Abstract
Tomato yellow leaf curl virus (TYLCV) (Geminiviridae: Begomovirus) is exclusively vectored by the whitefly Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). TYLCV transmission depends upon a 63-kDa GroEL protein produced by the vector's endosymbiotic bacteria. B. tabaci is a species complex comprising several genetically distinct biotypes that show different secondary-symbiont fauna. In Israel, the B biotype harbors Hamiltonella, and the Q biotype harbors Wolbachia and Arsenophonus. Both biotypes harbor Rickettsia and Portiera (the obligatory primary symbionts). The aim of this study was to determine which B. tabaci symbionts are involved in TYLCV transmission using B. tabaci populations collected in Israel. Virus transmission assays by B. tabaci showed that the B biotype efficiently transmits the virus, while the Q biotype scarcely transmits it. Yeast two-hybrid and protein pulldown assays showed that while the GroEL protein produced by Hamiltonella interacts with TYLCV coat protein, GroEL produced by Rickettsia and Portiera does not. To assess the role of Wolbachia and Arsenophonus GroEL proteins (GroELs), we used an immune capture PCR (IC-PCR) assay, employing in vivo- and in vitro-synthesized GroEL proteins from all symbionts and whitefly artificial feeding through membranes. Interaction between GroEL and TYLCV was found to occur in the B biotype, but not in the Q biotype. This assay further showed that release of virions protected by GroEL occurs adjacent to the primary salivary glands. Taken together, the GroEL protein produced by Hamiltonella (present in the B biotype, but absent in the Q biotype) facilitates TYLCV transmission. The other symbionts from both biotypes do not seem to be involved in transmission of this virus.
Tomato yellow leaf curl virus (TYLCV) is the name given to a group of single-stranded-DNA plant viruses of the genus Begomovirus in the family Geminiviridae that exhibit tissue tropism in the plant phloem; some of these viruses are effectively transmitted by the whitefly Bemisia tabaci (12, 13). While acquisition and transmission of TYLCV by B. tabaci have been studied in some detail (6, 7, 30, 36), the molecular interactions between this geminivirus (as well as others) and its insect vector are still poorly understood. Similar to other whitefly-borne geminiviruses, B. tabaci transmits TYLCV in a persistent-circulative nonpropagative manner (15), while some evidence points to viral transcriptional activity in the vector (38). During feeding, TYLCV virions are ingested through the stylet, taken up by midgut epithelial cells, and mobilized to the hemolymph, where they circulate until they enter the salivary glands, from which they are discharged into the plant (9, 11, 17, 24). TYLCV coat protein (CP) is the only virus-encoded protein required for vector-mediated transmission (1, 5, 19, 34).
Several lines of evidence indicate the direct involvement of a 63-kDa GroEL protein, produced by symbiotic bacteria of hemipterans, in phytovirus transmission (31, 32, 40, 41). First, GroEL is abundant in the hemolymph of the insect vector and exhibits binding affinity to TYLCV and Potato leafroll virus (PLRV) (Luteoviridae: Polerovirus) virions (20, 31). Second, feeding whiteflies with anti-Buchnera GroEL antiserum prior to acquisition of virions reduced TYLCV transmission to tomato test plants by more than 80%. In the hemolymph of these whiteflies, TYLCV DNA was reduced to amounts below the detection threshold for Southern blot hybridization (32). Third, it has been shown that TYLCV and PLRV particles that reach the hemolymph interact with GroEL on their way to the salivary glands, forming a complex that protects virions from rapid proteolysis (31, 41). Last, immunogold labeling with anti-Buchnera GroEL in B. tabaci B-biotype bacteriosomes suggests that this GroEL is produced by a secondary symbiont (32); however, the nature of this symbiont is not yet known.
B. tabaci is considered a species complex comprised of approximately 12 genetic groups that are well defined by DNA markers and is distinguished by several biological characteristics, such as dispersal, reproductive rate, and the ability to induce damage in plants (4). The most predominant and damaging biotypes are the B and Q biotypes, which differ considerably with regard to various fitness parameters: while the B biotype is defined by high fecundity and a wide host range (26, 28), the Q biotype is known to develop higher resistance to insecticides (22, 23). Interestingly, these two biotypes also vary in their associations with symbiotic bacteria. Both biotypes harbor Portiera aleyrodidarum, the obligatory primary symbiotic bacterium of whiteflies, as well as the facultative secondary symbiont Rickettsia (2, 18). On the other hand, the secondary symbiont Hamiltonella has been detected only in the B biotype, while Wolbachia and Arsenophonus have been found only in the Q biotype from Israel (8). As Hamiltonella has been found in all B-biotype populations tested, no B-biotype Hamiltonella-free populations have been found. Other secondary symbionts of B. tabaci have not been found in Israeli populations (8). These differences in the bacterial symbiont complements in the two B. tabaci biotypes raised the hypothesis that the efficiency of TYLCV transmission by the B and Q biotypes of B. tabaci depends on a specific symbiont.
In the study presented here, the abilities of both B and Q biotypes from Israel to transmit TYLCV were tested. Upon discovering that the B biotype was able to transmit the virus while the Q biotype was a poor transmitter, the hypothesis that these differences depend upon bacterial symbionts present in the respective biotypes was tested.
MATERIALS AND METHODS
Whiteflies and virus sources.
All B. tabaci populations used in this study were reared on cotton plants (Gossypium hirusutum cv. Akala) grown in insect-proof cages at 26°C ± 2°C as previously described (14). The populations of the B (Neg03E) and Q (AV04E) biotypes (23) used in this study harbored the primary symbiont Portiera. In addition, the Q biotype carried Arsenophonus and Rickettsia (termed Q-RA AV04E), whereas the B biotype harbored only Hamiltonella (termed B-H Neg03E). To maximize the difference in the secondary-symbiont profiles of the two biotypes, the previously described B-HRs (harboring Hamiltonella and Rickettsia with a scattered phenotype) and B-HRc (harboring Hamiltonella and Rickettsia with a confined phenotype) B-biotype isofemale strains, and the Q-A (harboring Arsenophonus) and Q-WA (harboring Arsenophonus and Wolbachia) Q-biotype isofemale strains were also used for virus transmission tests and immunocapture (IC)-PCR assays. The symbiotic complements of these populations were confirmed using genus-specific PCR amplification of an rRNA fragment and fluorescence in situ hybridization (FISH) as previously described (18). In contrast to the Israeli population, a Q-biotype population from Spain did contain Hamiltonella (E. Moriones, personal communication). B. tabaci Q-biotype samples from Spain were received from Koppert Biological System in 96% ethanol (La Mojonera, Almeria, Spain) in May 2006. An isolate of TYLCV from Israel (33) was maintained in tomato plants (Solanum lycopersicum cv. Daniela) by whitefly-mediated transmission. These plants served as the source for virus acquisition by whiteflies.
Acquisition and transmission of TYLCV by whiteflies from the B and Q biotypes.
All experiments were conducted in insect-proof cages kept at 26°C ± 2°C in an insect-proof growth chamber. Five to 8 days after emergence, insects from both biotypes were caged with a virus-infected tomato plant for 48 h. The whiteflies were then removed from the infected plant, and eight individuals were placed in a leaf cage on a virus-free tomato plant in at least 18 replicates for each biotype as previously described (15). After 3 to 4 weeks, the plants were visually monitored for the appearance of disease symptoms and subjected to PCR and dot blot analyses to detect the presence of the virus.
Dot blot and PCR analyses.
Detection of TYLCV in infected plants was performed using a digoxigenin-labeled probe as previously described (10). The probe was amplified from a plasmid bearing the full-length TYLCV genome with primers P1V (5′-ATACTTGGACACCTAATGGC-3′) and P5C (5′-AGTCACGGGCCCTTACAA-3′). The same primer set was used in PCR on tomato plant DNA extract and on whitefly DNA 48 h after virus acquisition as described previously (16).
Cloning and sequencing of TYLCV CP and GroEL genes in yeast plasmids.
A GroEL gene from the B biotype and the CP of TYLCV were previously cloned (31). Three contigs of the GroEL gene of Portiera were found in the sequences generated in the framework of the Whitefly Functional Genome Project (27). Using these sequences, the full-length sequence of this gene was amplified from genomic DNA using the primer pair PGEL-ATG (5′-ATGGCAGCAAAACAGATTAG-3′) and PGEL-STOP (5′-CGAAGATCTCATACCATTTACC-3′), with the introduction of an EcoRI restriction site on the 5′ primer and a XhoI restriction site on the 3′ primer. The full-length sequence of the B. tabaci Rickettsia GroEL gene was amplified based on the recently published genome sequence of Rickettsia bellii (35) using the primers RGEL-ATG (5′-ATGGCAACAAAACTTATTAAGC-3′) and RGEL-STOP (5′-TTAGAAGTCCATACCTCCCA-3′), with the introduction of an EcoRI restriction site on the 5′ primer and a XhoI restriction site on the 3′ primer. Both Portiera and Rickettsia GroEL genes were cloned in frame with the B42 activation domain into the EcoRI/XhoI sites of the Saccharomyces cerevisiae strain EGY48 plasmid pB42AD containing a TRPI marker. The TYLCV CP was cloned in frame with the LexA binding domain into the EcoRI/XhoI restriction sites of the yeast plasmid pLexA that contains a HIS3 marker (Clontech) (31).
Binding of TYLCV CP to symbiont GroELs in a yeast two-hybrid system.
This analysis of binding of TYLCV CP to symbiont GroEL proteins (GroELs) was performed according to the protocol described by Morin et al. (31). Hamiltonella, Portiera, and Rickettsia GroEL genes were cloned into the pB42AD plasmid, and the new clones were termed pB42AD-HamGroEL, pB42AD-PorGroEL and pB42AD-RickGroEL, respectively. These plasmids were individually introduced with pLexA containing TYLCV CP (pLexA-TYCP) into the yeast strain EGY48, using the lithium-acetate-mediated method (3). For a positive control, the Abutilon mosaic virus coat protein (ABMV CP) was introduced with the pB42AD-HamGroEL plasmid (31), and for a negative control, the plasmids pLexA-BD and pB42-AD were cotransformed into the yeast cells. In the EGY48 strain, the upstream activating sequences of the chromosomal LEU2 gene, required in the leucine biosynthetic pathway, are replaced with LexA operators (DNA-binding sites). In the first plating, which selects for yeast cells in which the two plasmids have been successfully introduced, transformed yeast cells were plated on complete minimal (CM) medium (39) lacking Ura, His, and Trp, with 2% glucose as the sugar source. In the second plating, which selects for yeast cells that contain interacting proteins, selected primary transformants were plated on CM medium lacking Ura, His, Trp, and Leu with 2% galactose or 1% raffinose as the sugar source.
To confirm that the fusion proteins had been synthesized, yeast cell crude lysate was prepared, fractionated by 10% SDS-PAGE, and electroblotted. Hamiltonella, Portiera, and Rickettsia GroEL fusion proteins expressed from pB42AD were immunodetected using antibodies raised against Buchnera GroEL from Myzus persicae (40). Proper in-frame cloning of Portiera and Rickettsia was also confirmed by sequencing and by yeast growth on CM medium. Growth in the absence or presence of Leu was evaluated by incubating the cells for 48 h at 28°C in a Leu-containing medium. The cells were then streaked (three 1-cm rows per cell line) on agar petri dishes containing medium with Leu and without Leu. Photos from the plates were taken after additional 48-h incubation.
Cloning of GroEL gene into an expression system and in vitro protein synthesis and pulldown assays.
GroEL genes from Portiera, Rickettsia, and Hamiltonella, or TYLCV CP were amplified with specific primers containing a XhoI restriction site on the 5′ primer and an EcoRI restriction site on the 3′ primer and were then cloned into p-RSETa cloning vector (Invitrogen) which includes a 6×His tag and a T7 promoter. The three GroEL proteins were synthesized in vitro from this vector using the TNT T7 quick coupled transcription/translation system (Promega) based on the reticulocyte lysate system and the Transcend tRNA system containing lysine conjugated to biotin (Promega). The proteins were also expressed in an expression vector that harbors a T7 promoter and lacks a histidine tag. The proteins were subjected to SDS-PAGE and blotted onto polyvinylidene difluoride (PVDF) membranes to confirm their production and size. Subsequent detection of the in vitro-synthesized proteins on Western blots and after pulldown assays was performed using streptavidin antibody linked to horseradish peroxidase (streptavidin-HRP) and enhanced chemiluminescence (ECL) reagent. Pulldown assays were performed using the ProFound poly(His) pulldown protein-protein interaction kit according to the manufacturer's instructions (Pierce), and were subjected to SDS-PAGE and blotted onto PVDF membranes. The transcription/translation reaction is using the Transcend tRNA and is composed of 40 μl master mix of reticulocyte lysate, 1 mM methionine, 1 μg of plasmid DNA containing the GroEL/TYLCV CP gene, and the Transcend biotin-lysyl-tRNA. After incubation at 37°C for 60 min, 20 μg of translated protein quantified using the bicinchoninic acid (BCA) protein assay reagent (Pierce) was used to verify production and size of the GroEL/TYLCV CP in a standard Western blot analysis. Approximately 100 μg of translated TYLCV CP or of the different GroEL proteins was used in pulldown analysis and artificial feeding. For pulldown assays, the in vitro-prepared poly(His)-tagged protein (TYLCV CP or the different GroELs) was used as “bait” and immobilized on cobalt agarose resin columns. Another 100 μg of the “prey” protein (TYLCV CP or the different GroELs) which lacked the poly(His) tag, was immobilized on the cobalt agarose resin, on which the “bait” protein was bound. The column contents were then eluted according to the manufacturer's instructions and subjected to SDS-PAGE, Western blotting, and protein detection using the streptavidin-HRP and ECL kit.
Artificial feeding of B. tabaci through membranes.
B. tabaci adults, 5 days after emergence, were fed through membranes on 100 μl of a 15% sucrose solution containing 40 μg of the translated GroEL protein. The insects were then transferred to TYLCV-infected tomato plants for an additional 48-h acquisition access period and subsequently transferred to cotton plants. After 6 h of incubation, they were subjected to IC-PCR as described below.
IC-PCR assay.
Interaction between GroELs and TYLCV was detected by IC-PCR (15) using an antibody raised against GroEL produced by M. persicae's primary symbiont Buchnera. The buffers used for IC-PCR are described by the manufacturer (Bioreba, Ebringen, Germany) and were used as instructed by the manufacturer. PCR tubes were filled with 200 μl of antiserum (1:1,000 diluted in coating buffer), incubated for 3 h at 37°C, and washed five times for 5 min each time with 200 μl washing buffer. Homogenates from 5 to 10 whiteflies, hemolymph from 50 whiteflies collected as described previously (32), or 100 primary salivary glands (17), from insects caged with TYLCV-infected tomato plants for 48 h were incubated for 18 h at 4°C in the coated PCR tubes in 200 μl of extraction buffer. The tubes were washed five times, 5 min each time with 200 μl washing buffer, and dried. PCR amplification of the viral DNA from the virions bound to the GroEL protein, which was itself bound to the antibody-coated tubes, was performed with the TYLCV-specific primers V61 and C473 (12). All experiments were replicated 3 to 5 times.
Nucleotide sequence accession numbers.
The Portiera and Rickettsia full-length GroEL gene sequences determined in this study were submitted to GenBank under access numbers EU435142 and EU435143, respectively.
RESULTS AND DISCUSSION
B and Q biotypes differ in their TYLCV transmission efficacy.
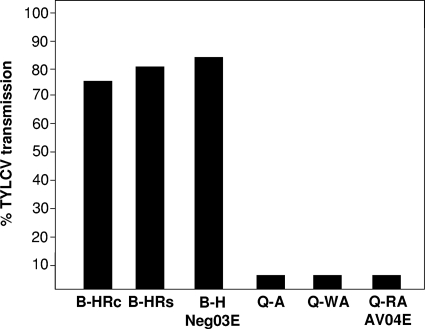
Previous tests with the B biotype from Israel have shown high levels of TYLCV transmission (15). At the time of this study, this B-biotype population tested positive for the presence of Hamiltonella and Rickettsia (not shown). A whitefly population from the Q biotype harbored Arsenophonus, Wolbachia, and Rickettsia. To elucidate possible differences in TYLCV transmission efficiency between the B and Q biotypes and correlate them to symbiotic content, three different independent transmission experiments with the B and Q biotypes with different symbiotic complements were conducted. Following a 24-h acquisition access period from infected plants, TYLCV was detected by PCR in all individual Q-biotype whiteflies. After the viruliferous Q-biotype whiteflies were caged with tomato seedlings, the virus could be detected by PCR and dot blot hybridization in only 6 of the 64 (9%) plantlets (Fig. 1). These six plants showed typical disease symptoms 4 weeks postinoculation. In contrast, the same analyses showed that in three independent transmission experiments with three different B-biotype B. tabaci populations harboring either Hamiltonella alone or Hamiltonella with Rickettsia, TYLCV was transmitted with a 80% efficacy (56 out of 70 seedlings) (Fig. 1). These results suggest that Hamiltonella, the only symbiont present in our B-biotype population and absent from the Q biotype in Israel, is most probably a secondary symbiont necessary for TYLCV transmission. Previous comparisons of transmission capabilities of B and Q biotypes in populations from Spain showed that both biotypes were able to transmit TYLCV, the Q biotype was even more efficient than the B biotype in this respect (25, 37). Interestingly, in contrast to the Q biotype from Israel, the Spanish Q-biotype population was found to harbor Hamiltonella (E. Moriones, personal communication). TYLCV transmission test results and symbiont fauna support the hypothesis that the GroEL produced by Hamiltonella is an important factor in facilitating TYLCV transmission.
FIG. 1.
TYLCV transmission experiments by different strains of B and Q biotypes of B. tabaci. B-HRc, B-HRs, and B-H Neg03E are B-biotype strains that harbor Hamiltonella and Rickettsia with the confined phenotype, Hamiltonella and Rickettsia with the scattered phenotype, and Hamiltonella, respectively. Q-A, Q-WA, and Q-RA AV04E strains are Q-biotype strains that harbor Arsenophonus, Wolbachia and Arsenophonus, and Rickettisa and Arsenophonus, respectively.
The Hamiltonella, but not the Portiera or Rickettsia, GroEL protein sequence is most similar to Buchnera GroEL from Myzus persicae.
To further assess whether Hamiltonella is the symbiont involved in viral transmission within the B biotype, the full-length sequences of the GroEL gene of the two other facultative B-biotype symbionts, Portiera and Rickettsia, were determined and compared with that of Hamiltonella and with other published GroEL sequences. Overall alignment showed various degrees of homology (Fig. 2): Hamiltonella and Buchnera proteins showed 78% homology, Hamiltonella and Portiera proteins showed 66% homology, Hamiltonella and Rickettsia proteins showed 54% homology, and Hamiltonella and Wolbachia proteins from Drosophila melanogaster showed 46% homology. Special attention was paid to the determinants shown to be essential for the interaction of the aphid M. persicae primary symbiont Buchnera GroEL with PLRV CP (20, 31). Five out of the six amino acids in the equatorial domain of GroEL that were shown to be essential for PLRV transmission, R13, K15, L17 in the N-terminal region and R441 and R445 in the C-terminal region, were found to be conserved between Hamiltonella and Buchnera GroEL proteins (Fig. 2). The only determinant different from the R18 in Buchnera was K18 in Hamiltonella. This, however, can be considered a minor substitution, since both Arg and Lys belong to the same group of positively charged and structurally similar amino acids. Portiera and Rickettsia GroEL proteins, however, showed more substitutions in these essential amino acids. In Portiera, K15, L17, R441, and R445 were replaced by R15, V17, K441, and K445, while in Rickettsia, K15, R18, R441, and R445 were replaced by Q15, E18, E441, and E445 (Fig. 2). The model suggested by Hogenhout et al. (21) predicts that these substitutions in Portiera and Rickettsia GroEL proteins will limit their ability to bind TYLCV CP. On the other hand, the overall similarity between the proteins, and specifically, the high similarity between the equatorial domains of Hamiltonella and Buchnera GroELs, both involved in virus transmission together with the specific in situ localization of GroEL within the B biotype (32), support the hypothesis that Hamiltonella GroEL might be responsible for the ability of TYLCV to survive the proteolytic environment in the B. tabaci circulative system (32).
FIG. 2.
Amino acid sequences of B. tabaci GroEL proteins from Hamiltonella, Portiera, and Rickettsia and comparison with GroELs from other symbiotic bacteria, including Buchnera from M. persicae and Wolbachia from Drosophila melanogaster. Identical amino acids are indicated by dashes. The equatorial domain that includes amino acids reported to be involved in PLRV binding (20) is shown on a gray background. The essential amino acids for binding to PLRV CP are indicated by vertical gray boxes.
Hamiltonella, but not Portiera and Rickettsia, GroEL interacts with TYLCV CP.
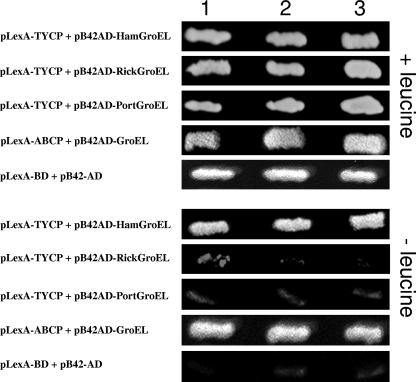
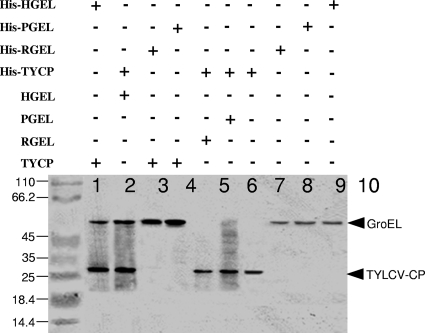
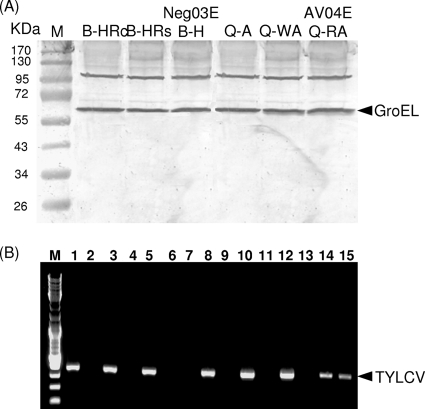
Interactions between Portiera, Rickettsia, and Hamiltonella GroELs with TYLCV CP were tested. First, a yeast two-hybrid system was used in which the pLexA-TYCP plasmid and either the pB42AD-RickGroEL, pB42AD-PorGroEL or pB42AD-HamGroEL plasmid were cointroduced into yeast cells. Yeast cells cotransformed with pLexA-TYCP or pLexA-ABCP as a positive control (CP of the Abutilon mosaic virus [ABMV]) with pB42AD-HamGroEL were able to grow on medium with and without Leu, indicating interaction between TYLCV CP or ABMV CP and the GroEL produced by Hamiltonella (Fig. 3). In contrast, when yeast cells were cotransformed with pLexA-TYCP and either pB42AD-PorGroEL or pB42AD-RickGroEL, they grew on medium containing Leu but showed limited growth or no growth on medium without this amino acid (Fig. 3). This limited growth or absence of growth is similar to the negative control in which yeast cells were cotransformed with empty vectors. These findings further support a specific interaction between TYLCV CP and the GroEL proteins produced by Hamiltonella. Second, Hamiltonella, Rickettsia, and Portiera GroEL genes were cloned into the pRSETa expression vector harboring a 6×His tag. The protein products of the three GroEL genes synthesized in vitro were used in pulldown assays to test for GroEL-TYLCV CP interactions. Figure 4 shows that while the Hamiltonella GroEL pulled down TYLCV CP (Fig. 4, lanes 1 and 2), Rickettsia and Portiera GroELs did not (Fig. 4, lanes 3 to 6), suggesting a lack of interaction. This result further supports the Hamiltonella GroEL-TYLCV CP interaction. Third, an IC-PCR assay performed with viruliferous whiteflies from the B and Q biotypes confirmed the specific Hamiltonella GroEL-TYLCV CP interaction in whole whiteflies and in the hemolymph. This analysis was based on an anti-GroEL antibody raised against M. persicae's primary symbiont Buchnera, which recognized GroELs from all whitefly strains used in this study (Fig. 5A). Extracts from three whole-fly B-biotype strains that harbored Hamiltonella and acquired TYLCV showed positive PCR results when incubated in a GroEL antibody-coated PCR tube, suggesting that a TYLCV-GroEL complex was bound to the GroEL antibody (Fig. 5B, lanes 1, 3, and 5). Since the antibody used recognized other GroEL genes, we tested Q-biotype populations with the same IC-PCR analysis. All PCRs performed after incubation with Q-biotype extracts that had acquired the virus showed negative PCR results, suggesting no interaction between GroELs from the Q biotype and TYLCV (Fig. 5B, lanes 7, 9, and 11). These results suggest that in vivo, GroEL proteins produced by Wolbachia, Arsenophonus, Rickettsia, and Portiera do not interact with TYLCV CP in the Q biotype and that only the GroEL produced by Hamiltonella interacts with TYLCV CP. It should be noted that we obtained a positive PCR result when we used extracts from a Spanish Q-biotype whitefly colony that had acquired TYLCV and been analyzed the same way (data not shown).
FIG. 3.
Interaction of TYLCV CP with B. tabaci GroEL proteins in yeast. (Top) Selection for growth in the presence of leucine as a control for the quality of the cells. The growth of yeast cells cotransfected with TYLCV CP-containing plasmid and the various GroEL plasmids (Hamiltonella, Portiera, Rickettsia, the GroEL gene reported by Morin et al. [31] as a positive control and the pLexA-BD + pB42-AD plasmids without any genes as a negative control) are shown. (Bottom) Growth of the same cotransfected yeast cell lines with medium lacking Leu. Growth or the lack of growth of three independent yeast colonies is shown for each cotransfection.
FIG. 4.
In vitro interaction between TYLCV CP (TYCP) and Hamiltonella GroEL (HGEL) in a pulldown assay. Polyhistidine and nonpolyhistidine in vitro-synthesized Hamiltonella (His-HGEL and HGEL, respectively), Portiera (His-PGEL and PGEL, respectively), Rickettsia (His-RGEL and RGEL, respectively), and TYLCV CP (His-TYCP and TYCP, respectively) were subjected to protein pulldown assays as described in Materials and Methods. Each pulldown assay was performed by binding one polyhistidine protein to a cobalt agarose column and immobilizing a second nonpolyhistidine protein to the column. After elution, proteins were subjected to an SDS-PAGE and Western blot analysis for detecting the proteins with anti-biotin streptavidin-HRP antibody. Hamiltonella GroEL interacted with TYLCV CP (lanes 1 and 2), but not with Rickettsia and Portiera GroELs (RGEL and PGEL, respectively) (lanes 3 to 6). Lanes 7 to 10 contain eluted 6×His-labeled recombinant proteins for size confirmation. The positions of molecular size markers (in kilodaltons) are shown to the left of the gel.
FIG. 5.
Immunocapture-PCR assay for detection of TYLCV CP-GroEL interactions. (A) Extracts from B-biotype strains B-HRc, B-HRs, and B-H Neg03E and from Q-biotype strains Q-A, Q-WA, and Q-RA AV04E (see the legend to Fig. 1) were subjected to SDS-PAGE, Western blotting, and immunodetection with anti-Buchnera GroEL antibody to confirm reactivity with all possible symbionts in whiteflies. The positions of molecular mass markers (lane M) (in kilodaltons) are indicated to the left of the gel. (B) PCR tubes coated with the GroEL-specific antibody and virus-specific primers were used to detect the interaction. Lane M contains 100-bp ladder marker. Lanes 1, 3, and 5 contain extracts from BHRc, BHRs, and B-H Neg03E populations, and lanes 2, 4, and 6 are no-antibody controls for each respective population. Lanes 7, 9, and 11 contain extracts from Q-A, Q-WA, and Q-RA AV04E populations, and lanes 8, 10, and 12 contain PCR mixtures from the three Q-biotype populations to verify TYLCV acquisition. Lane 13 is a negative no-template control, lane 14 is a positive PCR control plasmid (bearing the full TYLCV sequence), and lane 15 is a positive PCR control from a TYLCV-infected plant.
To address the question of whether differences in transmission efficacy between the two biotypes is an outcome of different acquisition abilities by the different whitefly strains or of the inability of the virus to cross the midgut barrier, we performed quantitative real-time PCR analyses to measure viral DNA from whole insects and from hemolymph of whiteflies that acquired the virus for 48 h in three replicates. We found that the levels of TYLCV DNA were similar between the two biotypes in whole insects and in the hemolymph. Since this analysis measured DNA concentration, we concluded that TYLCV was equally acquired by the B- and Q-biotype strains and that the virus crossed the midgut to the hemolymph with the same efficiency in both biotypes (data not shown).
Recombinant Hamiltonella GroEL interacts with TYLCV CP in the Q biotype.
Our results raised the question of whether Hamiltonella GroEL-TYLCV CP complex can form in the Q-biotype hemolymph after ingestion of this GroEL through artificial feeding. Thus, we supplemented the artificial diet of two Q-biotype populations (Q-RA AV04E and Q-WA) with in vitro-synthesized Hamiltonella GroEL protein and tested whether the interactions previously shown in the B-biotype populations between Hamiltonella GroEL and TYLCV CP could be duplicated. IC-PCR analysis showed positive interactions in the B-biotype populations as well as in the Q-biotype populations fed with Hamiltonella GroEL for 24 h (Fig. 6A), suggesting that the in vitro-synthesized Hamiltonella GroEL protein is ingested by whiteflies through the artificial membrane and crosses the midgut to reach the hemolymph, where it interacts with TYLCV CP. The same results were obtained in IC-PCR analysis performed on dissected hemolymph from the same Q-biotype populations used above after feeding on the in vitro-synthesized Hamiltonella GroEL (Fig. 6B).
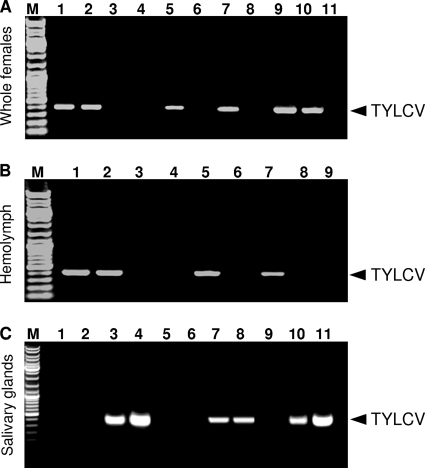
FIG. 6.
Recombinant Hamiltonella-GroEL interaction with TYLCV CP in whole whiteflies and hemolymph extracts but not in salivary glands. (A) Whole whiteflies (females). Lanes 1 and 2 show the results of IC-PCR with GroEL antibody and extracts from whole B-biotype viruliferous females from two strains (B-HRc and B-HRs, respectively), and lanes 3 and 4 are no-antibody controls. Lanes 5 and 7 show the results of IC-PCR analysis with GroEL antibody and extracts from two Q-biotype strains (Q-WA and Q-RA AV04E, respectively) that were fed for 24 h on in vitro-synthesized Hamiltonella GroEL, and lanes 6 and 8 are no-antibody controls. Lane 9 contains a positive PCR control plasmid (bearing the full TYLCV sequence), and lane 10 contains a positive PCR control from a TYLCV-infected plant. Lane 11 is a negative no-template control. Lane M contains molecular size markers. (B) Hemolymph. Lanes 1 and 2 show the results of IC-PCR with GroEL antibody and extracts of hemolymph from strains B-HRc and B-HRs, respectively, and lanes 3 and 4 are no-antibody controls. Lanes 5 and 7 show the results of IC-PCR analysis with GroEL antibody and hemolymph extracts from strains Q-WA and Q-RA AV04E, respectively, fed on recombinant Hamiltonella GroEL for 24 h, and lanes 6 and 8 are no-antibody controls. Lane 9 is a negative no-template control. (C) Salivary glands. Lanes 1 and 2 show the results of IC-PCR with GroEL antibody and extracts from salivary glands of viruliferous B-biotype (B-HRc and B-HRs, respectively) females. Lanes 3 and 4 show the results of IC-PCR with TYLCV CP antibody and extracts from salivary glands of the same viruliferous B-biotype females. Lanes 5 and 6 show the results of IC-PCR with GroEL antibody and extracts from salivary glands of viruliferous Q-biotype (Q-WA and Q-RA AV04E) females. Lanes 7 and 8 show the results of IC-PCR with TYLCV CP antibody and salivary glands extracts from the same viruliferous Q-biotype females. Lane 9 is a negative control (TYLCV-free plant). Lane 10 is a positive control (TYLCV-infected plant). Lane 11 contains plasmid (bearing the full TYLCV sequence) and is a positive control.
Protection in the hemolymph, as shown above, is not sufficient to ensure virus transmission. The virions must reach and enter the salivary glands in order to be discharged into the plant in subsequent feeding. Thus, we tested whether translocation of TYLCV into the salivary glands occurs. We performed IC-PCR analysis using tubes coated with GroEL antibody and extracts prepared from primary salivary glands dissected from Q-biotype females that had been fed through membranes on recombinant Hamiltonella GroEL for 24 h after TYLCV acquisition from infected plants and extracts from B-biotype females 24 h after TYLCV acquisition from infected plants. The results indicated that TYLCV virions are not associated with GroEL inside the salivary glands of either the B or Q biotype, as TYLCV-GroEL complexes were not found in the glands (Fig. 6C, lanes 1 and 2). To further support this hypothesis and test whether the virions are present in the salivary gland, we dissected salivary glands from B-biotype females that had acquired TYLCV for 48 h and subjected them to IC-PCR analysis using tubes coated with a TYLCV-CP-specific antibody. The results indicated the presence of TYLCV as a virion in the primary glands (Fig. 6C, lanes 3 and 4). IC-PCR with tubes coated with GroEL antibody and dissected salivary glands from the Q-biotype population fed on recombinant Hamiltonella GroEL protein did not detect TYLCV virions in the glands (Fig. 6C, lanes 5 and 6); however, a positive IC-PCR result was obtained when the assay was performed with tubes coated with TYLCV antibody (Fig. 6C, lanes 7 and 8). These results indicate that TYLCV in the Q biotype reaches the hemolymph and enters the salivary glands as in the B biotype. Therefore, our data support the hypothesis that reduced transmissibility of TYLCV by the Q biotype is a result of the lack of interaction between the virus and the appropriate Hamiltonella GroEL protein in the hemolymph.
Feeding Q biotype with recombinant Hamiltonella GroEL does not increase TYLCV transmission efficiency.
To test whether supplementing the artificial diet of whiteflies from the Q biotype with recombinant Hamiltonella GroEL could increase TYLCV transmission efficiency, we conducted transmission tests with the B-HRs and the Q-WA strains. Adults from the Q-biotype strain were fed through membranes for 24 h on a 15% sucrose solution containing ∼1 μg/μl recombinant Hamiltonella GroEL, while adults from the B-biotype strain were fed only with the sucrose solution for the same period of time. The adult whiteflies from both strains were then caged with TYLCV-infected plants for a 48-h virus acquisition access period and then were caged in groups of eight adults with a virus-free tomato plant for an additional 48-h inoculation access period. After 3 to 4 weeks, the plants were visually monitored for the appearance of disease symptoms and were subjected to PCR and dot blot analyses to detect the presence of the virus. The experiment was replicated twice for each whitefly strain, and 22 tomato plants were used in each replicate. Two of the 44 plants caged with Q biotype became infected (∼5%), compared with 28 of the 44 plants caged with the B biotype (63%). The whiteflies used in this experiment were collected from the test plants after 48 h in groups of 5 to 8 adults and tested for the formation of a TYLCV-Hamiltonella GroEL complex using IC-PCR. Eighty percent of the Q-biotype groups and 96% of the B-biotype groups tested positive for the TYLCV-Hamiltonella GroEL complex formation. The results from this experiment suggest that TYLCV-GroEL complexes were formed in the Q biotype; however, the transmission of TYLCV by this Q biotype did not increase. This result may be explained by the following: other unknown factors required for the transmission, insufficient amounts of protein in the artificial diet, or insufficient amounts of protein that crossed the midgut to the hemolymph. Therefore, further work is needed to confirm or negate these hypotheses.
In conclusion, the present study shows that the Q biotype from Israel, which does not harbor Hamiltonella but harbors three other secondary symbionts, scarcely transmits TYLCV, while the B biotype, which harbors Hamiltonella, is an efficient vector. The involvement of GroEL produced by Hamiltonella was demonstrated in in vivo and in vitro assays showing that interaction between GroEL and TYLCV CP occurs only in the B. tabaci B biotype. In addition, TYLCV could be detected in the hemolymph and salivary glands of both B biotype and Hamiltonella GroEL-supplemented Q biotypes, demonstrating that the virus follows the circulative pathway of transmission in both biotypes. Our results suggest that of the three symbionts present in the B biotype of B. tabaci, only Hamiltonella produces a GroEL protein that interacts with TYLCV CP and that this interaction is correlated with vectoring ability. The two other symbionts in the B biotype are probably not involved in TYLCV transmission; their GroELs, however, may be specifically involved in B. tabaci transmission of other virus families.
These results may well explain the inability of the Hamiltonella-free Israeli Q biotype to efficiently transmit TYLCV, in contrast to the Q biotype from Spain that carries Hamiltonella and exhibits high transmission efficiency (25, 37). The low transmission efficiency observed with the Israeli Q biotype suggests that in some cases, TYLCV virions may rapidly escape the proteolytic environment in the hemolymph to the salivary glands, where they can be transmitted to the plant. This can occur if a high titer of virions is acquired by B. tabaci, such that some will escape to the salivary glands without being attacked by the whitefly immune system. In an alternative scenario, which seems more reasonable, the midgut is often pushed into the thorax by a full abdomen in gravid females (17). In this case, the distance that virions have to pass from the midgut barrier to the salivary glands is minimal, and the hemolymph environment is mostly avoided. Since these cases are not common, they do not significantly contribute to the transmission efficiency of the Q biotype. TYLCV virions pass several barriers before they are transmitted to the plant, and protection by GroEL in the hemolymph is one step in the transmission process. However, crossing the barriers of the midgut and salivary glands requires other unknown vector factors. Thus, the genetic background of the insect may also play a major role in its ability to serve as an efficient vector for transmission. In this regard, it has been recently shown that at the DNA sequence level, both B and Q biotypes share levels of similarity that may reach up to 98% (29); thus, it is less likely that genetic background differences between both biotypes play a role in their ability to serve as vectors for plant viruses.
Acknowledgments
We thank Yedidya Gafni for providing yeast two-hybrid system plasmids and Alberto Urbaneja for supplying whitefly samples.
This research was supported by the High Council for Scientific and Technological Cooperation between France and Israel, program on sustainable agriculture CNOUS 05F14. This research was also partially supported by grant IS-4062-07 from the United States-Israel Binational Agricultural Research and Development Fund (BARD) to M.G. and H.C., by research grant 887/07 from the Israel Science Foundation to M.G. and H.C., and by grant 2001-353-2-10986 from the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service to E.Z.-F.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Azzam, O., J. Frazer, D. Delarosa, J. S. Beaver, P. Ahlquist, and D. P. Maxwell. 1994. Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology 204:289-296. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155-189. [DOI] [PubMed] [Google Scholar]
- 3.Becker, D. M., and V. Lundblad. 1997. Introduction of DNA into yeast cells, p. 13.7.3-13.7.10. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Wiley, New York, NY. [DOI] [PubMed]
- 4.Boykin, L. M., R. G. Shatters, Jr., R. C. Rosell, C. L. McKenzie, R. A. Bagnall, P. J. De Barro, and D. R. Frohlich. 2007. Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol. Phylogen. Evol. 3:1306-1319. [DOI] [PubMed] [Google Scholar]
- 5.Briddon, R. W., M. S. Pinner, J. Stanley, and P. G. Markham. 1990. Geminivirus coat protein gene replacement alters insect specificity. Virology 177:85-94. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. K., and H. Czosnek. 2002. Whitefly transmission of plant viruses, p. 65-100. In R. T. Plumb and J. A. Callow (ed.), Advances in botanical research. Academic Press, New York, NY.
- 7.Caciagli, P., D. Bosco, and L. Al-Bitar. 1995. Relationships of the Sardinian isolate of tomato yellow leaf curl geminivirus with its whitefly vector Bemisia tabaci Gen. Eur. J. Plant Pathol. 101:163-170. [Google Scholar]
- 8.Chiel, E., Y. Gottlieb, M. Inbar, E. Zchori-Fein, and M. Ghanim. 2007. Distribution of secondary symbionts in Israeli populations of Bemisia tabaci. Bull. Entomol. Res. 97:407-413. [DOI] [PubMed] [Google Scholar]
- 9.Cicero, J. M., E. Hiebert, and S. E. Webb. 1995. The alimentary canal of Bemisia tabaci and Trialeurodes abutilonea (Homoptera, Sternorrhyncha): histology, ultrastructure and correlations to function. Zoomorphology 115:31-39. [Google Scholar]
- 10.Crespi, S., G. P. Accotto, P. Caciagli, and B. Gronenborn. 1991. Use of digoxigenin-labelled probes for detection and host-range studies of tomato yellow leaf curl geminivirus. Res. Virol. 142:283-288. [DOI] [PubMed] [Google Scholar]
- 11.Czosnek, H., M. Ghanim, and M. Ghanim. 2002. The circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci—insights from studies with tomato yellow leaf curl virus. Ann. Appl. Biol. 140:215-231. [Google Scholar]
- 12.Czosnek, H., M. Ghanim, S. Morin, G. Rubinstein, V. Fridman, and M. Zeidan. 2001. Whiteflies: vectors, and victims (?), of geminiviruses. Adv. Virus Res. 56:291-322. [DOI] [PubMed] [Google Scholar]
- 13.Czosnek, H., and H. Laterrot. 1997. A worldwide survey of tomato yellow leaf curl viruses. Arch. Virol. 142:1391-1406. [DOI] [PubMed] [Google Scholar]
- 14.Ghanim, M., and S. Kontsedalov. 2007. Gene expression in pyriproxyfen-resistant Bemisia tabaci Q biotype. Pest Manag. Sci. 63:776-783. [DOI] [PubMed] [Google Scholar]
- 15.Ghanim, M., S. Morin, and H. Czosnek. 2001. Rate of tomato yellow leaf curl virus (TYLCV) translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathology 91:188-196. [DOI] [PubMed] [Google Scholar]
- 16.Ghanim, M., S. Morin, M. Zeidan, and H. Czosnek. 1998. Evidence for transovarial transmission of tomato yellow leaf curl virus by its vector the whitefly Bemisia tabaci. Virology 240:295-303. [DOI] [PubMed] [Google Scholar]
- 17.Ghanim, M., R. C. Rosell, L. R. Campbell, H. Czosnek, J. K. Brown, and D. E. Ullman. 2001. Microscopic analysis of the digestive, salivary and reproductive organs of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) biotype B. J. Morphol. 248:22-40. [DOI] [PubMed] [Google Scholar]
- 18.Gottlieb, Y., M. Ghanim, E. Chiel, D. Gerling, V. Portnoy, S. Steinberg, G. Tzuri, A. R. Horowitz, E. Belausov, N. Mozes-Daube, S. Kontsedalov, M. Gershon, S. Gal, N. Katzir, and E. Zchori-Fein. 2006. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 72:3646-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofer, P., I. D. Bedford, P. G. Markham, H. Jeske, and T. Frischmuth. 1997. Coat protein gene replacement results in whitefly transmission of an insect nontransmissible geminivirus isolate. Virology 236:288-295. [DOI] [PubMed] [Google Scholar]
- 20.Hogenhout, S. A., F. van der Wilk, M. Verbeek, R. W. Goldbach, and J. F. J. M. van den Heuvel. 1998. Potato leafroll virus binds to the equatorial domain of the aphid endosymbiotic GroEL homologue. J. Virol. 72:358-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogenhout, S. A., F. van der Wilk, M. Verbeek, R. W. Goldbach, and J. F. J. M. van den Heuvel. 2000. Identifying the determinants in the equatorial domain of Buchnera GroEL implicated in binding potato leafroll virus. J. Virol. 74:4541-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horowitz, A. R., S. Kontsedalov, and I. Ishaaya. 2004. Dynamics of resistance to the neonicotinoids acetamiprid and thiamethoxam in Bemisia tabaci (Homoptera: Aleyrodidae). J. Econ. Entomol. 97:2051-2056. [DOI] [PubMed] [Google Scholar]
- 23.Horowitz, A. R., S. Kontsedalov, V. Khasdan, and I. Ishaaya. 2005. Biotypes B and Q of Bemisia tabaci and their relevance to neonicotinoid and pyriproxyfen resistance. Arch. Insect Biochem. Physiol. 58:216-225. [DOI] [PubMed] [Google Scholar]
- 24.Hunter, W. B., E. Hiebert, S. E. Webb, J. K. Tsai, and J. E. Polston. 1998. Location of geminiviruses in the whitefly Bemisia tabaci (Homoptera: Aleyrodidae). Plant Dis. 82:1147-1151. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, Y. X., C. de Blas, I. D. Bedford, G. Nombela, and M. Muniz. 2004. Effects of Bemisia tabaci biotype on the transmission of tomato yellow leaf curl Sardinia virus (TYLCSV-ES) between tomato common weeds. Spanish J. Agric. Res. 2:115-119. [Google Scholar]
- 26.Kontsedalov, S., E. Zchori-Fein, E. Chiel, Y. Gottlieb, M. Inbar, and M. Ghanim. 2008. The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest Manag. Sci. 64:789-792. [DOI] [PubMed] [Google Scholar]
- 27.Leshkowitz, D., S. Gazit, E. Reuveni, M. Ghanim, H. Czosnek, C. McKenzie, R. G. Shatters, Jr., and J. K. Brown. 2006. Whitefly (Bemisia tabaci) genome project: analysis of sequenced clones from egg, instar, and adult (viruliferous and nonviruliferous) cDNA libraries. BMC Genomics 7:79-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S. S., P. J. De Barro, J. Xu, J. B. Luan, L. S. Zang, Y. M. Ruan, and F. H. Wan. 2007. Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318:1769-1772. [DOI] [PubMed] [Google Scholar]
- 29.Mahadav, A., S. Kontsedalov, H. Czosnek, and M. Ghanim. 2009. Thermotolerance and gene expression following heat stress in the whitefly Bemisia tabaci B and Q biotypes. Insect Biochem. Mol. Biol. 39:668-676. [DOI] [PubMed] [Google Scholar]
- 30.Mehta, P., J. A. Wyman, M. K. Nakhla, and D. P. Maxwell. 1994. Transmission of tomato yellow leaf curl geminivirus by Bemisia tabaci (Homoptera: Aleyrodidae). J. Econ. Entomol. 87:1291-1297. [DOI] [PubMed] [Google Scholar]
- 31.Morin, S., M. Ghanim, I. Sobol, and H. Czosnek. 2000. The GroEL protein of the whitefly Bemisia tabaci interacts with the coat protein of transmissible and nontransmissible begomoviruses in the yeast two-hybrid system. Virology 276:404-416. [DOI] [PubMed] [Google Scholar]
- 32.Morin, S., M. Ghanim, M. Zeidan, H. Czosnek, M. Verbeek, and J. F. J. M. van den Heuvel. 1999. A GroEL homologue from endosymbiotic bacteria of the whitefly Bemisia tabaci is implicated in the circulative transmission of tomato yellow leaf curl virus. Virology 256:75-84. [DOI] [PubMed] [Google Scholar]
- 33.Navot, N., E. Pichersky, M. Zeidan, D. Zamir, and H. Czosnek. 1991. Tomato yellow leaf curl virus: a whitefly-transmitted geminivirus with a single genomic component. Virology 185:151-161. [DOI] [PubMed] [Google Scholar]
- 34.Noris, E., A. M. Vaira, P. Caciagli, V. Masenga, B. Gronenborn, and G. P. Accotto. 1998. Amino acids in the capsid protein of tomato yellow leaf curl virus that are crucial for systemic infection, particle formation, and insect transmission. J. Virol. 72:10050-10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogata, H., B. La Scola, S. Audic, P. Renesto, G. Blanc, C. Robert, P.-E. Fournier, J.-M. Claverie, and D. Raoult. 2006. Genome sequence of Rickettsia bellii illuminates the role of Amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 22:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubinstein, G., and H. Czosnek. 1997. Long-term association of tomato yellow leaf curl virus (TYLCV) with its whitefly vector Bemisia tabaci: effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 78:2683-2689. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-Campos, S., J. Navas-Castillo, R. Camero, C. Soria, J. A. Diaz, and E. Moriones. 1999. Displacement of tomato yellow leaf curl virus (TYLCV)-Sr by TYLCV-Is in tomato epidemics in Spain. Phytopathology 89:1038-1043. [DOI] [PubMed] [Google Scholar]
- 38.Sinisterra, X. H., C. L. McKenzie, W. B. Hunter, C. A. Powell, and R. G. Shatters, Jr. 2005. Differential transcriptional activity of plant-pathogenic begomoviruses in their whitefly vector Bemisia tabaci Gennadius (Hemiptera: Aleyrodidae). J. Gen. Virol. 86:1525-1532. [DOI] [PubMed] [Google Scholar]
- 39.Treco, D. A., and V. Lundblad. 1997. Preparation of yeast media, p. 13.1.1-13.1.7. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Wiley, New York, NY. [DOI] [PubMed]
- 40.van den Heuvel, J. F. J. M., A. Bruyère, S. A. Hogenhout, V. Ziegler-Graff, V. Brault, M. Verbeek, F. van der Wilk, and K. Richards. 1997. The N-terminal region of the luteovirus readthrough domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J. Virol. 71:7258-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Heuvel, J. F. J. M., M. Verbeek, and F. van der Wilk. 1994. Endosymbiotic bacteria associated with circulative transmission of potato leafroll virus by Myzus persicae. J. Gen. Virol. 75:2559-2565. [DOI] [PubMed] [Google Scholar]