or MDCK-SIAT1 cells in 24-well plates were inoculated with viruses at a multiplicity of infection (MOI) of 5 TCID50/cell. Flasks were incubated at 35°C for 1 h to allow virus absorption. The cells were washed with 0.9% aqueous NaCl solution (pH 2.2) to remove any unattached infectious virus particles and then washed twice with PBS to adjust the pH (36). The cells were then overlaid with 1 ml maintenance medium. The flasks were incubated at 35°C in 5% CO2, and supernatant was collected 2, 4, 6, 8, and 10 h postinfection with each virus and stored at −70°C prior to determination of viral infectivity. For the multiple-step replication cycles, confluent MDCK or MDCK-SIAT1 cells in each T-25 (25-cm3) cell culture flask were inoculated with viruses at a multiplicity of infection (MOI) of 0.01 TCID50/cell. After initial incubation, as described for the single-step replication cycle, the infected cells were then overlaid with 10 ml maintenance medium and incubated at 35°C with 5% CO2. Three hundred microliters of supernatant was collected 6, 24, 30, 48, 54, 72, 78, 96, and 102 h postinfection for each virus and stored at −70°C for titration prior to determination of viral infectivity. To ensure consistency, viral infectivity assays were performed using the same cell line (either MDCK or MDCK-SIAT1 cells) from which the replication kinetics experiments were performed.
Ferret experiments.
To prepare the viruses for inoculation into ferrets, each of the four strains (R292 WT, R292K MUT, H274 WT, and H274Y MUT) was diluted to a standardized tissue culture infectivity titer of 1 × 105 TCID50/ml. These virus preparations, containing 1 × 105 TCID50/ml, were then used for inoculation of ferrets either as a pure population or for preparation of mixtures of wild-type and mutant strains. To prepare the different mutant and wild-type mixtures, the virus preparations (containing 1 × 105 TCID50/ml) were combined in three different proportions (80% WT-20% MUT, 50% WT-50% MUT, and 20% WT-80% MUT). For example, the 80% WT-20% MUT mixture was prepared by combining 8 ml of wild-type virus (at an infectivity titer of 1 × 105 TCID50/ml) with 2 ml of mutant virus (also at an infectivity titer of 1 × 105 TCID50/ml). As such, the two pure populations and three mixtures, now referred to as “virus groups,” all had an infectivity titer of 1 × 105 TCID50/ml. Female ferrets of approximately 6 months of age and weighing approximately 800 g were sourced from various ferret breeders in Victoria, Australia, and were microchipped, anesthetized, and pre-bled via the jugular vein to confirm the absence of preexisting antibodies to seasonal H1/H3/B influenza strains. For each of the five virus groups, two ferrets, referred to as “donor 1” (D1) and “donor 2” (D2), were anesthetized intramuscularly with 20 mg/ml Ilium Xylazil-20 (Troy Laboratories, NSW, Australia) and then dosed intranasally with 0.5 ml of virus (5 × 104 TCID50) (prepared following the methods detailed above). Donors 1 and 2 from the same virus group were housed together, but in separate HEPA filtered caging from the donor ferrets in other virus groups. Twenty-four hours postinfection (day 1), ferrets were nasal washed by dispensing 1 ml of nasal wash solution (CaMg-free PBS with 1% [wt/vol] bovine serum albumin [BSA], 100 μg/ml streptomycin, and 100 U/ml penicillin) using an Optiva (Medex) 20-gauge intravenous (i.v.) catheter and 1-ml syringe into the nostril of each ferret and collecting the returned nasal wash liquid in a sterile tube, which was stored at −70°C. After thorough cleaning of the cages, a naïve uninfected ferret, termed “recipient 1” (R1), was introduced into each of the five cages containing the donor ferrets from the different virus groups. Recipient 1 ferrets were nasal washed daily, until they were found to be influenza positive by a rapid point-of-care test (BinaxNOW Influenza A & B test kit; Inverness Medical, Waltham, MA), and then were removed and housed immediately in a new clean cage together with a naïve uninfected ferret termed “recipient 2” (R2). Nasal washes were collected either daily or every 2 days from all donor and recipient ferrets until day 11 (for the H274Y experiment) or day 18 (for the R292K experiment), after which they were given a terminal bleed.
Genetic analysis of nasal washes.
RNA was extracted as described earlier. To determine the relative proportions of wild-type and mutant influenza strains in ferret nasal wash samples, a TaqMan probe-based one-step real-time RT-PCR assay was conducted. The assay used the TaqMan one-step RT-PCR master mix reagents kit (Applied Biosystems) together with the ABI PRISM 7500 sequence detection system (Applied Biosystems) following the manufacturer's instructions. The primers and probes were kindly designed by Michael Tavaria (Applied Biosystems, Australia) based on NA sequences of the two pairs of viruses and following the manufacturer's design guidelines and software. For each pair of viruses, an assay was developed with two specific PCRs: one designed to detect the wild-type virus and a second to detect the mutant virus (Fig. 1). Following optimization to achieve the highest specificity for both wild-type and mutant reactions, primers and probes were used in the PCR assays at concentrations of 5 μM and 25 μM, respectively. RNAs extracted from the five virus groups used to infect ferrets (e.g., 100:0, 0:100, 80:20, 50:50, and 20:80) were included in each PCR assay as positive controls.
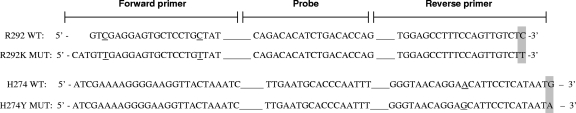
FIG. 1.
Primer and probe designs for R292K and H274Y real-time RT-PCR assays. Specific nucleotides highlighted in gray represent mutations associated with NAI resistance within the NA gene. The specific nucleotides underlined represent the location where sequence differs between the wild type and mutant but is not associated with residue mutation or NAI resistance.
Calculation of viral proportions in nasal washes.
The relative proportions of wild-type and mutant viruses in each nasal wash or positive control were determined by calculating the difference between the cycle threshold (CT) value for the wild-type PCR and the mutant PCR. CT is defined as the cycle at which a statistical increase in emission intensity of a specific strand crosses a predetermined fluorescence threshold. Based on the principle that a doubling of the quantity of PCR product occurs every cycle during the exponential phase of the reaction, it is inferred that a difference of one CT between samples indicates that there is twice the amount of starting RNA in one of the samples compared to the other. Therefore, a 50:50 mixture of wild-type and mutant viruses should give identical CT values for both the wild-type PCR and for the mutant PCR: i.e., no CT value difference between reactions indicates equal amounts of starting RNA. Following an adjustment based on the wild-type versus mutant CT values for the control viruses, the difference between the CT value for the wild-type reaction and that for the mutant reaction was calculated for each virus sample. The fold difference in RNA quantity between the WT and mutant viruses is therefore given by 2ΔCT, where ΔCT is the CT difference. The final estimated proportion (percentage) of WT to mutant virus in each sample was then calculated from the fold difference value. A CT value of greater than 40 cycles was considered to be a negative result. Assays were considered valid if the difference between the calculated WT/mutant proportion and the known proportions for any of the five mixture controls did not exceed 10 percentage points. For example, if calculations for the 80% WT-20% mutant control sample produced predicted proportions of 69% WT-31% mutant, then the assay was considered invalid and all samples from that assay would be retested.
Mathematical and statistical analysis.
To assess the within-host replication fitness of the R292K and H274Y mutant strains, we utilized an established technique previously used to asses the fitness of cytotoxic T-lymphocyte (CTL) escape mutants in chronic HIV infection (1). Briefly, we assumed both the wild type and mutant were recognized similarly by the immune system (both innate and adaptive components) and so the rate of removal, b, of the viruses was the same. The mutant has a different replication rate, a′, compared to that for the wild type, a. We let a, a′, and b vary over time to allow for the increase and then decrease in viral load, but made the assumption that a − a′ is constant in time. It follows that the proportion of the mutant in the viral population at any point in time is given by the equation
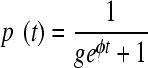 |
(1) |
where φ = a − a′ is a measure of replication fitness cost of the mutant virus and g is the ratio of wild type to mutant at the time of inoculation. Given this functional form for the changing mutant proportion over time, we made a model-dependent estimate of the replication fitness cost, φ, for the mutant virus in each ferret. This measure is the difference between two viral growth rates, a and a′, not a ratio of viral growth rates. We applied the Wilcoxon rank-sum test for difference between mutation types (R292K versus H274Y). Furthermore, to make inferences across strain type while accounting for the expected individual variation among ferrets, we performed a standard mixed-effects statistical analysis. A transformation of equation 1 yields a linear-model, ln {[1 − p(t)]/gp(t)} = φt, where a positive slope, φ, indicates a compromised mutant virus. We tested for evidence of a replication fitness cost for the R292K and H274Y mutants separately and for a difference in replication fitness cost between the two mutants, allowing for the random variation among individual ferrets in our analysis. Statistical analysis was performed in STATA/IC 10.1 2009. To assess transmission fitness costs—that is differences in the transmissibility of the mutant compared to that of the wild type—we used a graphical technique presented in the Results.
Nucleotide sequence accession number.
Sequences for each virus segment have been deposited in GenBank under the following accession numbers: A/Fukui/20/2004 (R292 WT), CY064955 to CY064962; A/Okayama/23/2004 (R292K MUT), CY064963 to CY064970; A/Brisbane/59/2007 (H274 WT), CY064971 to CY064978; and A/Sydney/142/2007 (H274Y MUT), CY064979 to CY064986.
RESULTS
In vitro replication kinetics.
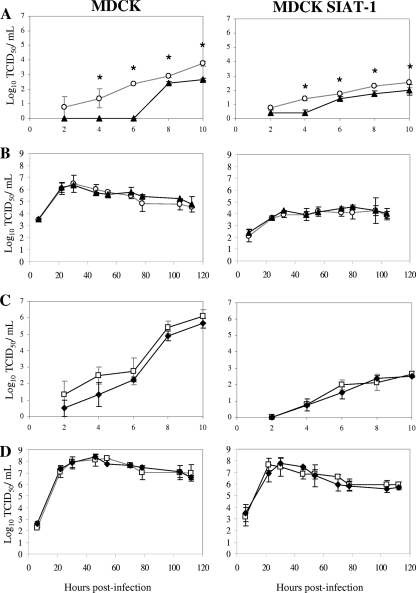
The replication kinetics of wild-type and mutant viruses in single- and multiple-step replication cycles were determined in MDCK and MDCK-SIAT1 cells. In the single-step growth studies, the growth rate of the oseltamivir-resistant R292K MUT virus was found to be significantly slower than that of the R292 WT virus (P < 0.05) in experiments conducted in both MDCK and MDCK-SIAT1 cells (Fig. 2 A), although in the multiple-step growth experiment, there was no significant difference between the growth curves of the two strains (Fig. 2B). Comparison of the in vitro replication kinetics for the H274 WT and H274Y MUT strains revealed no significant difference in either the single- or multiple-step experiments regardless of the cell line used (Fig. 2C and D). In both the MDCK and MDCK-SIAT1 multiple-step experiments, the H274 WT and H274Y MUT viruses achieved higher viral titers between the 20 and 120 h postinfection than those observed for the R292 WT and R292K MUT viruses during the same period.
FIG. 2.
In vitro replication kinetics of wild-type and mutant viruses in MDCK and MDCK-SIAT1 cells. Replication kinetics of the A/Fukui/20/2004 (R292 WT) (○) and A/Okayama/23/2004 (R292K mutant) (▴) viruses in single-step growth experiments (MOI of 5 TCID50/cell) (A) and multiple-step growth experiments (MOI of 0.01 TCID50/cell) (B) and the A/Brisbane/59/2007 (H274 WT) (□) and A/Sydney/142/2007 (H274Y mutant) (⧫) viruses in single-step growth experiments (C) and multiple-step growth experiments (D). Virus in the supernatant was titrated at the indicated time points postinfection (log10 TCID50/ml). Each data point represents the mean log10 TCID50 from triplicate experiments. *, statistically significant difference between WT and mutant virus (P < 0.05).
Validation of the real-time RT-PCR assays for estimating viral proportions.
Prior to testing the nasal wash samples, the ability of the R292K and H274Y real-time RT-PCR assays to correctly estimate the wild-type and mutant viral proportions in the control mixtures 80%:20%, 50%:50%, and 20%:80% was determined. Analysis of the R292K control mixtures in seven separate R292K real-time RT-PCR assays revealed that the estimated mixture proportion, as determined by the assay, differed from the actual proportions by a minimum of 0.2 and a maximum of 5.1 percentage points, with an absolute mean (± 1 standard deviation) difference of 2.2 (± 1.5) percentage points. Similarly, analysis of the H274Y control mixtures across seven H274Y real-time RT-PCR assays demonstrated that the estimated proportions differed from the actual proportions by a minimum of 0.1 and a maximum of 7.2 percentage points, with an absolute mean (± 1 standard deviation) difference of 2.7 (± 2.2) percentage points. To determine if the accuracy of estimating the viral proportions differed when analyzing mixtures with lower viral titers, the three R292K control mixtures (80%:20%, 50%:50%, and 20%:80%) were diluted (10−1 to 10−5) and tested in the R292K real-time RT-PCR assay. The absolute mean differences between the estimated and actual proportions for the three control mixtures were 1.8, 2.1, 2.4, and 2.0 percentage points for the undiluted and 10−1-, 10−2-, and 10−3-diluted controls, respectively. The control mixtures diluted to 10−4 or 10−5 could not be detected by the PCR assay. These results indicated the ability of the PCR assay to accurately estimate viral proportions, irrespective of viral load, within the limit of detection of the assay.
Analysis of viral proportions in ferret nasal washes.
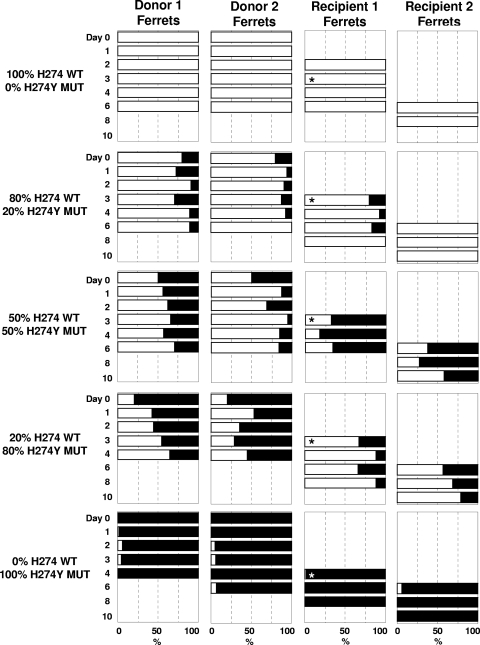
Figures 3 and 4 summarize the relative proportions of wild-type and mutant virus in each ferret over time for the R292K and H274Y experiments, respectively. We first present within-host viral replication data in the donor and recipient ferrets (replication fitness), before examining the characteristics of transmission of viral mixtures from donor ferrets to recipient ferrets (transmission fitness).
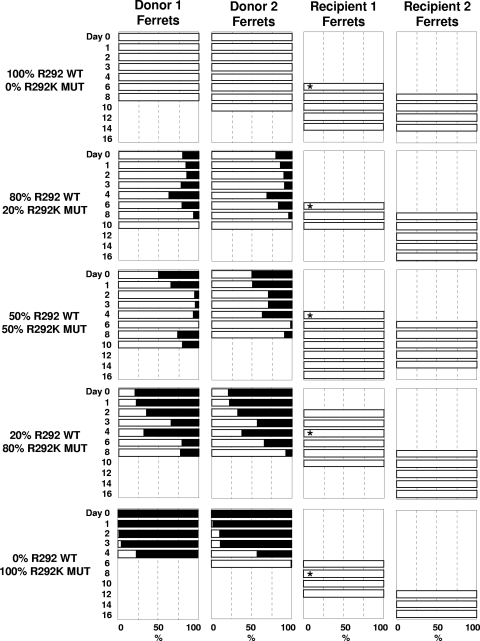
FIG. 3.
Infectivity and transmissibility of pure populations and mixtures of R292 WT and R292K MUT viruses in ferrets. The relative proportion of viral mixtures is indicated in the panels where the percentage of R292 WT virus is displayed using white bars and the R292K MUT proportion is displayed by black bars. No bars are present for samples when no virus was detected. R1 ferrets were introduced into the same cage as donor ferrets on day 1. Asterisks indicate the day when R1 ferrets were influenza positive according to the point-of-care rapid test and moved away from donor ferrets and housed with the R2 ferret. Retrospective RT-PCR analysis of nasal washes from the R1 ferret from the 20% WT-80% MUT group and the 0% WT-100% MUT group detected influenza virus 1 to 2 days prior to the date when ferrets were rapid test positive and housed with the R2 ferret.
Summary for the R292K experiment.
For the R292K experiment, we observed that in all donor ferrets the mutant virus (black) is outgrown by the wild-type virus (white) (Fig. 3). Both donor ferrets (D1 and D2) infected with 100% R292 WT virus shed virus until 8 or 10 days postinfection, with no detection of any mutant virus in any nasal washes during this period (Fig. 3). Recipient 1 (R1) became infected by day 6 of the experiment (5 days after being housed with the donor ferrets) and continued to shed detectable virus for another 4 days. Virus was detected in recipient 2 (R2) on the second day it was housed with R1 (day 8); R2 continued to shed virus until day 14 (8 days postexposure). The two donor ferrets infected with 100% R292K MUT virus shed detectable virus for 6 or 8 days, although the proportions of R292K MUT in the viral population, which started at 100%, were significantly reduced over the course of the infection to only 61% and 1% in D1 and D2, respectively, being replaced by R292 WT virus (Fig. 3). None of the R292K MUT virus was detected in the R1 ferret, and the R292 WT virus was found to persist for 6 days before R2 became infected, again with only the R292 WT virus (100%). A similar outcome was observed with the ferrets infected with the mixtures of mutant and wild-type virus. In each case, the proportion of the R292K MUT virus decreased over time in the donor ferrets and R292K MUT was not detectable in any of the R1 ferrets (Fig. 3).
Summary for the H274Y experiment.
Donor ferrets infected with a pure population of H274 WT virus demonstrated detectable virus shedding for 6 days postinfection (Fig. 4). R1 became infected within 24 h of being exposed to the infected donors and, after being rehoused on day 3, subsequently infected R2 by day 6. The two donor ferrets infected with a pure population of the H274Y MUT virus shed virus for 4 to 6 days postinfection. Although the proportion of H27Y MUT virus remained very high (93% to 100%) in both donor ferrets over the course of infection, the H274 WT virus was detected at low levels in nasal washes from various time points. Repeat real-time RT-PCR analysis of these samples confirmed these results. A low proportion of H274 WT virus was detected on occasions in nasal washes from R1 (day 4 nasal wash contained 1% H274 WT virus) and R2 (day 6 nasal wash contained 6% H274 WT virus).
FIG. 4.
Infectivity and transmissibility of pure populations and mixtures of H274 WT and H274Y MUT viruses in ferrets. The relative proportion of viral mixtures is indicated in the panels where the percentage of H274 WT virus is displayed using white bars and that of H274Y MUT is displayed by black bars. No bars are present for samples when no virus was detected. R1 recipient ferrets were introduced into the same cage as donor ferrets on day 1. Asterisks indicate the day when R1 ferrets were influenza positive according to the point-of-care rapid test and moved away from donor ferrets and housed with the R2 ferret. Retrospective RT-PCR analysis of nasal washes from the R1 ferret from the 100% H274 WT-0% H274Y MUT group detected influenza 1 day prior to the date when ferrets were rapid test positive and housed with the R2 ferret.
The within-host replication fitness cost for R292K and H274Y.
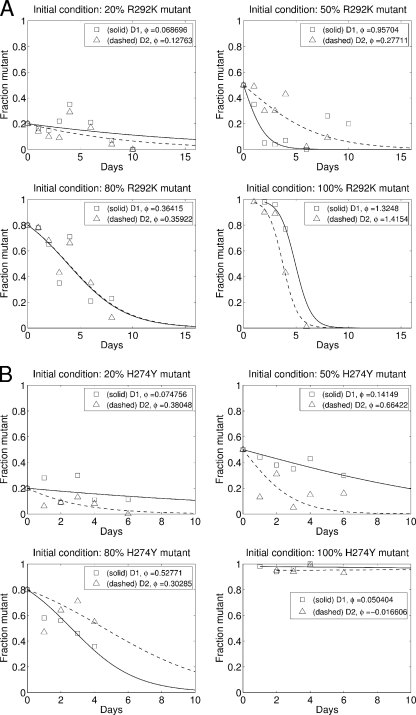
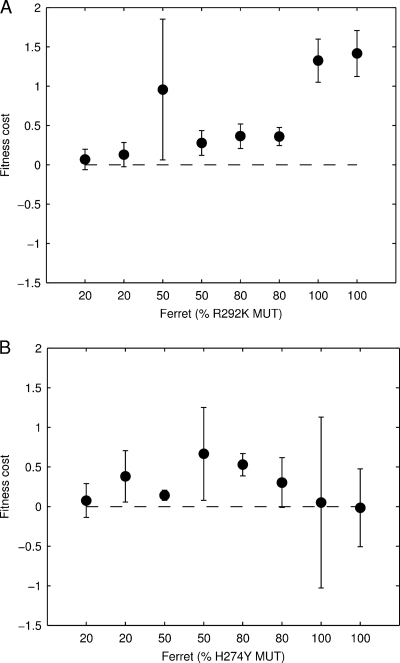
Figure 5 plots the mutant proportion over time and the best-fit model curve for each of the donor ferrets in the R292K experiment (Fig. 5A) and the H274Y experiment (Fig. 5B). A positive value for φ, the difference in viral growth rates, indicates that the mutant has a reduced replication fitness. Figure 6 plots the best-fit value for φ for each ferret and the model-fit-derived confidence interval for the estimate. Both the R292K (Fig. 6A) and H274Y (Fig. 6B) mutants demonstrate compromised replication fitness in the donor ferrets. The median replication fitness cost for R292K mutation was 0.36, and for the H274Y mutation, it was 0.22. The distributions of replication fitness costs for the two different mutant viruses were not significantly different (Wilcoxon's rank-sum test, P = 0.24).
FIG. 5.
The mutant proportion over time and the nonlinear model fit for each donor ferret. The replication fitness cost, φ, for the R292K mutant (A) and H274Y mutant (B) is estimated for each ferret separately. The initial proportion is used to fix g in the model for each ferret.
FIG. 6.
Summary of the within-host replication fitness of the mutant viruses in donor ferrets. Shown is a summary of the fitness cost, φ, for each donor ferret, for the R292K mutant (A) and H274Y mutant (B). The error bars indicate the 95% confidence interval for φ for each ferret, based on the asymptotic normal distribution for the parameter estimate.
Table 1 presents results from a mixed-effects linear regression of the transformed data: ln {[1 − p(t)}/gp(t)} = φt. We allow for a ferret- and strain-independent replication fitness cost, an additional replication fitness cost if the mutant is R292K (β292), and a random effect for each ferret in the estimation. That is, we concurrently fit, over all ferrets, i, the model φi = (β + ηiβ292 + βi)t + ɛi(t), where ηi is 0 for H274Y ferrets and 1 for R292K ferrets, βi has a mean of 0, and ɛi(t) is the ferret-specific time-dependent error. We see evidence for a reduced replication fitness for all mutant viruses (β = 0.25 [−0.01, 0.50]; P = 0.056). Although the point estimate for the replication fitness cost is approximately double for the R292K mutant compared to the H274Y mutant (β + β292 = 0.25 + 0.26 compared to β = 0.25), we do not have sufficient statistical power to definitively claim an increased replication fitness cost for the R292K mutant compared to the H274Y mutant. The effect of individual ferrets is substantial, as can be seen by the estimates for the variance of the random effect, βi. If we restrict the analysis to only the 100% MUT-0% WT infections, then we do find a statistically significant effect for the mutant type, in which the R292K mutation clearly has a greater replication fitness cost than the H274Y mutation (results not shown, but compare the lower right panels in Fig. 5A and B).
TABLE 1.
Results from the mixed-effects linear regression on the transformeda proportions data
| Predictor | Coefficient | 95% CIb | P value |
|---|---|---|---|
| β | 0.248 | (−0.007, 0.503) | 0.056 |
| β292 | 0.260 | (−0.095, 0.616) | 0.152 |
| Random-effects parameter | SD estimate | 95% CI on SD | |
|---|---|---|---|
| βic | 0.335 | (0.212, 0.528) | |
| Residual | 0.900 | (0.770, 1.054) |
See Materials and Methods for details.
95% CI, 95% confidence interval.
The random effect, βi, with a mean of zero and estimated standard deviation, allows for per-ferret random variation.
The transmission fitness for R292K and H274Y.
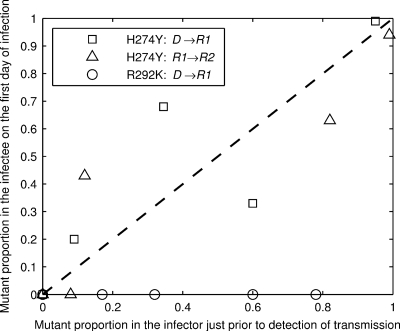
Figures 3 and 4 show a clear difference in the abilities of the R292K MUT and H272Y MUT viruses to transmit between ferrets. On no occasion was an R292K mutant transmitted (Fig. 3), even when the mutant dominated infection at the time of transmission (e.g., see Fig. 3, 20% WT-80% MUT donors, infection of R1 on day 2). In contrast, we see transmission of the H274Y mutant in all but one of the eight possible transmission events. Figure 7 summarizes the transmission observations presented in Fig. 3 and 4, plotting the infectee's mutant proportion as a function of the infector's mutant proportion. The H274Y data are consistent with a linear relationship with slope 1; that is, there is no evidence for a transmission fitness cost or advantage for the H274Y mutant, unlike the R292K mutant which did not transmit, as illustrated by the data points for that experiment lying on the y = 0 axis in Fig. 7.
FIG. 7.
Summary of the transmission fitness of mutant viruses. The abscissa shows the mutant proportion of the infecting ferret's viral load on the day preceding confirmed transmission. The ordinate shows the mutant proportion of the infected ferret's viral load within the first 24 h postinfection. For donor (D1 or D2)-to-R1 ferret transmission events, we take the average proportion in the two donors. Open circles (○) are the donor-to-R1 transmission events from the R292K experiment. Open squares (□) are the donor-to-R1 transmission events, and open triangles (Δ) are the R1-to-R2 transmission events in the H274Y experiment. Note that there are three points (1 circle, 1 square, and 1 triangle) at coordinates 0, 0. The dotted line shows the relationship y = x, a visual guide to help demonstrate the qualitative proportional-transmission relationship for the H274Y data.
The scatter about the line y = x for the H274Y strains in Fig. 7 indicates that the transmitted proportion, while qualitatively predicted by the infector's proportion, is subject to significant stochastic (random) variation. In particular, no mutant virus was detected in the R2 recipient ferret following infection of donor ferrets with a mixture containing 20% H274Y MUT virus [Fig. 4 and 7, triangle at (x, y) = (0.1, 0)]. A mixed-infection transmission chain has collapsed to a pure infection chain by chance, suggesting that the mutant may be competitively excluded from the ferret population over an extended number of generations. In contrast when donor ferrets were infected with a mixture containing either 50% or 80% H274Y MUT virus, the resistant virus remained present in the mixtures through to the R2 recipient ferret. For these two mixtures (Fig. 4 and Fig. 7), the two donor-to-R1 recipient transmission events show fluctuations in the proportion in opposite directions (60% down to 33% and 35% up to 68%). Potential sources for this variation in transmitted proportion include inherent stochastic variation in the transmitted inoculum and/or animal host variability.
DISCUSSION
The NAIs are an important class of antiviral drugs for the treatment and prevention of influenza virus infection. However, as is the case for many antiviral drugs, the emergence of resistant variants is a major concern. To accurately assess the risk that a resistant virus will emerge and spread throughout the community, it is important to determine the relative viral fitness of a strain. Previous studies have assessed viral fitness of NAI-resistant viruses using both in vitro and in vivo models (12) and have found that many NAI-resistant variants have compromised fitness, with impaired growth in vitro and reduced infectivity and transmissibility in animal experiments (22).
In vitro studies that determine the rate of viral replication and viral titers over time are relatively easy to perform, but previous studies suggest that these data do not accurately reflect the replication fitness of a strain in an animal model (7, 13, 29, 30, 36). This could be because in vitro replication kinetics experiments may not be impacted by the defective NA enzyme activity of mutant viruses as there are fewer host antiviral mechanisms present in cell lines (19). In the present study, the MDCK and MDCK-SIAT1 single-step experiments detected a significant difference between the growth of the R292 WT and R292K MUT viruses, but no difference was seen in the multiple-replication-cycle experiments. In contrast, the in vitro growth characteristics of the H274 WT and the H274Y MUT viruses were identical in both cell lines in single-step and multiple-replication assays.
Ferrets, in contrast to mice, have a type and distribution of human influenza receptors (α-2,6-glycosidic linkages) in their respiratory tracts similar to those in humans, and as such the ferret model is considered to be one of the best animal models to assess human influenza virus growth and transmissibility (28). Previous studies have assessed viral fitness of NAI-resistant viruses in a ferret model by infecting animals with a pure population of either a wild-type or mutant virus and comparing the minimum dose required to obtain an infection, the duration of infection, and how readily the viruses will transmit to other naïve ferrets. In this study, a novel method was used to assess viral replication and transmission fitness, in which ferrets were infected with either pure populations or various mixed populations of wild-type and mutant viruses. The resultant viral proportion of wild-type and mutant virus present in nasal washes in each animal was determined every 1 or 2 days during infection of both donor and recipient ferrets. A competitive-mixtures model such as this should mean that if one strain replicates at a faster rate than the other (i.e., it has an increased replication fitness), a larger number of progeny viruses will be produced and therefore infect a higher proportion of the finite number of cells available for infection prior to immune clearance, resulting in one strain outgrowing the other over time. Furthermore, we have used a simple mathematical model of the replication kinetics to capture this outgrowth behavior and then calculated (model-based) estimates of the replication fitness cost using a mixed-effects statistical model.
The competitive-mixtures model was able to clearly identify the compromised within-host replication fitness of both the R292K and H274Y mutant viruses. Within the animals infected with 100% R292K MUT virus, mutation during viral replication resulted in R292 WT viruses rather than R292K MUT strains being selected (due to the greater replication fitness of the wild-type virus compared to the mutant strain), which led to the dominance of the wild-type virus in the population. Because the transmission fitness of the R292K mutant was so low (our point estimate is that it is zero), only after the wild-type had begun to outgrow the mutant did we see transmission to recipient ferrets (e.g., in Fig. 3, 0% for R292 WT versus 100% for R292K MUT). Previous studies have also demonstrated the compromised transmission fitness of the R292K mutant (9), and as such for this type of mutant, there may be less need to conduct the competitive-mixtures experiment compared to using the standard ferret experiments that involve infection with only the pure viral populations. However, the competitive-mixtures model does appear to be a more powerful model when comparing two viruses that have similar within-host replication or transmission fitness. When ferrets were infected with pure populations of either H274 WT or H274Y MUT virus, both viruses were infectious in the donors, and the mutant demonstrated no detectable replication fitness cost (Fig. 5B, lower right panel). However, replication fitness differences did appear to be present when analyzing the proportions of the mutant and wild-type viruses in the ferrets infected with the mixtures (Fig. 6 and Table 1), suggesting that the H274Y MUT, like the R292K MUT virus, had a lower replication fitness cost than the wild type within the host. Our inability to identify a difference in replication fitness cost (φ) between R292K and H274Y may be attributable to a number of factors but is primarily due to a lack of statistical power.
The oseltamivir-resistant H274Y MUT virus used in this study is representative of the viruses that became the predominant circulating seasonal A(H1N1) strain during 2007 and 2008. The increased frequency of the H274Y mutant A(H1N1) viruses in the human population, compared to the H274 WT A(H1N1) viruses (11), suggests that these strains have a fitness level equal to or greater than that of the circulating wild type. In this study, we identified the mutant strain to have similar transmission fitness compared to the wild type but slightly reduced within-host replication fitness. These differences may be due to the limitations in the ferret model such as the relatively small numbers of ferrets that are able to be used in each experiment and animal-to-animal variability as, unlike mice, the ferrets used in these experiments are derived from an outbred population. However, a repeat of the H274Y competitive-mixtures experiment using the same strains generated results similar to those presented here (data not shown). Another limitation of this study and other similar studies is that particular wild-type and mutant strains are selected as being representative of their respective phenotypes/genotypes, and the potential exists that the choice of other representative strains may have produced slightly different results from those observed. As described previously, fitness differences between virus strains with the same resistance mutations have made in vivo comparisons of NAI-resistant mutant viruses difficult (36). To address this, future studies could use reverse genetics and site-directed mutagenesis to generate mutant and wild-type viruses that have an identical genome other than a single mutation in the NA gene that confers resistance. However, this approach also has its limitations, as it precludes the effects of compensatory mutations in other genes that may have occurred through natural evolution and may be necessary for maintaining viral fitness in humans.
The coinfection of ferrets with two different viruses as used in this study raises the possibility of gene segment reassortment leading to the generation of progeny viruses with genes derived from both of the parent strains. Previous studies, however, suggest this outcome is relatively rare. In ferrets coinfected with pandemic A(H1N1) 2009 and seasonal influenza A viruses, no reassortant viruses were detected (25), while a different study investigating coinfection of ferrets with A(H5N1) and A(H3N2) strains found that only 9% of recovered viruses contained genes from both parent viruses (15). Due to the high genetic similarity between the wild-type and mutant viruses used in the present study (the H274 WT and H274Y MUT strains differed by only nine amino acid differences across the entire genome), it is likely that any reassortants would not display a significant fitness difference from the parent strains, particularly given that none of the residue differences between the wild-type and mutant strains were located in sites known to affect growth, transmissibility, or virulence. The high genetic similarity between the progenitor strains also means that detection of reassortants is considerably more difficult than when analyzing progeny viruses from ferrets coinfected with two completely different strains [e.g., A(H5N1) and A(H3N2)]. It is also important to note that the degree of reassortment has no influence on the modeling component of this study. Mathematical models used to make assessments of antiviral deployment policy require information only on whether a strain is resistant or sensitive and the relative transmission fitness of that resistant mutant (compared to the wild-type sensitive strain) and do not require detailed genetic characterization (18, 21, 35).
This study has produced estimates for the replication fitness cost (φ) that will be used to determine the rate at which resistant-strain infections revert to sensitive-strain infections in mathematical models of influenza spread in human populations. Modeling studies to date have not allowed for this reversion process due to a lack of data. Furthermore, these models, while including analyses with a relative transmission fitness equal to 1, have also made inferences based on an assumed reduced transmission ability for the resistant strain. Figure 7 demonstrates that the H274Y MUT strain likely has a transmissibility equal to that of the wild type. Outcomes from existing models should be reevaluated in light of the findings presented here.
The replication and transmission fitness of NAI-resistant influenza A viruses was investigated in this study using a novel competitive-virus-mixtures ferret model, which enables the behavior of a mixed-virus population, comprising mutant and wild-type viruses, to be monitored during the course of infection and over several transmission events. This method appears to provide greater insight into the relative fitness of two different strains, compared to more traditional methods that infect ferrets with only pure populations of sensitive or resistant viruses. Given the importance of oseltamivir as the currently most prescribed NAI for the control of influenza infections, it is critical that the fitness of the H274Y variant and other NAI-resistant viruses be further investigated to assess their potential to spread widely throughout the community. For example, since the emergence of the A(H1N1) pandemic in 2009, over 200 A(H1N1) pandemic strains have been shown to contain the H274Y mutation conferring oseltamivir resistance (34). Although there has been some evidence of transmission of these resistant strains, at this stage it appears to occur only between close contacts (8, 17). It would be of significant value to test these viruses for transmission efficiency in models such as the one described here. The results from this preliminary study suggest that the competitive-mixtures ferret model is a valuable tool to compare the relative fitness of two influenza virus strains and can provide real insights into the infectivity and transmissibility of NAI-resistant strains.
Acknowledgments
The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health and Ageing.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Asquith, B., C. T. Edwards, M. Lipsitch, and A. R. McLean. 2006. Inefficient cytotoxic T lymphocyte-mediated killing of HIV-1-infected cells in vivo. PLoS Biol. 4:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouvier, N. M., A. C. Lowen, and P. Palese. 2008. Oseltamivir-resistant influenza A viruses are transmitted efficiently among guinea pigs by direct contact but not by aerosol. J. Virol. 82:10052-10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins, P. J., L. F. Haire, Y. P. Lin, J. Liu, R. J. Russell, P. A. Walker, J. J. Skehel, S. R. Martin, A. J. Hay, and S. J. Gamblin. 2008. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453:1258-1261. [DOI] [PubMed] [Google Scholar]
- 4.Dharan, N. J., L. V. Gubareva, J. J. Meyer, M. Okomo-Adhiambo, R. C. McClinton, S. A. Marshall, K. St. George, S. Epperson, L. Brammer, A. I. Klimov, J. S. Bresee, and A. M. Fry. 2009. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 301:1034-1041. [DOI] [PubMed] [Google Scholar]
- 5.Gerloff, N. A., J. R. Kremer, J. Mossong, M. Opp, and C. P. Muller. 2009. Genomic diversity of oseltamivir-resistant influenza virus A (H1N1), Luxembourg, 2007-08. Emerg. Infect. Dis. 15:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubareva, L. V., M. N. Matrosovich, M. K. Brenner, R. C. Bethell, and R. G. Webster. 1998. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 178:1257-1262. [DOI] [PubMed] [Google Scholar]
- 7.Gubareva, L. V., M. J. Robinson, R. C. Bethell, and R. G. Webster. 1997. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J. Virol. 71:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulland, A. 2009. First cases of spread of oseltamivir resistant swine flu between patients are reported in Wales. BMJ 339:b4975. [DOI] [PubMed] [Google Scholar]
- 9.Herlocher, M. L., J. Carr, J. Ives, S. Elias, R. Truscon, N. Roberts, and A. S. Monto. 2002. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res. 54:99-111. [DOI] [PubMed] [Google Scholar]
- 10.Herlocher, M. L., R. Truscon, S. Elias, H. L. Yen, N. A. Roberts, S. E. Ohmit, and A. S. Monto. 2004. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J. Infect. Dis. 190:1627-1630. [DOI] [PubMed] [Google Scholar]
- 11.Hurt, A. C., J. Ernest, Y. Deng, P. Iannello, T. G. Besselaar, C. Birch, P. Buchy, M. Chittaganpitch, S. C. Chiu, D. E. Dwyer, A. Guigon, B. Harrower, I. P. Kei, T. Kok, C. Lin, K. McPhie, A. Mohd, R. Olveda, T. Panayotou, W. Rawlinson, L. Scott, D. Smith, H. D'Souza, N. Komadina, R. Shaw, A. Kelso, and I. G. Barr. 2009. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res. 83:90-93. [DOI] [PubMed] [Google Scholar]
- 12.Hurt, A. C., H. T. Ho, and I. Barr. 2006. Resistance to anti-influenza drugs: adamantanes and neuraminidase inhibitors. Expert Rev. Anti Infect. Ther. 4:795-805. [DOI] [PubMed] [Google Scholar]
- 13.Ives, J. A. L., J. A. Carr, D. B. Mendel, C. Y. Tai, R. Lambkin, L. Kelly, J. S. Oxford, F. G. Hayden, and N. A. Roberts. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leaves virus severely compromised both in vitro and in vivo. Antiviral Res. 55:307-317. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, H. C., N. Roberts, Z. M. Wang, and R. Belshe. 2000. Management of influenza: use of new antivirals and resistance in perspective. Clin. Drug Invest. 20:447-454. [Google Scholar]
- 15.Jackson, S., H. N. Van, L. M. Chen, T. R. Maines, N. J. Cox, J. M. Katz, and R. O. Donis. 2009. Reassortment between avian H5N1 and human H3N2 influenza viruses in ferrets: a public health risk assessment. J. Virol. 83:8131-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiso, M., K. Mitamura, Y. Sakai-Tagawa, K. Shiraishi, C. Kawakami, K. Kimura, F. G. Hayden, N. Sugaya, and Y. Kawaoka. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759-765. [DOI] [PubMed] [Google Scholar]
- 17.Le, Q. M., H. F. Wertheim, N. D. Tran, H. R. van Doorn, T. H. Nguyen, and P. Horby. 2010. A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. N. Engl. J. Med. 362:86-87. [DOI] [PubMed] [Google Scholar]
- 18.Lipsitch, M., T. Cohen, M. Murray, and B. R. Levin. 2007. Antiviral resistance and the control of pandemic influenza. PLoS Med. 4:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matrosovich, M., T. Matrosovich, J. Carr, N. A. Roberts, and H. D. Klenk. 2003. Overexpression of the α-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J. Virol. 77:8418-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCaw, J. M., J. G. Wood, E. S. McBryde, T. M. Nolan, J. T. Wu, M. Lipsitch, and J. McVernon. 2009. Understanding Australia's influenza pandemic policy on the strategic use of the antiviral drug stockpile. Med. J. Aust. 191:136-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCaw, J. M., J. G. Wood, C. T. McCaw, and J. McVernon. 2008. Impact of emerging antiviral drug resistance on influenza containment and spread: influence of subclinical infection and strategic use of a stockpile containing one or two drugs. PLoS One 3:e2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKimm-Breschkin, J. L. 2000. Resistance of influenza viruses to neuraminidase inhibitors—a review. Antiviral Res. 47:1-17. [DOI] [PubMed] [Google Scholar]
- 23.McVernon, J., J. M. McCaw, and T. M. Nolan. 2010. Modelling strategic use of the national stockpile during the CONTAIN and SUSTAIN phases of an Australian pandemic influenza response. Aust. N. Z. J. Public Health 34:113-119. [DOI] [PubMed] [Google Scholar]
- 24.Meijer, A., A. Lackenby, O. Hungnes, B. Lina, S. van der Werf, B. Schweiger, M. Opp, J. Paget, J. van de Kassteele, A. Hay, and M. Zambon. 2009. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerg. Infect. Dis. 15:552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez, D. R., E. Sorrell, M. Angel, J. Ye, D. Hickman, L. Pena, G. Ramirez-Nieto, B. Kimble, and Y. Araya. 2009. Fitness of pandemic H1N1 and seasonal influenza A viruses during co-infection: evidence of competitive advantage of pandemic H1N1 influenza versus seasonal influenza. PLoS Curr. Influenza August:RRN1011. [DOI] [PMC free article] [PubMed]
- 26.Rameix-Welti, M. A., V. Enouf, F. Cuvelier, P. Jeannin, and S. van der Werf. 2008. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog. 4:e1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 28.Smith, H., and C. Sweet. 1988. Lessons for human influenza from pathogenicity studies with ferrets. Rev. Infect. Dis. 10:56-75. [DOI] [PubMed] [Google Scholar]
- 29.Staschke, K. A., J. M. Colacino, A. J. Baxter, G. M. Air, A. Bansal, W. J. Hornback, J. E. Munroe, and W. G. Laver. 1995. Molecular basis for the resistance of influenza viruses to 4-guanidino-Neu5Ac2en. Virology 214:642-646. [DOI] [PubMed] [Google Scholar]
- 30.Tai, C. Y., P. A. Escarpe, R. W. Sidwell, M. A. Williams, W. Lew, H. Wu, C. U. Kim, and D. B. Mendel. 1998. Characterization of human influenza virus variants selected in vitro in the presence of the neuraminidase inhibitor GS 4071. Antimicrob. Agents Chemother. 42:3234-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tannock, G. A., J. A. Paul, R. Herd, R. D. Barry, A. L. Reid, M. J. Hensley, R. S. Gillett, S. M. Gillett, P. Lawrance, and R. L. Henry. 1989. Improved colorimetric assay for detecting influenza B virus neutralizing antibody responses to vaccination and infection. J. Clin. Microbiol. 27:524-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, M. Z., C. Y. Tai, and D. B. Mendel. 2002. Mechanism by which mutations at His274 alter sensitivity of influenza A virus N1 neuraminidase to oseltamivir carboxylate and zanamivir. Antimicrob. Agents Chemother. 46:3809-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitley, R. J., F. G. Hayden, K. S. Reisinger, N. Young, R. Dutkowski, D. Ipe, R. G. Mills, and P. Ward. 2001. Oral oseltamivir treatment of influenza in children. Pediatr. Infect. Dis. J. 20:127-133. [DOI] [PubMed] [Google Scholar]
- 34.WHO. Accessed 20 November 2009. Pandemic (H1N1) 2009—update. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/swineflu/laboratory20_11_2009/en/index.html.
- 35.Wu, J. T., G. M. Leung, M. Lipsitch, B. S. Cooper, and S. Riley. 2009. Hedging against antiviral resistance during the next influenza pandemic using small stockpiles of an alternative chemotherapy. PLoS Med. 6:e1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yen, H. L., L. M. Herlocher, E. Hoffmann, M. N. Matrosovich, A. S. Monto, R. G. Webster, and E. A. Govorkova. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob. Agents Chemother. 49:4075-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]