Abstract
The alphaherpesvirus proteins UL31 and UL34 and their homologues in other herpesvirus subfamilies cooperate at the nuclear membrane in the export of nascent herpesvirus capsids. We studied the respective betaherpesvirus proteins M53 and M50 in mouse cytomegalovirus (MCMV). Recently, we established a random approach to identify dominant negative (DN) mutants of essential viral genes and isolated DN mutants of M50 (B. Rupp, Z. Ruzsics, C. Buser, B. Adler, P. Walther and U. H. Koszinowski, J. Virol 81:5508-5517). Here, we report the identification and phenotypic characterization of DN alleles of its partner, M53. While mutations in the middle of the M53 open reading frame (ORF) resulted in DN mutants inhibiting MCMV replication by ∼100-fold, mutations at the C terminus resulted in up to 1,000,000-fold inhibition of virus production. C-terminal DN mutants affected nuclear distribution and steady-state levels of the nuclear egress complex and completely blocked export of viral capsids. In addition, they induced a marked maturation defect of viral capsids, resulting in the accumulation of nuclear capsids with aberrant morphology. This was associated with a two-thirds reduction in the total amount of unit length genomes, indicating an accessory role for M53 in DNA packaging.
Our understanding of herpesvirus morphogenesis is mainly derived from studies of Alphaherpesvirinae, such as herpes simplex virus type 1 (HSV-1) and pseudorabies virus (PrV). A faster replication cycle and a more productive infection in tissue culture aided genetic analysis of alphaherpesvirus morphogenesis. In addition, deletion mutants of key morphogenesis genes in alphaherpesviruses often maintain basic replication capacity, whereas the mutations of their homologues in Betaherpesvirinae or Gammaherpesvirinae mostly result in a lethal phenotype (for the UL31 and the UL34 family, see references 3, 6, 9-11, 16, 20, 21, and 42). These genes became amenable to comprehensive genetic analysis in betaherpesviruses only after their genomes were cloned as infectious bacterial artificial chromosomes (BACs), which obviated the need to generate replication-competent intermediates or complementing cell lines (3, 21, 23). BAC-based mutagenesis allowed viability screens mapping essential genes (8, 43) or even functional sites of essential genes in cytomegaloviruses (3, 21). However, these approaches cannot easily be applied to reveal the null phenotypes in the context of virus replication, as mutant viruses are not easily reconstituted. In addition, deletion of an essential viral gene can reveal the null phenotype of only the first of perhaps several essential functions during virus morphogenesis. This problem can be addressed to some extent by using dominant negative (DN) mutations (36). DN mutants are loss-of-function mutants that induce a null phenotype in the presence of the wild-type (wt) allele (14). Analysis of phenotypes induced by DN mutants proved to be extremely useful in genetics and cell biology, signaling, and biochemistry. Such inhibitory mutants of cellular proteins are often designed based on knowledge on the structural or functional role of a well-characterized protein domain. Unfortunately, we lack the structural information that would allow knowledge-based design of viral DN mutants for the majority of herpesvirus gene products. Thus, we established a random screen consisting of three steps to identify mutants of viral genes with DN potential (36): (i) a library of mutants is generated by random insertion of 5 amino acids (aa) or a stop codon into the open reading frame (ORF) of interest using transposon mutagenesis, (ii) nonfunctional mutants are identified by cis complementation of the respective deletion mutant mouse cytomegalovirus (MCMV) BAC, and (iii) nonfunctional mutants are tested for their inhibitory potential upon reconstitution of the wt BAC cloned genomes. In the last screen, mutants that have a specific inhibitory effect on the activity of the wt allele are selected. The specific phenotype obtained upon induction of the inhibitory mutants in the context of virus replication is then verified and further characterized using a tetracycline (Tet) regulon-based viral conditional expression system (36, 37).
One intriguing aspect of herpesvirus morphogenesis is the transition of capsids from the nuclear to the cytoplasmic phase of virus morphogenesis. Two conserved nonstructural proteins, the homologues of the membrane protein pUL34 and its nuclear partner protein pUL31, form a nuclear egress complex (NEC) (18, 27, 42), which is required for primary envelopment and export of nuclear capsids to the cytoplasm (reviewed in references 24 and 25). Recent studies have revealed that the homologues of alphaherpesvirus pUL34 and pUL31, the M50 and the M53 gene products of the betaherpesvirus MCMV (pM50 and pM53, respectively) and the BFRF1 and the BFLF2 gene products of the gammaherpesvirus Epstein-Barr virus (EBV), apparently share the major functions of these two proteins. The lack of one or both proteins of the NEC generally results in the retention of viral capsids in the nucleus. This is lethal for beta- and gammaherpesvirus production (3, 9-11, 16, 18, 21, 27, 35, 42).
The details of the mechanisms by which the NEC proteins mediate capsid export through the nuclear envelope are poorly understood. We (3, 21, 36, 38) and others (1, 19, 34) have started to dissect details of the NEC function using a genetic approach based on subtle mutagenesis of the respective genes. Analysis of the MCMV M50 gene by comprehensive mutagenesis localized two different functional sites. They were the M53 binding site within the N-terminal domain of M50, as well as the transmembrane region at its C terminus (3). Liang and Baines located the respective binding site in HSV-1 UL34 at aa 137 to 181 (19). Our approach, based on screens for DN mutants, identified a proline-rich sequence (aa 179 to 207) in the M50 gene product as an additional essential region (36). A recombinant virus expressing an M50 mutant lacking this site was defective in capsid egress from the nucleus despite the presence of the wt M50 protein. Consequently, the production of infectious particles after infection was reduced by more than 2 orders of magnitude. The UL34 homologues of alpha- and gammaherpesviruses lack a similar polyproline motif, but the result was confirmed by mutating the human cytomegalovirus (HCMV) homologue UL50 at the corresponding region, which is conserved within betaherpesviruses (36). The M50 mutants lacking the proline-rich motif still bind and colocalize to their respective NEC partner, pM53. Interestingly, Bjerke and coworkers also provided genetic evidence for the existence of at least one additional, yet-unknown, but essential functional entity in pUL34 of HSV-1, besides its known pUL31 binding activity, using a screen based on charged-cluster mutations (1). Further analysis of one of the noncomplementing charged-cluster mutants carrying the defect in the N-terminal domain of pUL34 also revealed a DN activity and suggested a new functional site involved in membrane curvature formation, together with the C-terminal domain of UL31 (34).
The genetic analysis of M53 by Tn7-based linker scanning mutagenesis, followed by a cis complementation assay, localized the M50-binding site between aa 112 and 137 within the first of the four conserved regions (CRs) shared among the herpesvirus UL31 homologues (21). This analysis, together with a study we performed for further characterization of pM50/pM53 interaction, revealed that the large C-terminal part of pM53, comprising CR2 to -4, must carry at least one additional, yet-unknown, but essential functional site (21, 38).
Here, we screened loss-of-function mutants of the MCMV M53 gene to retrieve M53 alleles with DN activity to localize this new functional domain. Mutants with a very strong inhibitory potential accumulated within CR4 of pM53 close to its C terminus. These CR4 mutants induced a block of capsid export from the nucleus. In addition, we could associate these mutations with the induction of a defect in capsid maturation and/or DNA packaging. These data suggested that pM53 is not only crucial for nuclear egress, but also involved in earlier steps of MCMV morphogenesis.
MATERIALS AND METHODS
Cells and viruses.
Mouse embryonic fibroblasts (MEFs), NIH 3T3 fibroblasts (ATCC CRL-1658), M2-10B4 cells (a bone marrow-derived stromal cell line; ATCC CRL-1972), and SVEC 4-10 endothelial cells (ATCC CRL-2181), were propagated as described previously (22). Mouse mammary epithelial C127 cells (ATCC CRL-1616) and 293 cells (ATCC CRL-1573) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) (36). The pSM3fr-FRT BAC carries a target site for Flp recombinase (FRT site) between the m16 and m17 genes. MCMV reconstituted from this BAC showed growth properties identical to those of wt MCMV strain Smith in vitro and in vivo (4) and therefore served as the wt MCMV control. All mutant viruses generated in this study were based on the pSM3fr-FRT BAC and were reconstituted in duplicate by transfection of subconfluent MEFs on a 6-cm dish with 1.5 μg of the respective BAC DNA using Superfect transfection reagent (Qiagen), in accordance with the manufacturer's instructions. Twenty-four hours posttransfection, the cells were replated onto 10-cm dishes, and the medium was replaced weekly. The reconstituted viruses were propagated, and virus stocks were prepared on M2-10B4 cells as described previously (37). Virus titers were quantified by a plaque assay of MEFs as described previously (31).
The infection experiments described in this study were carried out without centrifugal enhancement, and the multiplicity of infection (MOI) was calculated directly as a function of the determined PFU of the virus stocks used and the cell numbers on the day of infection.
Plasmids and construction of mutant M53 alleles.
Forty-two M53 mutants cloned in the pori6k-ie, vector (3), which were described in our previous study (21), were tested for their capacities to inhibit virus reconstitution after insertion into the pSM3fr-FRT BAC. They included 7 stop mutants (s168, s185, s233, s290, s294, s309, and s331) and 35 insertion mutants (i115, i128, i131, i138, i146, i154, i161, i168, i182, i191, i195, i198, i201, i207, i212, i217, i220, i227, i234, i236, i244, i251, i259, i264, i266, i272, i281, i292, i303, i307, i313, i321, i324, i325, and i326). In addition, 4 new M53 mutants were generated from wt M53 cloned in the pori6k-ie vector. One insertion mutation at histidine 262 of the M53 ORF was generated by PCR with overlapping primers (H262Sfor1/H262Srev1 and H262Sfor2/H262Srev2) (Table 1). The assembled PCR products were inserted into pori6k-ie-M53 (21) by ApaI/KpnI, generating po6k-ie-i262. To generate three other stop mutants within CR4 of the M53 ORF, stop codons were introduced at positions 303, 321, and 327 using specific reverse primers (D303rev1, D321rev1, and D327rev1) (Table 1) and a common forward primer (D303for1) to amplify the C-terminal region of M53 with the respective mutations. All PCR products were reinserted into pori6k-ie-M53 using BspMI/NsiI, resulting in pori6k-ie-s303, -s321, and -s327.
TABLE 1.
Primer sequences used in construction of the expression vectors
| Primer name | Size (bp) | Tma(°C) | Sequence |
|---|---|---|---|
| s309for | 31 | 77.6 | 5′ GCG CAT GCA TCC ATG TTT AGG AGC CCG GAG G 3′ |
| s309rev | 29 | 73.2 | 5′ GCG CCT CGA GTT AAA CAA GCA TCA GCA CG 3′ |
| T309for | 30 | 72.1 | 5′ AAA AGG CGC GCC AAG CTT GGT ACC ATG TTT 3′ |
| T309rev | 29 | 71.8 | 5′ GGG AAA AAG CC ACCT TCC TCT GCA GCA TT 3′ |
| H262Sfor1 | 25 | 69.5 | 5′ GGG GGG TAC CAT GTT TAG GAG CCC G 3′ |
| H262Srev1 | 29 | 68.1 | 5′ CCC CAT CGA TAC CGT CTC CTT CTC GAA AA 3′ |
| H262Sfor2 | 26 | 71.1 | 5′ GGG GAT CGA TCC CGT CCC AGT GCA TC 3′ |
| H262Srev2 | 27 | 68.0 | 5′ GGG GGC ATC AGG CGA AAT TGT AAA CGG 3′ |
| D303for1 | 29 | 72.3 | 5′ GGG GAA AAC ACA CGT GCC TGG ACC TCT CC 3′ |
| D303rev1 | 30 | 73.6 | 5′ GGG GAT GCA TCA GCT CGA CTC TAG CCG CAG 3′ |
| D321rev1 | 30 | 72.2 | 5′ GGG GAT GCA TCA GAT CTC GAG CTC GTC CAC 3′ |
| D327rev1 | 30 | 72.2 | 5′ GGG GAT GCA TCA CTC GTT CGT CTC GTT GGG 3′ |
| Sak1for | 30 | 75.0 | 5′ GGG GAC CGG TCG AGA AGA GGA GGA AGG AGG 3′ |
| Sak1rev | 30 | 70.9 | 5′ GGG GAC CGG TGC TCC TAA ACT TAT CGT CGT 3′ |
| SakFlagfor | 39 | 79.9 | 5′ GGG GAC CGG TCG AGA AGA TGC TCC TAA ACT TAT CGT CGT 3′ |
| SVTinsfor | 38 | 84.0 | 5′ GGG GGG GGC GCG CCA AGC TTG GTA CCA TGT TTA GGA GC 3′ |
| CMVSak | 19 | 67.4 | 5′ GCC CAA CGA CCC CCG CCC A 3′ |
| M53for1 | 19 | 61.0 | 5′ CAC ACG TGC CTG GAC CTC T 3′ |
| M53rev1 | 19 | 61.0 | 5′ AGA GGT CCA GGC ACG TGT G 3′ |
| Pack1for | 20 | 67.0 | 5′ TGC ATC GAC GGT CCC AGC CA 3′ |
| Pack1rev | 20 | 65.0 | 5′ CCG CGG TGG TCC CCA TTG TG 3′ |
Tm, melting temperature.
In order to detect the s309 gene products in the presence of the wt M53, a Flag tag was introduced at the N terminus of the s309 ORF. The Flag tag coding sequence was fused to the extreme N terminus of the M53 ORF by gene synthesis and subcloned into pori6k-ie-M53, resulting in pori6k-ie-Flag-M53. The pori6k-ie-Flag-M53 plasmid was treated with BspMI/NsiI endonucleases, and the internal BspMI/NsiI fragment was replaced by the respective BspMI/NsiI fragment derived from pori6k-ie-s309, generating pori6k-ie-Flag-s309. The coding sequence for the epitope that is recognized by the M53 polyclonal antiserum (27) was deleted from the s309 ORF using PCR with overlapping primers (CMVsak/Sak1rev and Sak1for/D303rev1) (Table 1). The assembled PCR products were inserted into the NdeI/BspMI-treated pori6k-ie-Flags309, generating pori6k-ie-Flag-ΔN-s309.
Generation of virus mutants from recombinant MCMV BACs.
To insert the pori6k-ie-based loss-of-function mutants of M53 into the wt MCMV BAC, Escherichia coli strain DH10B (Invitrogen) containing the pSM3fr-FRT BAC and the temperature-sensitive Flp recombinase expression plasmid pCP20 (7) was transformed with pori6k-ie constructs containing different M53 mutants one by one and treated as described previously (3). The correct recombinants were verified by NotI restriction analysis. During virus reconstitution, three MCMV BACs were reconstituted as controls: the original pSM3fr-FRT BAC; the ΔM53MCMV BAC, comprising a pSM3fr-FRT BAC derivative in which the M53 gene was deleted (21); and the ΔM53MCMV-EK128A BAC, in which the M53 deletion is rescued by ectopic insertion of an attenuated point mutant (21). A virus reconstitution experiment was considered valid if (i) plaques of wt MCMV BAC (pSM3fr-FRT BAC)-derived virus occurred after 1 week and grew exponentially over 2 more weeks (positive control), (ii) transfection with ΔM53MCMV-EK128A BAC resulted in plaque formation between the second and fourth weeks (attenuated virus control), and (iii) no plaques were observed for the ΔM53MCMV BAC transfection after a total of 6 weeks (negative control).
Conditional gene expression.
The viral ORFs from pori6k-ie-M53 and selected constructs carrying inhibitory mutants of M53 (i207, s309, i313, and i321) or their tagged derivatives were cloned into the Tet-regulated expression cassette controlled by the simian virus 40 (SV40) early enhancer previously described (37). All conditional constructs reported here were generated on the basis of the pO6SVT-TR vector (37). The s309 ORF was amplified by PCR using the T309for and T309rev primers (Table 1). The PCR product was inserted into po6k-ie-SVTe by AscI/BsiWI, generating the po6k-SVT-s309 construct. The i313 and i321 insertion mutants were first cloned into the pL-M53 Litmus plasmid (21) by BamHI/BspEI, resulting in pL-i313 and pL-i321. Next, the insertion mutants were recloned into po6k-ie-SVTe by AscI/StuI, generating po6k-ie-SVTi313 and po6k-ie-SVTi321. The mutant M53 fragment from i207 was inserted into po6k-ie-SVT-M53 by BspMI/BsiWI, replacing the wt sequences and generating po6k-ie-SVT-i207. The Flag-M53, Flag-s309, and Flag-ΔN-s309 ORFs were amplified from the respective po6k-ie constructs by PCR using the SakFlagfor and D303rev1 primers (Table 1). The PCR products were inserted into po6k-ie-SVT-M53 and po6k-ie-SVT-s309 by AscI/BspMI, generating po6k-ie-SVT-Flag-M53, po6k-ie-SVT-Flag-s309, and po6k-ie-SVT-Flag-ΔN-s309, respectively. To insert the conditional expression cassettes into the MCMV genome, E. coli strain DH10B (Invitrogen) containing the pSM3fr-FRT BAC and the temperature-sensitive Flp recombinase expression plasmid pCP20 was transformed with the po6k-ie-SVTe-based constructs and propagated as described above. This procedure resulted in recombinant viruses that carried a regulation cassette expressing the respective M53 derivatives. The recombinant viruses were named to indicate the promoter and the regulated mutants they carry and were labeled with an “R” for the regulation cassette at the end, for example, MCMV-SVT-M53R.
Analysis of viral growth kinetics.
For analysis of virus replication under multistep growth conditions, MEFs or NIH 3T3, M2-10B4, SVEC 4-10, or C127 cells were infected in duplicate at an MOI of 0.1 with the BAC-derived wt and recombinant MCMVs. The inoculum was removed after 1 h, and normal medium or medium supplemented with 1 μg/ml doxycycline (dox) was added. On day 3, dox was added a second time. The supernatants of infected cells were harvested on days 0, 1, 3, and 5 postinfection (p.i.), and virus titers were determined by plaque assay on MEFs in the absence of dox. For analysis of virus replication under single-step growth conditions, M2-10B4 or C127 cells were infected at an MOI of 1 both in the absence and in the presence of dox. Supernatants were collected on days 0, 1, 2, and 3 p.i., and virus titers were quantified as described above.
Transmission electron microscopy.
NIH 3T3 cells were grown on carbon-coated sapphire discs and infected with viruses conditionally expressing wt M53 or the s309 mutant at an MOI of 0.5 with or without addition of 1 μg/ml dox for 72 h. The cells were fixed by high-pressure freezing using an HPF 01 (Engineering Office M. Wohlwend GmbH, Switzerland), freeze substituted, and embedded in plastic as described previously (36).
Southern blot analysis.
NIH 3T3 cells were infected with wt BAC-derived MCMV or recombinant viruses that conditionally expressed the wt M53 or one of the s309, i313, and i321 mutants at an MOI of 2, in both the presence and absence of dox. Cells were harvested at 48 h p.i., and total nuclear DNA was extracted using a DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's instructions. All samples were digested with SphI endonuclease at 37°C and separated on a 0.8% agarose gel. The fragments were transferred to Hybond-N membranes (Amersham Biosciences) and hybridized with a probe specific for the left terminus of the MCMV genome, called PACK, according to the manufacturers’ protocol. The PACK probe was generated and labeled with digoxigenin by PCR using the PCR Dig probe Synthesis Kit (Roche). Pack1for and Pack1rev primers (Table 1) were used to amplify the respective region of the pSM3fr wt MCMV BAC. The specific hybridization was visualized with the CSPD Luminescent detection kit for nucleic acids (Roche).
Western blot analysis.
Semiconfluent NIH 3T3 cells in 6-cm dishes were infected with the viruses of interest in the absence and in the presence of 1 μg/ml of dox at an MOI of 0.5. After 24 h, the NIH 3T3 fibroblasts were washed with phosphate-buffered saline (PBS), scratched from the plates, and resuspended in PBS. The cell suspension was lysed directly on ice in loading buffer (62.5 mM Tris, pH 6.8, 2% [vol/vol] sodium dodecyl sulfate [SDS], 10% [vol/vol] glycerol, 6 M urea, 5% [vol/vol] β-mercaptoethanol, 0.01% [vol/vol] bromphenol blue). Soluble proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto Hybond-P membranes (GE Healthcare) by semidry blotting in the presence of blotting buffer (25 mM Tris, 192 mM glycine, 20% [vol/vol] methanol, pH 8.3). The membranes were blocked in Tris-buffered saline (0.05% Tween 20) (TBS-T) containing 5% nonfat dry milk overnight at 4°C. For pp89 (ie1) (89 kDa) detection, the membrane was incubated at 4°C with TBS-T containing a specific mouse monoclonal antibody (croma 101; kindly provided by S. Jonjic, University of Rijeka, Rijeka, Croatia). Next, the membrane was washed with TBS-T and incubated at room temperature (RT) with goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Dianova). To detect M86 (150 kDa) or M53 (38 kDa), specific polyclonal rat antisera were used (21, 27) (primary antibodies). Goat anti-rat horseradish peroxidase-conjugated antibody (Dianova) served as a secondary antibody. For the detection of M50 (35 kDa), a polyclonal rabbit antiserum was used (27). Here, a donkey anti-rabbit horseradish peroxidase-conjugated antibody (Dianova) was used as a secondary antibody. To detect the Flag-tagged proteins, Flag-M53 and Flag-ΔN-s309, an anti-Flag M2 monoclonal mouse antibody (Sigma) was used as the primary antibody and a goat anti-mouse horseradish peroxidase-conjugated antibody (Dianova) as the secondary antibody. The proteins under study were visualized with the ECL-Plus Western blot detection system (GE Healthcare).
HA tag-specific IP-Western analysis.
Subconfluent 293 cells in 6-cm dishes were cotransfected with expression plasmids encoding hemagglutinin (HA)-tagged M50 and M53 mutants. Twenty-four hours after transfection, the cells were scratched from the plates and resuspended in PBS. The cells were then collected by centrifugation, and the pellets were lysed in immunoprecipitation (IP) lysis buffer (10 mM Tris-HCl, pH 8.0, 400 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, and 1× Complete protease inhibitor from Roche). The lysates were cleared by centrifugation at 14,000 × g. The cleared lysates were loaded onto high-affinity HA-agarose columns (Roche) and processed according to the manufacturer's instructions. The bound proteins were eluted in loading buffer (62.5 mM Tris, pH 6.8, 2% [vol/vol] SDS, 10% [vol/vol] glycerol, 6 M urea, 5% [vol/vol] β-mercaptoethanol, 0.01% [vol/vol] bromphenol blue) and processed as described above for Western blots.
Confocal laser scanning microscopy.
NIH 3T3 cells grown on glass coverslips were infected with the MCMV-SVT-Flag-M53R and MCMV-SVT-Flag-ΔN-s309R viruses at an MOI of 0.5 for 28 h with or without addition of 1 μg/ml dox. The cells were washed once with PBS and fixed by paraformaldehyde as described previously (21). M53 localization was visualized with a specific polyclonal rat antiserum as the primary antibody and a fluorescein isothiocyanate (FITC)-conjugated goat anti-rat immunoglobulin G (Dianova) as the secondary antibody. The intracellular distributions of the M50 and Flag-tagged proteins were visualized with specific polyclonal rabbit antiserum (anti-M50) and with anti-Flag polyclonal rabbit antibody (Cell Signaling), respectively, as primary antibodies and Texas red-conjugated goat anti-rabbit immunoglobulin G (Dianova) as the secondary antibody. Stained cells were analyzed by using a Zeiss Axiovert 200 M microscope and the Zeiss LSM510 Meta laser system.
RESULTS
Genetic screen for inhibitory M53 mutants that block virus production.
DN alleles encode mutant proteins that inhibit the function of the wt gene product. Here, we tested the general applicability of our procedure described for the M50 gene (36) to recover inhibitory mutants for its partner, M53. The collection of nonfunctional mutants of the M53 gene (21) were inserted via Flp recombination one by one into the FRT site of the wt pSM3fr-FRT BAC. The resulting recombinants contained the intact M53 locus, together with the M53 mutant, which was expressed under the control of the constitutive HCMV immediate-early promoter in the intergenic region between the nonessential genes m16 and m17. Virus reconstitution after transfection of MEFs by BAC DNA was assessed by recording plaque formation for a period of 6 weeks after transfection. We tested a total of 42 random and 4 targeted M53 mutants, altogether comprising 10 mutants that contained stop codons and 36 in-frame insertions of 5 aa. Two independent clones for each 46 loss-of-function mutants of M53 were tested in duplicate for virus reconstitution. Constructs which did not lead to viral plaque formation upon transfection were reisolated from bacteria and retested four times in total. The results of this genetic screen are summarized in Fig. 1. Four stop mutants (s303, s309, s321, and s327) and nine insertion mutants (i259, i272, i292, i307, i313, i321, i324, i325, and i326), accumulating within CR4, prevented plaque formation. Another set of M53 mutants in which the 5-aa insertions located within CR2 (i182, i191, i195, i198, i207, i217, and i220) and one in CR3 (i281) were not lethal but resulted in severely delayed plaque formation. In the presence of mutants with this phenotype, the first plaques took as long as 4 weeks to appear rather than the usual 4 to 5 days after transfection. These plaques did not grow, and the supernatants contained little, if any, infectious virus progeny, which could not be retrieved for further virus propagation.
FIG. 1.
Screening for inhibitory mutants of the MCMV M53 gene. The amino acid sequence encoded by the M53 gene of MCMV is displayed, including the four conserved regions (CR1 to -4; underlined), and the positions of the different mutations are indicated. Transposon insertions (arrowheads) resulting in introduction of 5 aa (depicted above the sequence) or stop codons (depicted below the sequence) are indicated along the ORF. Mutants preventing virus reconstitution after insertion into the wt MCMV BAC are labeled by black symbols, and mutants which did not interfere with virus reconstitution are depicted by white symbols. The intermediate phenotype (see the text) is indicated by gray symbols.
Conditional expression of the inhibitory mutants of M53.
As constitutive expression of the DN allele results in a block of virus reconstitution, conditional expression of the mutants of interest is required to study the inhibitory phenotype in the context of virus replication (33, 36). Thus, virus reconstitution is allowed in the “off” state while the DN phenotype is analyzed upon induction. We constructed recombinant MCMV BACs for conditional expression of a selected set of the inhibitory mutants of M53. The regulation cassette we used (37) contained two transcription units: one expressed the mutant of interest under the control of a promoter that contained inhibitory tetO sites, while the other expressed the Tet repressor (tetR) (Fig. 2A). Upon insertion of this cassette into the MCMV genome via Flp recombinase, it was, by default, set in the “off” state, as the constitutive HCMV promoter resulted in prompt and strong expression of the repressor, which immediately interfered with the expression of the tetO-containing transcription unit. In the presence of dox, which inhibits tetR binding to tetO sites, the regulated cassette is released from this repression and the regulated gene of interest is expressed according to the characteristics of the promoter/enhancer (33, 36, 37). The wt M53 ORF and four mutants (i207, s309, i313, and i321) were inserted into the regulation cassette, in which the expression of the mutants was driven by the SV40 early enhancer. The i207 mutant is representative of DN insertion mutants within CR2, while the s309, i313, and i321 mutants represent the group of CR4 DN mutants. As a control, we also constructed a dox-inducible expression cassette for the wt M53 gene. The resulting constructs were integrated into the MCMV genome one by one by site-specific recombination at the FRT site between the m16 and m17 genes (Fig. 2A). The recombinant viruses (MCMV-SVT-i207, -s309R, -i313R, -i321R, and -M53R) were reconstituted by transfection of MEFs in the absence of dox and propagated in M2-10B4 cells.
FIG. 2.
Effect of conditional expression of inhibitory M53 mutants on virus growth. (A) Schematic representation of the conditional cassette used for regulated expression of the M53 mutants. The M53 mutants (M53 alleles) were cloned into the SV40 enhancer-based conditional expression cassette of a vector plasmid carrying an FRT site (modified after Rupp et al. [37]). These constructs were delivered to the MCMV BAC by Flp recombination at an FRT site located between the m16 and m17 genes within the HindIII A fragment as described previously (36, 37). The wt M53 gene is located within the HindIII H fragment. Permissive cells are transfected with the recombinant BACs to reconstitute the viruses carrying the regulation cassettes of M53 mutants. The M53 mutants are not expressed during virus reconstitution, because the constitutively expressed tetracycline repressor (tetR) blocks their transcription. Upon dox administration, tetR is inactivated and the DN M53 mutants are expressed under the control of the SV40 promoter (SV40e-tetO2). (B) Effects of the M53 inhibitory mutants on the production of viral progeny. M2-10B4 cells were infected for 5 days at an MOI of 0.1 in either the absence or the presence of 1 μg/ml dox. Wt MCMV was used as a positive control for infection and a negative control for dox toxicity. In addition, the virus overexpressing the wt M53 allele represented the control for M53 overexpression. Cell supernatants were collected on the indicated days postinfection, and the released infectious units were determined by titration on MEFs. The balcony diagram shows the regulation of virus growth in response to dox administration 3 days or 5 days after infection with the indicated constructs. The titer reduction induced by dox is shown as a ratio between the titer in the absence of dox and the titer in the presence of dox. Only titer reductions of >10-fold were considered significant. Each mutant was analyzed in duplicate in at least two independent experiments. The error bars indicate standard deviations.
The replication of MCMV-SVT-i207, -s309R, -i313R, -i321R, and -M53R was analyzed under multistep growth conditions in the absence and presence of dox. In the absence of dox, all of the recombinant viruses grew to titers indistinguishable from that of the control virus (data not shown). Strong dox-induced expression of a second gene copy of wt M53 had no significant effect on the replication of the control virus (Fig. 2B, M53R). In contrast, induction of the s309, i313, and i321 mutants completely prevented virus growth, i.e., it resulted in up to 1,000,000-fold reduction of virus titers at 5 days p.i. (Fig. 2B). Expression of the i207 mutant resulted in a reduction of viral titers of about 100-fold (Fig. 2B, i207R). Previously, we had observed cell-type-specific limitations on conditional expression of the DN fusion protein of MCMV SCP in C127 cells. These included a lower dynamic range of induction and higher background expression levels of the DN allele (37). Therefore, we tested the inhibitory potential of the s309 mutant in NIH 3T3 fibroblasts, the endothelial cell line SVEC 4-10, and, particularly, the epithelial cell line C127. In contrast to the SCP DN, which failed to regulate virus production in C127 cells, dox-dependent inhibition by M53-derived s309 was strong and similar in all cell lines tested (data not shown).
CR4 mutants affect the steady-state levels and distribution of wt M53.
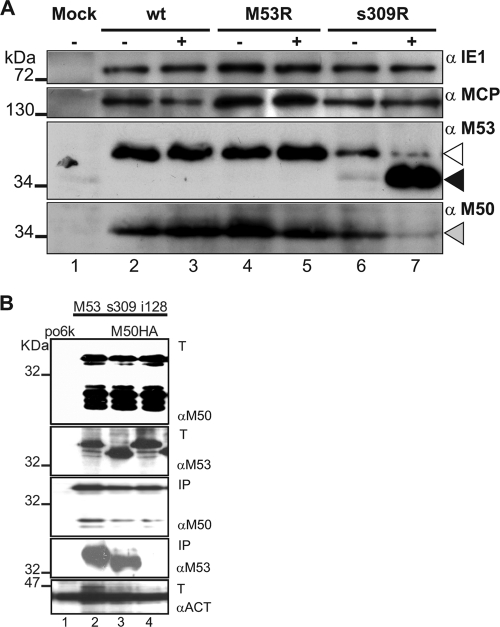
One mode of action of a DN mutant is to directly compromise the function of the wt protein. Therefore, we first tested the distribution and steady-state levels of wt pM53 in the presence of the DN mutants. To this end, NIH 3T3 fibroblasts were infected at an MOI of 0.5, with either MCMV-SVT-M53R or MCMV-SVT-s309R, in the absence and presence of dox. Wt-MCMV-infected cells served as an additional control. The infected cells were lysed 24 h after infection, and samples were analyzed by SDS-PAGE, followed by Western blots probed with specific antibodies to a selected set of viral proteins (Fig. 3A). No dox-dependent changes in expression of the immediate-early gene 1 product (pp89/89-kDa protein) (Fig. 3A, top blot) and of the late major capsid protein (MCP) gene product (M86/150 kDa) (Fig. 3A, second blot from top) were observed in cells infected with MCMV-SVT-M53R or MCMV-SVT-s309R. As expected, dox treatment resulted in enhanced expression of wt M53 in MCMV-SVT-M53R. Some basal expression of s309 in the MCMV-SVT-s309R recombinant was already seen in the off setting, whereas after dox induction, strong expression of s309 was evident (Fig. 3A, third blot from top, lanes 6 and 7). Notably, the amount of wt M53 protein decreased in the presence of s309 overexpression. In addition, the M50 signal also appeared to be weaker after induction of s309. Dox treatment alone did not affect the levels of the wt NEC proteins (Fig. 3A, lanes 3 and 4). Therefore, expression of the s309 DN mutant caused the destabilization of M53, or perhaps even of both wt NEC proteins.
FIG. 3.
Analysis of viral-protein expression upon induction of DN s309 overexpression. (A) NIH 3T3 cells were mock treated (lane 1) or infected at an MOI of 0.5 with wt MCMV (lanes 2 and 3), with MCMV mutants overexpressing the M53 wt allele (M53R; lanes 4 and 5), or with the DN s309 allele (s309R; lanes 6 and 7) in the absence (−; lanes 1, 2, 4, and 6) and presence (+; lanes 3, 5, and 7) of 1 μg/ml dox. Protein expression was analyzed 24 h after infection by Western blotting with specific antibodies against pp89 (immediately-early protein, 89 kDa; IE1; top), M86 (late protein, 150 kDa; MCP; second blot from top), M53 (38 kDa), and M50 (35 kDa) (NEC proteins; third and fourth blots from top, respectively). The dox-induced s309 (≅35 kDa; black arrowhead) expression decreased the native M53 signal (open arrowhead) and also reduced levels of M50 (gray arrowhead). (B) The DN mutant s309 binds to M50. 293 cells were transfected with the vector pori6-ie (po6k; lane 1) or cotransfected with plasmids expressing HA-tagged M50 (lanes 2 to 4) and the indicated M53 derivatives: wild type M53 (lane 2), s309 (lane 3), and i128 (lane 4). Thirty percent of the transfected cells were lysed in IP lysis buffer, and one-third of these lysates were loaded (upper two gels and bottom gel, indicated by T) and Western blotted with rabbit anti-M50 antiserum (αM50), rat anti-M53 antiserum (αM53), and rabbit anti-actin antiserum (αACT), respectively. The remaining 70% of the lysates were processed by an HA pulldown. The eluates from the HA columns were split 1:9 and Western blotted with rabbit anti-M50 antiserum and rat anti-M53 antiserum, respectively (third and fourth gels from top, respectively, indicated by IP). The expected molecular mass of the M50 protein is 35 kDa; the wt M53 and the mutant i128 protein migrate at about 38 kDa and s309 at about 34 to 35 kDa.
It has been reported that the levels of HSV-1 UL31 protein and its homologues depend on their interaction with the UL34 homologues (42). In order to study whether DN s309 still bound to M50 like wt M53 did (27), we performed a pulldown assay. 293 cells were cotransfected with plasmids expressing HA-tagged M50 (HA-M50) and either wt M53 or the s309 mutant. As a negative control, the cells were cotransfected with HA-M50 and i128, a well-characterized mutant of M53 that does not bind to M50 (21). As a negative control for the Western blotting, 293 cells were transfected with the po6k-ie vector. Twenty-four hours after transfection, the cells were lysed, and the cleared lysates were either subjected to a control Western blot directly or to an HA tag-specific immunoprecipitation using anti-HA-agarose columns (Roche). The HA tag-specific immunoprecipitates were analyzed with Western blots specific for either M50 or M53. The s309 bound HA-M50 as the wt M53 did (Fig. 3B, compare lanes 2 and 3 in the fourth gel from the top). In contrast, as expected, the i128 mutant did not react (Fig. 3B, lane 4 in the fourth blot from the top). We concluded that the s309 mutants of M53 can interact with M50 and are recruited into the wt NEC via M50.
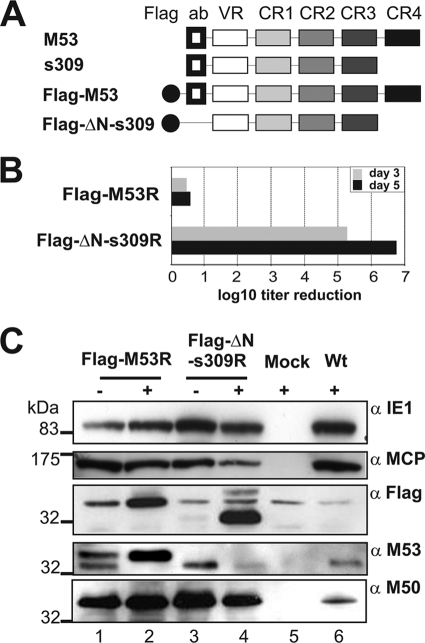
Next, we asked whether the expression of s309 influenced the distribution of the NEC in the virus context. To distinguish s309 from the wt M53 protein and to trace its effect on the subcellular localization of the wt NEC proteins, we replaced the nonessential N-terminal 15-aa epitope (N) of s309, which is recognized by the anti-M53 antiserum (21), by a 9-aa Flag tag, resulting in the Flag-ΔN-309 construct. This construct permitted discrimination between mutant and wt proteins by Flag-specific and M53-specific antibodies. In addition, the electrophoretic mobility of this M53 derivative is also different from that of M50. To assess nonspecific effects of the Flag tag insertion, the wt M53 ORF was changed accordingly (Fig. 4A). These modified ORFs were introduced into the regulation cassette and flipped into pSM3fr-FRT as described above. The reconstituted viruses MCMV-SVT-Flag-ΔN-s309R and MCMV-SVT-Flag-M53R were analyzed upon dox induction in M2-10B4 cells. While expression of Flag-M53 did not affect virus replication, Flag-ΔN-s309 still prevented virus production (Fig. 4B). The regulation of Flag-ΔN-s309 by dox-induced expression was confirmed by Western blotting (Fig. 4C). Basal expression of both the wt and the mutant proteins was detectable in the off settings, but strong induction upon dox treatment was evident, similar to MCMV-SVT-s309R (Fig. 3). Destabilization of wt pM53 upon overexpression of the DN mutant was confirmed, whereas the expression of pM50 was not detected in this setting. As the two proteins have similar migratory properties in this gel (Fig. 3A, fourth gel from top, lane 7), overexpression of s309 may affect the representation of the M50 protein.
FIG. 4.
Flag tagging does not affect the inhibitory features of DN s309. (A) Schematic representation of the tagging strategy. Depicted are the wt M53 ORF domains. The N-terminal epitope recognized by the M53-specific serum (ab), the variable region (VR), and the C-terminal four conserved regions (CR1 to -4) are indicated. The N-terminal end of the s309 mutant was replaced by a Flag tag (black circle). This construct (Flag-ΔN-s309) is not recognized by the anti-M53 serum. (B) Flag tagging does not affect the inhibitory features of s309. M2-10B4 cells were infected at an MOI of 0.1 in either the absence or the presence of 1 μg/ml dox with Flag-M53R and Flag-DN-s309R viruses. Cell supernatants were collected on the indicated days postinfection, and the released infectious units were determined by titration on MEFs. The balcony diagram shows the regulation of virus growth in response to dox administration 3 days or 5 days after infection. The titer reduction induced by dox is the ratio between the titer in the absence of dox and the titer in the presence of dox. The results of one representative experiment set up by duplicates of both viruses are shown. (C) NIH 3T3 cells were infected at an MOI of 0.5 with the FlagM53R (lanes 1 and 2) and the Flag-ΔN-s309R (lanes 3 and 4) viruses in the absence (−; lanes 1 and 3) and presence (+; lanes 2 and 4) of 1 μg/ml dox. Uninfected cells (lane 5, +) and wt infection (lane 6, +) were used as negative and positive controls, respectively. Protein expression was analyzed 24 h after infection by Western blotting using antibodies against pp89 (immediate-early protein, 89 kDa; IE1), M86 (late protein, 150 kDa; MCP), M53 (38 kDa), and M50 (35 kDa) (NEC proteins); and Flag tag. pp89 and M86 did not show any significant difference after dox was added to the system. The native M53 protein (38 kDa) was detected in the absence of dox, together with Flag-M53 protein (≥40 kDa). Accumulation of Flag-tagged wt M53 (Flag-M53; ≥40 kDa) and s309 (Flag-ΔN-s309; ∼32 to 33 kDa) upon dox induction was associated with a reduction in the native M53 protein levels.
Strong expression of the Flag-tagged wt protein also destabilized the native M53 protein, indicating that wt M53 that is not bound in an M50/M53 complex is degraded. This mutant has no growth phenotype, because the native protein is replaced by a functional derivative. This result confirms observations in HSV, namely, that the interaction with pUL34 affects the stability of the pUL31 homologue (42).
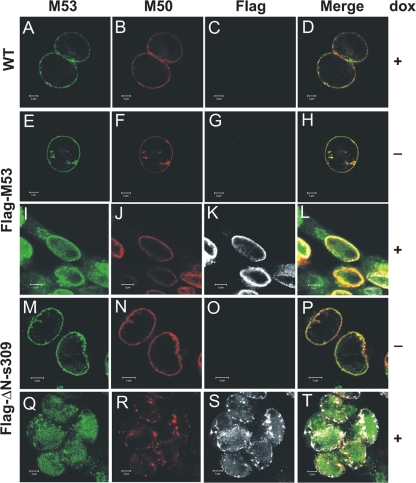
Next, MCMV-SVT-Flag-M53R and MCMV-SVT-Flag-ΔN-s309R were used to study the subcellular distributions of DN and wt NEC proteins. NIH 3T3 cells infected at an MOI of 0.5 were analyzed at 28 h after infection by immunofluorescence microscopy. The typical colocalization of the M53 and M50 proteins at the nuclear rim (21, 27) was observed when NIH 3T3 cells were infected with wt MCMV (Fig. 5A, B, and D), MCMV-Flag-M53R (Fig. 5E, F, and H), or MCMV-Flag-ΔN-s309R in the absence of dox (Fig. 5M, N, and P). In dox-treated cells, the Flag antibody detected the strong expression of both wt M53 and a mutant s309 derivative (Fig. 5, compare G to K and O to S, respectively). Dox-induced overexpression of Flag-M53 resulted in nucleosolic staining, in addition to the expected localization at the nuclear rim, a distribution also seen after isolated expression of the protein in the absence of M50 (Fig. 5I, K, and L). Since the M53-specific antiserum detects both wt M53 and Flag-M53, it was not clear whether wt M53 was also distributed to the nucleosol. Dox-induced expression of Flag-ΔN-s309 confirmed the nucleosolic localization of both wt M53 and Flag-ΔN-s309 (Fig. 5Q and S). In addition, the typical dispersed and homogeneous nuclear-rim staining of wt M53 was changed to a discrete aggregated and clumped distribution. M50 was also redistributed and showed the same aggregates at or beneath the nuclear rim as wt M53 (Fig. 5Q, R, S, and T). Altogether, expression of the inhibitory s309 DN mutant of M53 had fundamental effects on the subcellular distributions of both NEC proteins.
FIG. 5.
Subcellular distribution of the wt NEC protein upon expression of DN s309. NIH 3T3 cells were infected at an MOI of 0.2 with Flag-M53R (E to L) and Flag-ΔN-s309R (M to T) in the absence (E to H and M to P) and presence (I to L and Q to T) of 1 μg/ml dox. Analysis of wt MCMV infection (A to D, in the presence of dox) was used as a control. Twenty-four hours after infection, the localization of the M53, M50, Flag-M53, and Flag-ΔN-s309 proteins was analyzed by confocal immunofluorescence microscopy using specific primary antibodies, followed by Alexa fluorochrome-coupled secondary antibodies (Alexa 488, M53; Alexa 555, Flag tag; Alexa 633, M50). The scale bars indicate 5 μm.
DN s309 expression blocks capsid export from the nucleus.
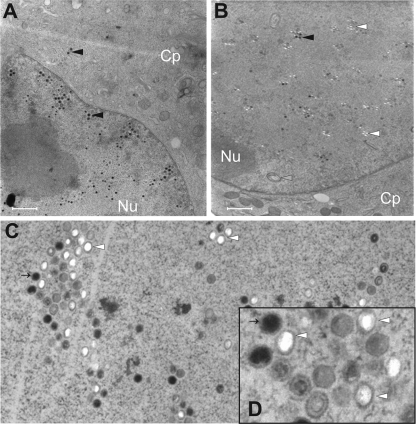
M53 and its homologues play crucial roles in nuclear egress of capsids. Thus, we predicted that a DN inhibitor of M53 should interfere with nuclear export of capsids. To test this, NIH 3T3 cells were infected with MCMV-SVT-M53R or MCMV-SVT-s309R, in the absence and in the presence of dox. Wt-MCMV-infected cells served as an additional control. Seventy-two hours postinfection, the intracellular distribution of viral capsids was analyzed by transmission electron microscopy of high-pressure frozen and freeze-substituted samples. Wt MCMV capsids were found in the nucleus and the cytoplasm (data not shown) (36). The same distribution of capsids was found for MCMV-SVT-M53R (data not shown) and MCMV-SVT-s309R in the absence of dox (Fig. 6A). However, in dox-treated MCMV-SVT-s309R-infected cells, a primary envelopment was not detected, and capsids remained strictly confined to the nucleus (Fig. 6B). In addition, expression of s309 caused accumulation of capsids with unusual morphology (Fig. 6C and D). About 50% of the capsids had regular icosahedral morphology but lacked density inside their shells and were dissimilar to the regular ring structure that is characteristic of B capsids. In addition, there were only a few mature C capsids, characterized by their high-density cores. A core without density is typical for A capsids lacking DNA. As it was previously shown that dox treatment alone does not induce a maturation defect in MCMV (36), our data showed that the overexpression of an s309-induced phenotype was characterized not only by a block of capsid export, but also by a defect in nuclear capsid maturation.
FIG. 6.
DN s309 prevents viral-capsid egress from the cell nucleus and induces accumulation of aberrant capsids in the nucleus. NIH 3T3 cells were infected for 72 h at an MOI of 3 with viruses that conditionally expressed s309 in the absence (A) or in the presence (B, C, and D) of 1 μg/ml doxycycline. The cells were fixed by high-pressure freezing, freeze-substituted, embedded in plastic, and analyzed by electron microscopy. The black arrowheads point at mature C-type viral capsids, and the white arrowheads point at capsids with unusual morphology. The gray arrowheads point at intranuclear membrane accumulation. (D) Electronic zoom of panel C; the upper left capsid is indicated by a thin arrow to aid orientation. The scale bars indicate 1 μm.
DN s309 M53 interferes with cleavage of unit length genomes.
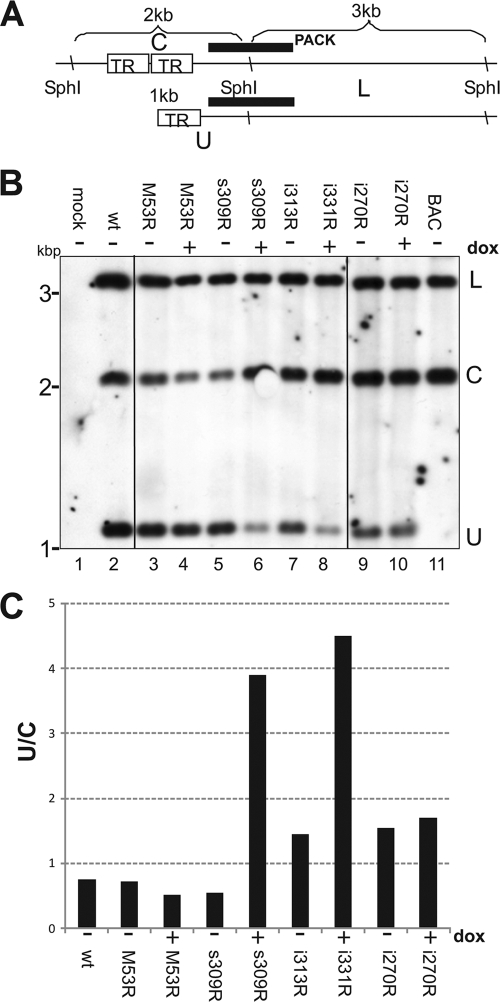
The nonuniform shape of intranuclear capsids was suggestive of accumulations of intermediates or by-products of the genome-packaging process (29). To test whether s309 induces a capsid maturation defect by interfering with the DNA-packaging machinery, we analyzed the cleavage of viral genomes. Unit length genomes are released from concatemeric replication intermediates during DNA packaging. In herpesviruses, this reaction is coupled with the transfer of viral DNA into procapsids (2, 29). To this end, NIH 3T3 cells were infected at an MOI of 2 in the absence or presence of dox with viruses which conditionally express the wt M53 or the s309, i313, and i270 mutants. DNA was extracted from infected cells 48 h after infection, treated with SphI restriction endonuclease, and subjected to Southern blot analysis (Fig. 7). SphI cleavage of the linear genome results in an approximately 1-kb terminal fragment representing the nucleotide positions (nt) 1 to 1298, according to the reference sequence (30) (GenBank NC_004065), and an ∼3-kb subterminal fragment (nt 1299 to 4521, predicting a 3,222-bp fragment). A probe hybridizing with both fragments can visualize the extent of DNA cleavage (a schematic representation is shown in Fig. 7A), because in the concatemeric form of viral genomes, the terminal 1,298-nt fragment is fused to the other terminal fragment (nt 229385 to 230238, representing 894 bp). The fragment comprising both viral termini is detected as an ∼2-kb fragment after SphI restriction (the left terminal 1,298-bp fragment plus the right terminal 894-bp fragment). During genome packaging, the intensity of the 2-kb fragment representative of concatemers (C) should decrease and the intensity of the 1-kb fragment representative of the abundance of unit length genomes (U) should increase. The abundance of the 3-kb subterminal fragment, however, does not change during the cleavage/packaging reaction and represents a control for the relative genome load. The circular form of the pSM3fr-16FRT17 BAC mimics the concatemeric form, showing only the 2-kb and 3-kb fragments (Fig. 7B, lane 11).
FIG. 7.
DN mutants of M53 CR4 inhibit the generation of unit length genomes. (A) Schematic representation of the Southern blot assay for analysis of genome cleavage/packaging during MCMV infection. The terminal regions of MCMV genomes are depicted as they formed concatemers after replication (top) or as the genome ends were released as a consequence of a cleavage/packaging reaction (bottom). In the concatemers, the genomes are connected via the short terminal repeats (TR) of the MCMV. The hybridization probe used in this study (black bar) overlapped with both the left terminal and the next right subterminal fragments of the SphI-digested MCMV genome. The subterminal fragment (L; ∼3 kbp) was detected in all genomic forms. Therefore, it served as a measure for the genome load. In the concatemeric form, the left terminal fragment is connected to the right terminal fragment. Thus, the same probe recognizes the concatemeric genome ends as an ∼2-kb fragment (C). In the unit length genomes, the left terminal fragment is separated from the other terminal fragment and an ∼1-kb fragment (U) is recognized by the same probe. (B) Southern blot analysis of a genome cleavage/packaging reaction upon induction of DN M53 mutants. NIH 3T3 cells were mock infected (lane 1) or infected with wt MCMV (lane 2), MCMV-M53R (lanes 3 and 4), -s309R (lanes 5 and 6), -i313R (lanes 7 and 8), and -i207R (lanes 9 and 10) at an MOI of 2 both in the absence (−; lanes 1, 2, 3, 5, 7, and 9) and in the presence (+; lanes 4, 6, and 10) of dox. Purified wt MCMV BAC DNA (lane 11) was used as a hybridization control. Forty-eight hours after infection, the nuclear DNA was extracted and digested with SphI. Then, the DNA fragments were separated by agarose gel electrophoresis and analyzed by Southern blotting using a digoxigenin-tagged “PACK” probe, depicted in panel A. The detected fragments are indicated on the right of the blot according to the labeling specified for panel A (L, C, and U). (C) Quantification of the inhibition of the cleavage/packaging induced by DN M53 mutants. The density of each band of the Southern blot shown in panel B was quantified using the ImageJ Gel Analysis Tool (http://rsb.info.nih.gov/ij/). The densities of the C and U fragments were normalized to the absolute densities measured for the L fragments in the corresponding lanes. The ratios of the normalized densities of the concatemeric C fragment to the normalized densities of the respective U fragments representing the unit length genomes are plotted (U/C).
Total genomic DNA derived from wt-MCMV-infected cells contains all three fragments (Fig. 7B, lane 2). Quantification of the respective bands detected about 60% of the viral DNA in the concatemeric, uncleaved form (Fig. 7C, lane 2). The same pattern was observed when cells were infected with MCMV-SVT-M53R in the absence or presence of dox (Fig. 7B and C, lanes 3 and 4, respectively) and s309R and i313 in the absence of dox (Fig. 7B and C, lanes 5 and 7). In contrast, dox-induced expression of s309R and i313R reduced the intensity of the 1-kb fragment derived from the unit length genomes to about one-third (Fig. 7B and C, lanes 6 and 8). Notably, dox-induced expression of DN i207 did not alter the DNA cleavage pattern of this mutant (compare Fig. 7B and C, lanes 9 and 10). These data suggest that mainly the DN CR4 mutants of M53 affect the DNA cleavage/packaging process.
DISCUSSION
Dominant negative mutants are valuable tools to dissect functional properties of multifunctional proteins. Recently, we reported on the identification of DN mutants of the MCMV NEC protein M50 (36). Here, we report a similar screen for M53, the other partner of the MCMV NEC. Our results confirm and expand earlier results by showing that our random-mutagenesis approach to isolate viral DN mutants is reproducible and generally applicable. The number of identified DN alleles and the quality of the screens we performed during this study were superior to those of the previous one. In this screen, the coverage and number of individual insertions increased, allowing detailed analysis of small coding stretches. We believe this was due to improved procedures for generating mutant libraries (3, 21). These improvements now make it feasible to test collections of viral-gene mutants in the future.
Here, we identified inhibitory M53 mutants accumulating in two regions of the ORF: (i) insertions near the C-terminal end of the M53 ORF, almost fitting the borders of CR4 (n = 10), exerted a strong inhibitory effect on MCMV growth; (ii) insertions accumulating within CR2 (n = 7) resulted in weaker but still highly significant inhibition and need further study. Only a few additional inhibitory mutants were identified in the rest of the M53 ORF, which were found in close proximity to CR4, namely, in the C-terminal end of the predicted CR3 or in the spacer region between CR3 and CR4. Conditional expression of the strong CR4 DN mutants revealed that they mildly affect capsid maturation and strongly block nuclear export of capsids. This suggests that the functional C-terminal domain of M53 expands over the borders of CR4 annotated by homology prediction. It seems remarkable, however, that a simple alignment of conserved sequences in this ORF nicely correlates with the functional relevance determined by a random mutational screen. It will be interesting to test whether this is predictive of the results of a targeted screen of the CR4 domains of other herpesvirus UL31 ORFs. Interestingly, while studying a DN mutant of UL34 in HSV-1, a new functional activity of UL31 was localized recently to the CR3 of that protein, which may be involved, together with the N-terminal region of UL34, in induction of membrane curvature (34). We did not retrieve inhibitory mutants from the CR3 of M53. However, these data suggest that the i259 and i272 mutants merit further study.
Overexpression of the CR4 mutant most likely induced degradation of the wt M53 and relocalized the NEC. It is possible that CR4 mutants affect a yet-unknown protein-protein interaction by pM53 that is involved in the correct distribution or functional structure of the NEC. Interestingly, there was no detectable primary envelopment in the presence of the DN M53, suggesting that a defect induced by this mutation was affecting an initial step of the primary envelopment or that the link between the primary envelopment and previous steps of the nuclear morphogenesis was broken.
Studies on the functionality of the conserved viral proteins pUL31 and pUL34 preferentially addressed their roles in the primary envelopment of virus capsids at the nuclear membrane and their effects on the nuclear lamina (26-28, 32, 39). There is a preference for primary envelopment of DNA-containing capsids over envelopment of immature capsid forms that lack viral DNA (5, 13). However, so far there has been no link between capsid morphogenesis and the viral mediators of capsid export at the inner nuclear membrane (namely, the NEC) (25). Here, we have shown a strong effect of a DN M53 on capsid export, which explains the lack of viral progeny in the supernatant. In addition, we propose that the members of the UL31 family connect nuclear capsid maturation with the recruitment of mature capsids to the inner nuclear membrane. Interference with this process by a DN reduced, but did not completely prevent, capsid maturation.
Such a mild DNA-packaging defect of the UL31-deficient HSV-1 was noted already when the first deletion mutants for this family of herpesvirus proteins were analyzed (6). The focus of the research on the UL31 family members then shifted to their pivotal roles in nuclear capsid export. Recently, others again observed a connection between the egress of capsids from the nucleus and DNA packaging (12, 15, 17). In EBV, the packaging defect and the accumulation of A and B particles was apparent after deletion of the UL31 homologue BFLF2 (12). This mutant has a defect in intracellular capsid migration and DNA packaging, whereas there is still export of defective viral particles. This finding was interpreted as an evolutionary divergence of the gammaherpesvirus version of the UL31 family, which resulted in acquisition of related but distinct functions, in addition to its role in primary envelopment. Indeed, the effect of UL31 deletion on DNA packaging is just traceable in alphaherpesviruses (6). Here, we report on an inhibitory phenotype of DN mutants of M53, a betaherpesvirus UL31 homologue, which is associated with inhibition of the cleavage/packaging process. The number of capsids lacking DNA was only ∼50%, and there was a 3-fold reduction in the number of unit length genomes. Apparently the effect on DNA cleavage/packaging is not responsible for the strength of the DN effect on virus replication, which is explained by the complete block of capsid export, since this process is essential in MCMV. Nevertheless, these data support the notion of a link between capsid maturation and nuclear export exerted by UL31 family members, which is apparently conserved in all three subfamilies of herpesviruses.
Specific detection of the DN and wt M53 made it possible to follow the fate of the wt protein in excess of the DN mutant. The s309 DN mutant of M53 binds M50 and thereby displaces the native M53 from its interaction with M50. This results in destabilization of the wt M53 protein. The failure in binding the UL34 homologue as a cause for degradation of UL31 has also been observed in an alphaherpesvirus (42) and indicates a conserved property. We believe that s309 and the other DN CR4 mutants act by displacing the wt protein from the NEC.
The processes that link capsid maturation with capsid egress at the nuclear membrane are so far unknown. Our data could be interpreted to mean that M53 might sense capsid maturation. Under steady-state conditions, the UL31 homologues are clearly localized at the nuclear envelope. It is attractive to hypothesize that either the process of inner nuclear membrane invagination already connects the NEC with mature capsids or that members of the UL31 family shuttle between the nucleosol and the inner nuclear membrane. Thus, they might escort mature capsids to the inner nuclear membrane docking sites formed by UL34 homologues. A simple model would be that the M53 (UL31 family) proteins sense a structural change on the capsid once packaging is completed or even contribute to completion of this process. The signal could be provided by the portal protein, one of the terminase subunits, or even accessory proteins involved in DNA packaging or stabilization of DNA-containing capsids, because capsids of double-stranded DNA (dsDNA) viruses change their structural properties during maturation, which affects the affinity of binding proteins (40). In yeast two-hybrid assays, NEC proteins interact with the members of the UL33 family (M51 in MCMV) (41). Also, a UL25 deletion mutant of PrV showed a defect in nucleocapsid export. This gene product is considered to be involved in DNA packaging (15) and indicates a connection between DNA packaging and nuclear egress, but so far, the interaction between UL33, UL25, and the NEC has not been studied in the context of virus replication.
In summary, our data demonstrate once again that viral DN mutants can be successfully retrieved by random procedures. This allows identification of DN mutants of viral genes without knowledge of the structures or functions of their products. Our DN mutants of the two NEC proteins M50 (36) and M53 (present study), as well as novel DNs of additional viral proteins, should represent useful tools to trace and to dissect the viral-protein complexes involved in viral morphogenesis. The detailed study of the DN mutants of M53 CR4 showed that (i) in addition to binding to pM50, which is an essential activity of the M53 protein localized in its CR1, there is a second, yet-unknown vital activity that is clearly separated from its binding activity and located in its CR4. In addition, our results studying MCMV, together with other published data on herpesviruses from other subfamilies, stress that there is a second conserved function of the UL31 family proteins, namely, their accessory role in capsid maturation/DNA packaging, which merits further attention.
Acknowledgments
We thank Sigrid Seelmeir, Simone Boos, and Natalie Röder for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through SFB 455, “Viral Functions and Immune Regulation,” and the priority research program SPP1175, “Dynamics of Cellular Membranes and Their Exploitation by Viruses.”
Footnotes
Published ahead of print on 7 July 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Bjerke, S. L., J. M. Cowan, J. K. Kerr, A. E. Reynolds, J. D. Baines, and R. J. Roller. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601-7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogner, E. 2002. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev. Med. Virol. 12:115-127. [DOI] [PubMed] [Google Scholar]
- 3.Bubeck, A., M. Wagner, Z. Ruzsics, M. Lotzerich, M. Iglesias, I. R. Singh, and U. H. Koszinowski. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J. Virol. 78:8026-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bubić, I., M. Wagner, A. Krmpoti, T. Saulig, S. Kim, W. M. Yokoyama, S. Jonji, and U. H. Koszinowski. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buser, C., P. Walther, T. Mertens, and D. Michel. 2007. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J. Virol. 81:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the U(L)31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H. J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 79:3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonnella, R., A. Farina, R. Santarelli, S. Raffa, R. Feederle, R. Bei, M. Granato, A. Modesti, L. Frati, H. J. Delecluse, M. R. Torrisi, A. Angeloni, and A. Faggioni. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 79:3713-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granato, M., R. Feederle, A. Farina, R. Gonnella, R. Santarelli, B. Hub, A. Faggioni, and H. J. Delecluse. 2008. Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 82:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herskowitz, I. 1987. Functional inactivation of genes by dominant negative mutations. Nature 329:219-222. [DOI] [PubMed] [Google Scholar]
- 15.Klupp, B. G., H. Granzow, G. M. Keil, and T. C. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn, J., T. Leege, B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2008. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J. Virol. 82:5725-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lake, C. M., and L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99-106. [DOI] [PubMed] [Google Scholar]
- 19.Liang, L., and J. D. Baines. 2005. Identification of an essential domain in the herpes simplex virus 1 UL34 protein that is necessary and sufficient to interact with UL31 protein. J. Virol. 79:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, L., M. Tanaka, Y. Kawaguchi, and J. D. Baines. 2004. Cell lines that support replication of a novel herpes simplex virus 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 329:68-76. [DOI] [PubMed] [Google Scholar]
- 21.Lötzerich, M., Z. Ruzsics, and U. H. Koszinowski. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 80:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ménard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U. S. A. 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2009. Herpesvirus assembly: an update. Virus Res. 143:222-234. [DOI] [PubMed] [Google Scholar]
- 25.Mettenleiter, T. C., and T. Minson. 2006. Egress of alphaherpesviruses. J. Virol. 80:1610-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mou, F., E. G. Wills, R. Park, and J. D. Baines. 2008. Effects of lamin A/C, lamin B1, and viral US3 kinase activity on viral infectivity, virion egress, and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane. J. Virol. 82:8094-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 28.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellet, P. E., and B. Roizman. 2007. The family Herpesviridae: a brief introduction, p. 2479-2499. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 30.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddehase, M. J., F. Weiland, K. Munch, S. Jonjic, A. Luske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robbins, S. H., G. Bessou, A. Cornillon, N. Zucchini, B. Rupp, Z. Ruzsics, T. Sacher, E. Tomasello, E. Vivier, U. H. Koszinowski, and M. Dalod. 2007. Natural killer cells promote early CD8 T cell responses against cytomegalovirus. PLoS Pathog. 3:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roller, R. J., S. L. Bjerke, A. C. Haugo, and S. Hanson. 2010. Analysis of a charge cluster mutation of herpes simplex virus type 1 UL34 and its extragenic suppressor suggest a novel interaction between pUL34 and pUL31 that is necessary for membrane curvature around capsids. J. Virol. 84:3921-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rupp, B., Z. Ruzsics, C. Buser, B. Adler, P. Walther, and U. H. Koszinowski. 2007. Random screening for dominant-negative mutants of the cytomegalovirus nuclear egress protein M50. J. Virol. 81:5508-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rupp, B., Z. Ruzsics, T. Sacher, and U. H. Koszinowski. 2005. Conditional cytomegalovirus replication in vitro and in vivo. J. Virol. 79:486-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnee, M., Z. Ruzsics, A. Bubeck, and U. H. Koszinowski. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 80:11658-11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson-Holley, M., R. C. Colgrove, G. Nalepa, J. W. Harper, and D. M. Knipe. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79:12840-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trus, B. L., W. W. Newcomb, N. Cheng, G. Cardone, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uetz, P., Y. A. Dong, C. Zeretzke, C. Atzler, A. Baiker, B. Berger, S. V. Rajagopala, M. Roupelieva, D. Rose, E. Fossum, and J. Haas. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239-242. [DOI] [PubMed] [Google Scholar]
- 42.Ye, G. J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. U. S. A. 97:11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]