Abstract
MK-2048 represents a prototype second-generation integrase strand transfer inhibitor (INSTI) developed with the goal of retaining activity against viruses containing mutations associated with resistance to first-generation INSTIs, raltegravir (RAL) and elvitegravir (EVG). Here, we report the identification of mutations (G118R and E138K) which confer resistance to MK-2048 and not to RAL or EVG. These mutations were selected in vitro and confirmed by site-specific mutagenesis. G118R, which appeared first in cell culture, conferred low levels of resistance to MK-2048. G118R also reduced viral replication capacity to approximately 1% that of the isogenic wild-type (wt) virus. The subsequent selection of E138K partially restored replication capacity to ≈13% of wt levels and increased resistance to MK-2048 to ≈8-fold. Viruses containing G118R and E138K remained largely susceptible to both RAL and EVG, suggesting a unique interaction between this second-generation INSTI and the enzyme may be defined by these residues as a potential basis for the increased intrinsic affinity and longer “off” rate of MK-2048. In silico structural analysis suggests that the introduction of a positively charged arginine at position 118, near the catalytic amino acid 116, might decrease Mg2+ binding, compromising enzyme function and thus leading to the significant reduction in both integration and viral replication capacity observed with these mutations.
Selective pressure exerted by antiretroviral drugs, in conjunction with high viral mutation rates, promotes the inevitable emergence of drug-resistant HIV-1 variants. This necessitates an ongoing search for novel antiretroviral compounds that either have novel mechanisms and inhibit different stages of viral replication or inhibit targets that have acquired resistance to existing drugs. In the latter case, such newer next-generation agents should ideally display resistance profiles which are distinct and nonoverlapping with those of the first-generation drugs.
Integration of viral cDNA into the host cell genome is a distinct feature of retroviral replication, and inhibitors of HIV-1 integrase have recently been added to the arsenal of clinically approved antiretroviral drugs. Raltegravir (RAL) was the first integrase strand transfer inhibitor (INSTI) to be approved by the U.S. Food and Drug Administration (FDA) after clinical trials showed that this drug promoted a rapid and sustained antiviral effect (13). Elvitegravir (EVG), another integrase inhibitor, is currently in phase III clinical trials (27). Resistance mutations common to both of these first-generation integrase inhibitors have been reported and can result in high levels of drug resistance (26). Mutations which engender cross-resistance between RAL and EVG have been reported in clinical trials, cell culture studies, and biochemical assays (9, 26). This has prompted the search for second-generation integrase inhibitors that might display novel patterns of resistance, allowing their use in patients who have failed therapy with RAL or EVG. MK-2048 (28) is a prototype second-generation INSTI that retains potency against viruses containing common single and double mutations observed in the clinic with first-generation agents with a 95% inhibitory concentration (IC95) in the nM range. MK-2048 has been previously reported to be active against viruses resistant to RAL and EVG (28, 29). Given common mechanisms of action among INSTIs and a lack of structural information on integrase inhibitor complexes with resistance mutations, an understanding of resistance to second-generation agents such as MK-2048 is important.
This article describes the selection of resistance to MK-2048 in tissue culture and the characterization of mutations associated with such resistance, G118R and E138K. The identification of distinct mutations which appear to confer resistance to MK-2048 and not to either RAL or EVG has potential implications for understanding the structural basis for the second-generation profile of this compound as well as future drug discovery and development efforts focused on this mechanism.
MATERIALS AND METHODS
Selections for resistance.
Cell culture selections were performed as previously described using both wild-type virus as well as a clinical isolate termed 4742 that contains a number of baseline polymorphisms in integrase at positions V72I, Q95P, T125A, V201I, and I203M (18, 24). Briefly, wild-type clinical isolates were serially passaged in cord blood mononuclear cells (CBMCs) or MT-2 cells in the presence of increasing concentrations of RAL, EVG, or MK-2048 (Fig. 1), for as long as 50 weeks. Potential resistance-associated mutations were identified by sequencing viral RNA as described previously (1).
FIG. 1.
Clinical structures of RAL, EVG, and MK-2048.
Site-directed mutagenesis and production of clonal virus.
The HIV-1 molecular clone pNL4-3 was engineered to contain the G118R and E138K mutations, singly or in combination, by site-directed mutagenesis using the following primers: G118R Forward (CAGACAATCGCAGCAATTTC), G118R Reverse (GAAATTGCTGCGATTGTCTG), E138K Forward (GGATCAAGCAGAAATTTGGCATTCC), and E138K Reverse (GGAATGCCAAATTTCTGCTTGATCC). For production of clonal virus, 293T cells were transfected using Lipofectamine 2000 (Invitrogen); viruses were collected after 48 h and passed through a 0.45-μm-pore filter. Enzyme-linked immunosorbent assays (ELISAs) were performed to quantify production of viral p24 protein.
Determination of drug 50% effective concentrations (EC50s) in cell culture.
A total of 75,000 PM1 cells per well were infected with clonal NL4-3 virus containing the integrase mutations G118R, E138K, or G118R and E138K or with wild-type viruses (45 ng p24 virus per well) by spinoculation at 1,200 × g for 2 h at 37°C. Cells were washed twice to remove unbound virus and then resuspended in RPMI growth medium alone or containing various concentrations of MK-2048, RAL, or EVG, ranging between 0.0256 nM and 2 μM. Three independent experiments were performed, each in duplicate. Supernatant reverse transcriptase (RT) activity was measured at 72 h postinfection as an indicator of virus replication. Data were normalized based on uninfected and no-drug controls included in each experiment. EC50s were calculated using GraphPad Prism 5.0 software by fitting data to sigmoidal dose-response (variable slope) curves. Viral replication capacity was determined in PM1 cells using the same methodology as that used to determine EC50s, by comparing levels of supernatant RT activity of each mutant virus to those of wild-type virus in the absence of drug.
qRT-PCR for viral RNA and qPCR for viral DNA.
A total of 75,000 PM1 cells per well were infected with clonal NL4-3 virus containing the integrase mutations G118R, E138K, or G118R and E138K or with wild-type viruses (45 ng p24 of virus per well) by spinoculation at 1,200 × g for 2 h at 37°C. Cells were washed twice to remove unbound virus and then resuspended in RPMI growth medium containing 1 μM darunavir to ensure that only one cycle of viral replication would occur. Real-time quantitative PCR (qPCR) was performed as previously described (31) with certain modifications (7).
(i) Early and late reverse transcripts and 2-LTR circles.
Cellular DNA was extracted with a DNeasy blood and tissue kit (Qiagen). PCR was performed with Platinum qPCR SuperMix-UDG (Invitrogen) on a Corbett Rotor-Gene 6000 thermocycler. The samples were normalized for their β-globin DNA contents and quantified against cloned standards that were diluted with DNA from uninfected cells. The cycling conditions were 50°C for 2 min, 95°C for 1 min, and 45 cycles at 95°C for 3 s and 60°C for 30 s, with 65 ng template being used per reaction mixture. The primers and probes used for the early reverse transcripts were primers ERT2F and ERT2R (22) and probe ERT (5′-6-carboxyfluorescein [FAM]-ACTAGAGATCCCTCAGACCCTTTT-BHQ1-3′). For the late reverse transcripts, primer total F, primer total R, and total probe were used (30). For two-long terminal repeat (2-LTR) circles, primer circle F, primer circle R, and circle probe were used (30). For β-globin, primer betaGlo-F (5′-GGTACGGCTGTCATCACTTAGAC-3′), primer betaGlo-R (5-AACGGCAGACTTCTCCTCAG-3′), and the betaGlo-probe (5′-FAM-CTCACCCTGTGGAGCCACACC-BHQ1-3′) were used.
(ii) Integrated DNA.
DNA was extracted and normalized as described above. A previously described Alu-gag PCR (31) was used with the following modifications. The first-round reaction was performed with undiluted samples (65 ng template) and 1:10 dilutions of each sample (6.5 ng template diluted with uninfected DNA, with 65 ng DNA total) in the presence of 2 mM MgCl2 and 200 μM deoxynucleoside triphosphates (dNTPs). Nine microliters of the resulting first-round product was used as the template for the second round of the nested reaction in the presence of 5 mM MgCl2 (final concentration, including the carryover from the first round) and 200 μM dNTPs; only the wild-type probe was used (31). The second-round cycling conditions were as described above for 2-LTR circles. To generate a standard curve for the relative quantification of integrated DNA, the Alu-gag PCR was first performed with a dilution series of DNA from infected PM1 cells (diluted with DNA from uninfected cells).
Structural analysis.
A crystal structure derived from a partial integrase sequence, Protein Data Bank identifier 1QS4 (11), was used to perform an in silico mutagenesis assay with PyMol (http://www.pymol.org) (4) and Chimera software (http://www.cgl.ucsf.edu/chimera/) (3).
RESULTS
Susceptibility of G118R- and E138K-containing viruses to integrase inhibitors.
Our initial selection studies involved a variety of clinical isolates that were subjected to selection pressure with MK-2048, RAL, or EVG. Potential resistance-associated mutations were identified by sequencing of the full-length IN gene following breakthrough of virus replication. A variety of resistance mutations that have already been described in the literature were selected with RAL and EVG within 8 to 10 weeks (5, 21). In contrast, exposure to MK-2048 led to the selection of G118R as a possible novel resistance mutation after 19 weeks (Table 1). Continued pressure with MK-2048 subsequently led to an additional substitution after 29 weeks at position E138K, within the IN gene, suggesting that the latter might be a secondary mutation. These mutations were identified in each of two separate series of selection experiments. Neither RAL nor EVG selected for the G118R mutations, and mutations at position 118 have not previously been observed against either RAL or EVG in either cell culture or clinical studies. The presence of a glycine (G) at this position is conserved across multiple HIV-1 subtypes, with polymorphisms not yet reported (25).
TABLE 1.
MK-2048 concentration during selection of virus strain 4742 resistance mutations in CBMCs
| wk | MK-2048 concn (μM) | Mutation(s) acquired |
|---|---|---|
| 1 | 0.001 | None |
| 19 | 0.05 | G118R |
| 29 | 0.25 | G118R and E138K |
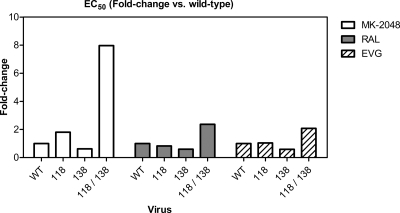
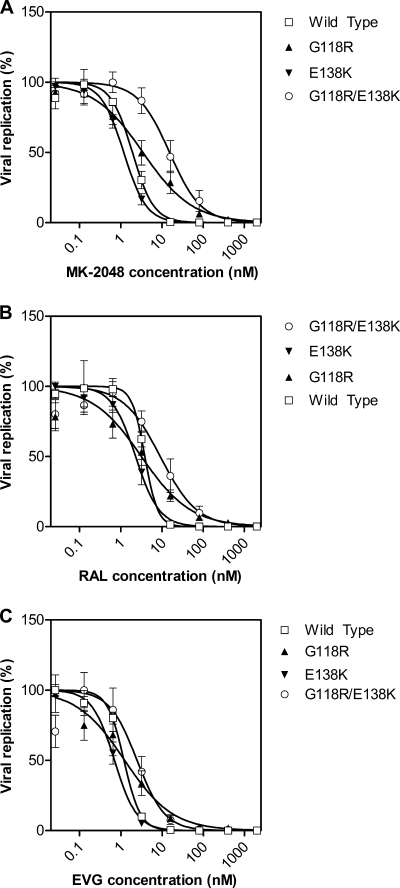
The doubly mutated selected virus containing both G118R and E138K showed reduced susceptibility to MK-2048 but remained susceptible to both RAL and EVG (Table 2.). In order to confirm the possible biological relevance of these substitutions to MK-2048 resistance, G118R and E138K were introduced, individually and in combination, into the pNL4-3 subtype B molecular clone by site-directed mutagenesis, and drug susceptibility and replication capacity were assessed. The results in Fig. 3 show that the G118R mutation conferred approximately 2-fold resistance to MK-2048 but no loss of susceptibility to either RAL or EVG. The E138K mutation on its own seemed to confer slightly increased susceptibility to all three integrase inhibitors compared to wild-type virus. In contrast, the G118R E138K double mutant conferred approximately 2-fold resistance to both RAL and EVG and about 8-fold resistance to MK-2048 (Fig. 2 and 3 and Table 2).
TABLE 2.
EC50s for MK-2048, RAL, and EVG against clonal mutant viruses
| Virus | EC50 in μM (95% confidence interval) |
||
|---|---|---|---|
| MK-2048 | RAL | EVG | |
| NL4-3 wt | 0.0019 (0.0016-0.0023) | 0.0038 (0.0033-0.0047) | 0.0012 (0.0010-0.0014) |
| NL4-3 G118R | 0.0035 (0.0021-0.0057) | 0.0032 (0.0016-0.0061) | 0.0012 (0.0007-0.0023) |
| NL4-3 E138K | 0.0012 (0.0010-0.0013) | 0.0023 (0.0018-0.0029) | 0.0007 (0.0006-0.0008) |
| NL4-3 G118R E138K | 0.0153 (0.0100-0.0234) | 0.0091 (0.0055-0.0150) | 0.0025 (0.0015-0.0042) |
FIG. 3.
Fold change in EC50s versus the wild type. The fold change in EC50 values for each drug is shown in terms of fold change versus wild-type virus for that drug (wild type = 1).
FIG. 2.
Dose-response curves for MK-2048, RAL, and EVG against resistant viruses. Dose-response curves for MK-2048 (A), RAL (B), and EVG (C) are shown for wild-type, G118R, E138K, and G118R E138K viruses. Data are fitted to sigmoidal dose-response (variable slope) curves, from which EC50 values were calculated. Data are from three experiments, and error bars represent the standard error of the mean (SEM).
Significantly impaired replication capacity of resistant viruses.
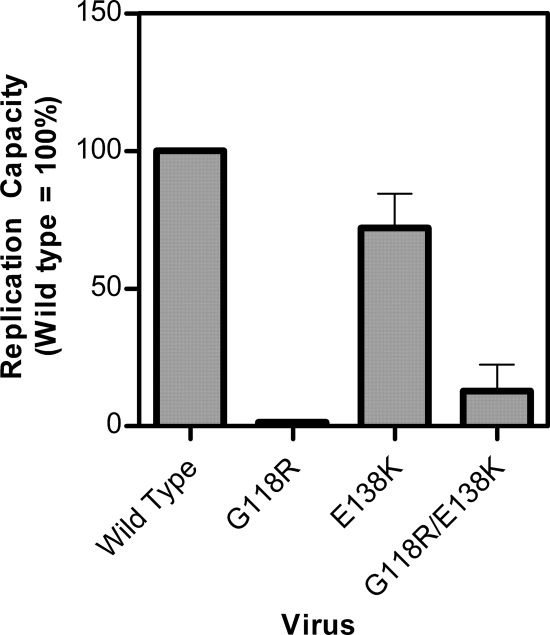
Although the G118R mutation alone conferred only slight resistance to MK-2048 but not to RAL or EVG, its presence resulted in a dramatic reduction in viral replication capacity compared to wild-type NL4-3. In the absence of antiviral drugs, production of 118R-containing virus at 72 h after infection was only 1% that of wild-type virus, as measured by reverse transcription assay of culture fluids, despite the fact that equal levels of viral p24 antigen (Ag) had been used to initiate infection (Fig. 4).
FIG. 4.
Replication capacity of wild-type and mutant viruses. Replication capacity was determined by comparing levels of RT activity in culture supernatants between mutated and wild-type viruses at 72 h postinfection. Error bars represent SEM.
The E138K substitution has been previously reported to be a secondary mutation with regard to both RAL and EVG that has only minimal impact on viral replication capacity (20). Introduction of E138K alone into NL4-3 conferred a slight reduction in replication capacity, but the addition of E138K to viruses containing G118R partially compensated for the G118R replication defect and enhanced replication capacity to levels approximately 9-fold higher than those of viruses containing G118R alone (Fig. 4). This notwithstanding, the replication capacity of the G118R E138K double-mutant virus remained significantly impaired: i.e., about 13% of that of wild-type viruses (Fig. 4). Thus, E138K both partially restored viral replication capacity and also contributed to increased levels of resistance against MK-2048. Of course, the clinical relevance of tissue culture viral replication capacity studies remains to be shown.
Assessment of reverse transcription and impairment of integration capacity with resistant viruses.
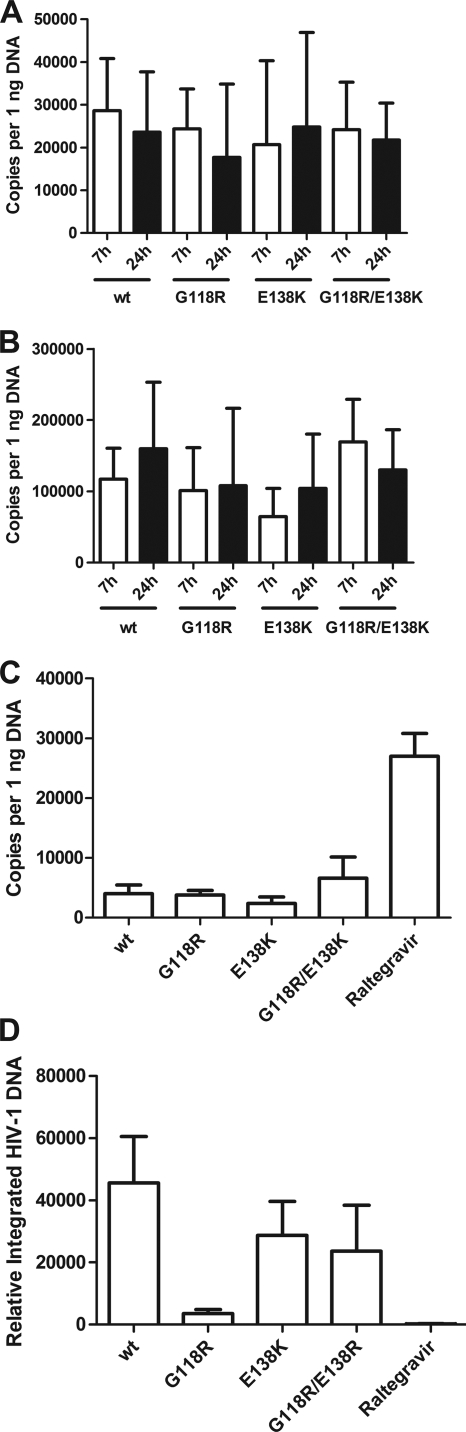
Early and late reverse transcription in wt viruses as well as in singly and doubly mutated resistant viruses was evaluated by real-time PCR. No effect of mutations at positions G118R and E138K in IN was observed (Fig. 5 A and B). Integration was assessed by examining accumulation of 2-LTR circles and integrated DNA product (Fig. 5C and D). The G118R-containing virus was associated with a significant decrease in integration of viral DNA. Consistent with the observed compensatory effect on replication, the E138K mutation was also able to partially restore integration capacity (Fig. 5D).
FIG. 5.
Reverse transcription and integration evaluated by real-time PCR. (A) Early reverse transcription products at 7 and 24 h postinfection; (B) late reverse transcription products at 7 and 24 h postinfection; (C) 2-LTR circle accumulation at 24 h postinfection; (D) integrated viral DNA at 24 h postinfection.
Structural analysis of G118R and E138K mutant integrases.
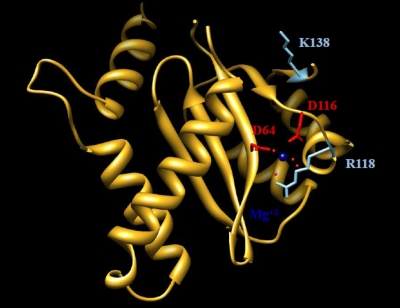
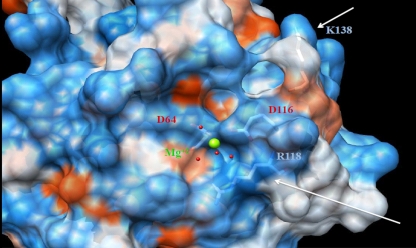
Although there is no full-length structure of the HIV-1 integrase, previously published structures of the catalytic core domain (11, 12) and the recently described structure of the full-length foamy virus intasome permit an examination of the potential effects that mutations might have on integrase enzymatic activity as well as on the organization and structure of the catalytic core and potential interactions with inhibitors. Amino acid 118 is very close to the catalytic triad of integrase, in particular to amino acid 116, one of the three amino residues that form the strictly conserved D(64)D(116)E(152) motif. This motif is conserved among all integrase enzymes and is essential for their catalytic function because of its role in the coordination of Mg2+ cations. Representation of the catalytic core domain as a ribbon diagram (Fig. 6) shows that a change from the flexible glycine (G) at position 118 to a positively charged arginine (R) and a change from a negatively charged glutamate (E) at position 138 to a positively charged lysine (K) might lead to alterations in the organization and structure of this domain. The location of amino acid 138 in proximity to a flexible loop makes it a candidate for a role in interactions with the target DNA (2, 23). In addition, these changes have the potential to alter the charge distribution across the catalytic core of the protein (Fig. 7). The position 118 residue is conserved in the foamy virus enzyme structure, suggesting its importance in these critical interactions.
FIG. 6.
Ribbon representation of the integrase catalytic core. Shown are G118R and E138K. Dark blue, Mg2+ cation; red, catalytic amino acids 64 and 116; light blue, amino acids 118 and 138.
FIG. 7.
Electrostatic surface representation of the integrase catalytic core. (A) G118R and E138K structure. On the surface, red represents negatively charged elements, white represents neutral elements, and blue represents positively charged elements. Green, Mg2+ cation. Underneath the semitransparent surface, side chains and amino acids 64, 116, 118, and 138 are shown. The arrow indicates amino acids 118 and 138 and the area of charge change as a result of the amino acid substitution.
DISCUSSION
As a result of cross-resistance between RAL and EVG, the need for second-generation integrase inhibitors with nonoverlapping resistance profiles is obvious. Resistance against MK-2048 has not previously been reported, but several pathways for resistance have been described for each of the RAL and EVG integrase inhibitors in both clinical and cell culture studies. Here we demonstrate the selection of G118R E138K double-mutated viruses, after a prolonged period of selection with MK-2048 but not RAL or EVG. Site-directed mutagenesis has confirmed that this doubly mutated virus is about 8-fold resistant to MK-2048, compared to the wild-type, but not to RAL or EVG (Fig. 3).
A recent clinical study showed that switching patients away from RAL-based treatment regimens because of resistance quickly resulted in nondectability of the RAL-resistant viral quasispecies, suggesting that the “fitness” cost of resistance mutations in integrase is high (4). This finding is consistent with the notion that a fitness-compromising mutation such as G118R might then require HIV-1 to accumulate secondary mutations in order to partially restore replication capacity. Previous work has documented that this is also true with regard to each of three different primary mutations that are associated with resistance to both RAL and EVG: i.e., 143, 148, and 155 (5, 6, 10, 23). Whether the removal of MK-2048 from culture medium would likewise lead to reversions and the possible reemergence of more replication-fit wild-type virus is currently under examination.
Mutations in integrase can affect various stages in the viral replication cycle (8). Class I mutants are blocked at the integration step, while class II mutants exhibit a more pleiotropic effect on various preintegration steps, such as reverse transcription and preintegrational nuclear import, or on postintegration steps, such as viral particle assembly (8, 19, 20). Until now, class II mutants have not been observed in the context of integrase inhibitor resistance. We used real-time PCR to evaluate reverse transcription and integration. The results showed that neither early nor late-stage reverse transcription was affected by either the G118R or E138K mutation, although integration was markedly affected and correlated with the effects of these mutations on overall replication capacity (Fig. 5). Assessment of integration efficiency suggests that G118R behaves like an IN class I mutation, affecting integration but not early or late reverse transcription (Fig. 5A to C). However, the absence of accumulation of 2-LTR circles suggests that G118R might also compromise integration for other reasons, as has been suggested for class II integrase mutations (19). Additional biochemical and cell culture studies are under way to determine whether G118R and E138K individually display a class I or class II mutational profile. We are also evaluating the effects of both of these mutations, alone and in combination, on integrase biochemical activity.
The active site of integrase, located in the enzyme catalytic core, is highly flexible (17). A partial crystal structure of the enzyme reveals only a single cation, yet it has been proposed that two cations are probably coordinated by the DDE motif: one by D64 and D116 and a second by D64 and E152 (11). INSTIs have been shown to chelate these critical cations in the enzyme active site and have high affinity for integrase only in the cDNA-bound state (15). Recent cocrystal structures of RAL and EVG with the intasome of prototype foamy virus (PFV) have confirmed the interaction of the pharmacophores of these compounds with two divalent metals at the active site as well as an interaction between the benzyl group and the bound viral DNA end (16). The overall conservation of these key structural interactions provides a plausible rationale for the broad cross-resistance between RAL and EVG as well as many other structurally diverse INSTIs but may not readily explain the unique resistance properties of second-generation compounds such as MK-2048.
The identification of a unique resistance pattern that can apparently confer resistance selectively to a prototypical second-generation INSTI can facilitate an understanding of the molecular activity of such molecules as MK-2048. Conceivably, G118R might confer resistance to MK-2048 as a result of conformational change induced by the addition of the positive charge at position 118 in the catalytic pocket. G118R might also result in a conformational change of the amino acids responsible for Mg2+ coordination, which might decrease the ability of the drug to chelate the cation. E138, which is close to the flexible loop comprised of amino acids 139 to 147, is thought to be involved in binding of DNA. Hence, an E138K substitution might affect the flexibility of this interaction, altering the conformation of integrase to a structure that is suboptimal for high-affinity INSTI interaction. The half-life of the integrase-RAL complex is 7.3 h, whereas that of MK-2048 is 32 h (14). Therefore, the increased affinity of MK-2048 for integrase might permit strong affinity even for mutated integrases derived from RAL-resistant viruses. The identification, as shown here, of a distinct resistance mutation not previously identified for first-generation integrase inhibitors raises the possibility that MK-2048 might exploit a different site(s) of interaction with integrase from those of RAL or EVG.
Using the cocrystal structure of RAL as a guide, modeling of MK-2048 suggests that G118R is uniquely selected with MK-2048, potentially because of the close hydrophobic interaction between the ethyl group (not present in RAL or EVG) and G118 (Jean-Francois Truchon, personal communication). The extended pharmacophore in MK-2048, as compared to RAL or EVG, might facilitate this unique interaction that could contribute to the enhanced affinity and longer off rate.
We have shown that the G118R mutation does not confer resistance to either RAL or EVG. This suggests that resistance against MK-2048, and possibly other second-generation INSTIs that have similar structures, may follow different pathways from those involved in resistance against either EVG or RAL.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (CIHR) and Merck, Inc. R.D.S. is the recipient of a postdoctoral fellowship from the CIHR Canadian HIV Trials Network and the Canadian Foundation for AIDS Research (CANFAR). D.A.D. is the recipient of a predoctoral fellowship from the Fonds de la Recherche en Santé du Québec (FRSQ).
We thank Yudong Quan and Bluma Brenner for helpful discussions, Susan Colby-Germinario for technical assistance, and Estrella Moyal and Bonnie Spira for assistance with digital artwork.
D.J.H. is an employee of Merck, Inc.
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Bar-Magen, T., R. Sloan, V. Faltenbacher, D. Donahue, B. Kuhl, M. Oliveira, H. Xu, and M. Wainberg. 2009. Comparative biochemical analysis of HIV-1 subtype B and C integrase enzymes. Retrovirology 6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, X., M. Tsiang, F. Yu, M. Hung, G. S. Jones, A. Zeynalzadegan, X. Qi, H. Jin, C. U. Kim, S. Swaminathan, and J. M. Chen. 2008. Modeling, analysis, and validation of a novel HIV integrase structure provide insights into the binding modes of potent integrase inhibitors. J. Mol. Biol. 380:504-519. [DOI] [PubMed] [Google Scholar]
- 3.Couch, G. S., D. K. Hendrix, and T. E. Ferrin. 2006. Nucleic acid visualization with UCSF Chimera. Nucleic Acids Res. 34:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLano, W. L. 2002. The PyMOL Molecular Graphics System.
- 5.Delelis, O., I. Malet, L. Na, L. Tchertanov, V. Calvez, A.-G. Marcelin, F. Subra, E. Deprez, and J.-F. Mouscadet. 2009. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 37:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delelis, O., S. Thierry, F. Subra, F. Simon, I. Malet, C. Alloui, S. Sayon, V. Calvez, E. Deprez, A. G. Marcelin, L. Tchertanov, and J. F. Mouscadet. 2010. Impact of Y143 HIV-1 integrase mutations on resistance to raltegravir in vitro and in vivo. Antimicrob. Agents Chemother. 54:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donahue, D. A., R. D. Sloan, B. D. Kuhl, T. Bar-Magen, S. M. Schader, and M. A. Wainberg. 2010. Stage-dependent inhibition of HIV-1 replication by antiretroviral drugs in cell culture. Antimicrob. Agents Chemother. 54:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman, A. 1999. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 52:411-426. [DOI] [PubMed] [Google Scholar]
- 9.Ferns, R. B., S. Kirk, J. Bennett, I. Williams, S. Edwards, and D. Pillay. 2009. The dynamics of appearance and disappearance of HIV-1 integrase mutations during and after withdrawal of raltegravir therapy. AIDS 23:2159-2164. [DOI] [PubMed] [Google Scholar]
- 10.Fransen, S., S. Gupta, R. Danovich, D. Hazuda, M. Miller, M. Witmer, C. J. Petropoulos, and W. Huang. 2009. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J. Virol. 83:11440-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldgur, Y., R. Craigie, G. H. Cohen, T. Fujiwara, T. Yoshinaga, T. Fujishita, H. Sugimoto, T. Endo, H. Murai, and D. R. Davies. 1999. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl. Acad. Sci. U. S. A. 96:13040-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldgur, Y., F. Dyda, A. B. Hickman, T. M. Jenkins, R. Craigie, and D. R. Davies. 1998. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. U. S. A. 95:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grinsztejn, B., B. Y. Nguyen, C. Katlama, J. M. Gatell, A. Lazzarin, D. Vittecoq, C. J. Gonzalez, J. Chen, C. M. Harvey, and R. D. Isaacs. 2007. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet 369:1261-1269. [DOI] [PubMed] [Google Scholar]
- 14.Grobler, J. A., P. M. McKenna, S. Ly, K. A. Stillmock, C. M. Bahnck, R. M. Danovich, G. Dornadula, D. J. Hazuda, and M. D. Miller. 2009. HIV integrase inhibitor dissociation rates correlate with efficacy in vitro. 18th International HIV Drug Resistance Workshop. Antiviral Ther. 14(Suppl. 1):A27. [Google Scholar]
- 15.Grobler, J. A., K. Stillmock, B. Hu, M. Witmer, P. Felock, A. S. Espeseth, A. Wolfe, M. Egbertson, M. Bourgeois, J. Melamed, J. S. Wai, S. Young, J. Vacca, and D. J. Hazuda. 2002. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. U. S. A. 99:6661-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hare, S., S. S. Gupta, E. Valkov, A. Engelman, and P. Cherepanov. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazuda, D. J., N. J. Anthony, R. P. Gomez, S. M. Jolly, J. S. Wai, L. Zhuang, T. E. Fisher, M. Embrey, J. P. Guare, M. S. Egbertson, J. P. Vacca, J. R. Huff, P. J. Felock, M. V. Witmer, K. A. Stillmock, R. Danovich, J. Grobler, M. D. Miller, A. S. Espeseth, L. Jin, I. W. Chen, J. H. Lin, K. Kassahun, J. D. Ellis, B. K. Wong, W. Xu, P. G. Pearson, W. A. Schleif, R. Cortese, E. Emini, V. Summa, M. K. Holloway, and S. D. Young. 2004. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc. Natl. Acad. Sci. U. S. A. 101:11233-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loemba, H., B. Brenner, M. A. Parniak, S. Ma'ayan, B. Spira, D. Moisi, M. Oliveira, M. Detorio, and M. A. Wainberg. 2002. Genetic divergence of human immunodeficiency virus type 1 Ethiopian clade C reverse transcriptase (RT) and rapid development of resistance against nonnucleoside inhibitors of RT. Antimicrob. Agents Chemother. 46:2087-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, R., A. Limon, E. Devroe, P. A. Silver, P. Cherepanov, and A. Engelman. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735-12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, R., A. Limon, H. Z. Ghory, and A. Engelman. 2005. Genetic analyses of DNA-binding mutants in the catalytic core domain of human immunodeficiency virus type 1 integrase. J. Virol. 79:2493-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McColl, D., S. Fransen, S. Gupta, N. Parking, N. Margot, S. Chuck, A. Cheng, and M. Miller. 2007. Resistance and cross-resistance to first generation integrase inhibitors: insights from a phase II study of elvitegravir (GS-9137), abstr. 9. Antiviral Ther. 12:S11. [Google Scholar]
- 22.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. U. S. A. 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakahara, K., C. Wakasa-Morimoto, M. Kobayashi, S. Miki, T. Noshi, T. Seki, M. Kanamori-Koyama, S. Kawauchi, A. Suyama, T. Fujishita, T. Yoshinaga, E. P. Garvey, B. A. Johns, S. A. Foster, M. R. Underwood, A. Sato, and T. Fujiwara. 2009. Secondary mutations in viruses resistant to HIV-1 integrase inhibitors that restore viral infectivity and replication kinetics. Antiviral Res. 81:141-146. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira, M., B. G. Brenner, and M. A. Wainberg. 2009. Isolation of drug-resistant mutant HIV variants using tissue culture drug selection. Methods Mol. Biol. 485:427-433. [DOI] [PubMed] [Google Scholar]
- 25.Rhee, S. Y., T. F. Liu, M. Kiuchi, R. Zioni, R. J. Gifford, S. P. Holmes, and R. W. Shafer. 2008. Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology 5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serrao, E., S. Odde, K. Ramkumar, and N. Neamati. 2009. Raltegravir, elvitegravir, and metoogravir: the birth of “me-too” HIV-1 integrase inhibitors. Retrovirology 6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimura, K., E. Kodama, Y. Sakagami, Y. Matsuzaki, W. Watanabe, K. Yamataka, Y. Watanabe, Y. Ohata, S. Doi, M. Sato, M. Kano, S. Ikeda, and M. Matsuoka. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J. Virol. 82:764-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vacca, J., W. J. Fisher, T. Embrey, M. Hazuda, D. Miller, M. Felock, P. Witmer, M. Gabryelski, and L. T. Lyle. 2007. Discovery of MK-2048—subtle changes confer unique resistance properties to a series of tricyclic hydroxypyrrole integrase strand transfer inhibitors, abstr. WEPEA088. 4th IAS Conf., Sydney, Australia.
- 29.Wai J., T. F., M. Embrey, M. Egbertson, J. Vacca, D. Hazuda, M. Miller, M. Witmer, L. Gabryelski, and T. Lyle. 2007. Next generation of inhibitors of HIV-1 integrase strand transfer inhibitor: structural diversity and resistance profiles, abstr. 87. Conf. Retroviruses Opportunistic Infect., 2007, Los Angeles, CA.
- 30.Yamamoto, N., C. Tanaka, Y. Wu, M. O. Chang, Y. Inagaki, Y. Saito, T. Naito, H. Ogasawara, I. Sekigawa, and Y. Hayashida. 2006. Analysis of human immunodeficiency virus type 1 integration by using a specific, sensitive and quantitative assay based on real-time polymerase chain reaction. Virus Genes 32:105-113. [DOI] [PubMed] [Google Scholar]
- 31.Yu, J. J., T. L. Wu, M. K. Liszewski, J. Dai, W. J. Swiggard, C. Baytop, I. Frank, B. L. Levine, W. Yang, T. Theodosopoulos, and U. O'Doherty. 2008. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology 379:78-86. [DOI] [PMC free article] [PubMed] [Google Scholar]