Abstract
Human immunodeficiency virus type 1 (HIV-1) gp41 plays a critical role in the viral fusion process, and its N- and C-terminal heptad repeat domains serve as important targets for developing anti-HIV-1 drugs, like T-20 (generic name, enfuvirtide; brand name, Fuzeon). Here, we conducted a yeast two-hybrid screening on a human bone marrow cDNA library using the recombinant soluble gp41 ectodomain as the bait and identified a novel gp41 core-binding molecule, designated P20. P20 showed no homology with a current HIV fusion inhibitor, T-20, but had sequence homology to a human protein, troponin I type 3 interacting kinase (TNNI3K)-like protein. While it could bind to the six-helix bundle core structure formed by the N- and C-terminal heptad repeats, P20 did not interrupt the formation of the six-helix bundle. P20 was effective in blocking HIV-1 Env-mediated syncytium formation and inhibiting infection by a broad spectrum of HIV-1 strains with distinct subtypes and coreceptor tropism, while it was ineffective against other enveloped viruses, such as vesicular stomatitis virus and influenza A virus. P20 exhibited no significant cytotoxicity to the CD4+ cells that were used for testing antiviral activity. Among the 11 P20 mutants, four analogous peptides with a common motif (WGRLEGRRT) exhibited significantly reduced anti-HIV-1 activity, suggesting that this region is the critical active site of P20. Therefore, this peptide can be used as a lead for developing novel HIV fusion inhibitors and as a probe for studying the membrane-fusogenic mechanism of HIV.
Human immunodeficiency virus type 1 (HIV-1) is an enveloped virus, and its envelope protein (Env) complex controls the key processes by which HIV-1 delivers its replicative material into target cells. Specifically, the Env surface subunit, gp120, binds the cellular receptor CD4 and a coreceptor, CCR5 or CXCR4, which triggers conformational changes of the transmembrane subunit, gp41 (8). The N-terminal heptad repeat (NHR) in the gp41 ectodomain interacts with its C-terminal heptad repeat (CHR) to form a trimer of hairpins, or six-helix bundle (6-HB; also known as the gp41 fusion core) (38, 51), which brings the viral and target cell membranes into close proximity and promotes membrane fusion (3, 51). Therefore, the gp41 6-HB core plays an important role in viral fusion and may serve as an attractive target for the development of HIV fusion/entry inhibitors (20).
In the early 1990s, a number of peptides derived from the gp41 NHR and CHR regions were discovered to exhibit highly potent anti-HIV-1 activity by binding to the corresponding region of gp41 at the fusion-intermediate state (22, 23, 38, 52, 53) and blocking gp41 6-HB core formation (4, 9, 32, 47). One of the CHR-peptides, T-20 (generic name, enfuvirtide; brand name, Fuzeon), was licensed by the FDA as the first member of a new class of anti-HIV drugs, the HIV fusion inhibitors (33, 53). Although T-20 is very effective in inhibiting infection by a broad spectrum of HIV-1 strains, especially those resistant to current antiretroviral therapies (26), T-20 itself also can easily induce drug resistance in T-20-treated patients, resulting in virologic failure (36, 46, 50, 55). Therefore, it is essential to identify and develop novel HIV-1 fusion inhibitors having a mechanism of action or target different from that for T-20 and with improved drug resistance profiles.
Here, we sought to screen a human bone marrow cDNA library in a yeast two-hybrid screening assay using the recombinant soluble gp41 ectodomain (rsgp41e) as the bait in hopes of identifying a novel HIV fusion inhibitor with sequence homology to a human protein and low immunogenicity to humans to avoid its rapid clearance by specific human antibodies (1). We identified a 32-mer peptide, designated P20, with sequence homology to human troponin I type 3 interacting kinase (TNNI3K)-like protein. P20 could specifically bind to the gp41 6-HB core and strongly blocked HIV-1 Env-mediated membrane fusion. It potently inhibited infection by a number of laboratory-adapted HIV-1 strains, including T-20-resistant variants, and a broad spectrum of primary HIV-1 isolates. These results suggest that P20 has the potential to be developed further as a novel anti-HIV-1 therapeutic and can be used as a probe to study the role of the HIV-1 gp41 6-HB core in the membrane fusion process.
MATERIALS AND METHODS
Cells and viruses.
3T3 cells stably transduced with murine leukemia virus MX-CD4 and MX-CXCR4 vectors (3T3.T4.CXCR4) were cultured in Dulbecco's modified Eagle medium (DMEM) complemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 IU/ml streptomycin (Invitrogen, Carlsbad, CA). CHO cells stably transfected with either the HIV-1HXB2 Env-expressing vector pEE14 (CHO-WT) or control pEE14 vector (CHO-EE) were cultured in glutamine-deficient minimal essential medium containing 400 μM methionine sulfoximine (Sigma, St. Louis, MO). The cells, including MT-2 and TZM-bl cells; the viruses, including HIV-1 strains IIIB, Bal, NL4-3, NL4-3(36G)N42S (T-20 sensitive), NL4-3(36G)V38A/N42D, and NL4-3(36G)V38E/N42S (T-20 resistant), and primary HIV-1 isolates; and the plasmids, including pHEF-VSVG and pNL4-3.luc.RE, were obtained from the NIH AIDS Research and Reference Reagent Program. The vesicular stomatitis virus glycoprotein (VSV-G) and influenza A virus hemagglutinin (IAV HA) pseudovirus were produced by cotransfecting 293T cells with pNL4-3.luc.RE and pHEF-VSVG or the plasmid encoding HA of IAV H5N1, respectively, using Lipofectamine 2000 (Invitrogen) as previously described (5, 16).
Yeast two-hybrid system.
The BD Matchmaker yeast two-hybrid system (BD Bioscience) was employed in our study, and all of the procedures were followed according to the manufacturer's instructions. First, the rsgp41e (amino acids [aa] 539 to 684) of HIV-1HXB2 was amplified and cloned into the pGBKT7 vector (encoding the Gal4 DNA-binding domain [BD]) as the bait plasmid. The yeast strain AH109 was transformed with the bait plasmid pGBKT7-rsgp41e and mated with yeast Y187, which contains the human bone marrow cDNA library (constructed in vector pGADT7-Rec, which encodes the Gal4 activation domain [AD]). The mated yeasts were screened on dropout medium deficient in Try, Leu, and His (−Try/−Leu/−His). Primary His-positive colonies were rescreened on −Try/−Leu/−His/−Ade/5-bromo-4-chloro-3-indolyl-â-d-galactopyranoside (X-Gal) plates to confirm protein interaction. The library plasmids from the blue colonies were recovered and retested to eliminate false positives.
Recombinant protein expression and purification.
PGEX-6p-1 vector, glutathione Sepharose 4 fast-flow beads, and PreScission protease from Amersham Pharmacia Biotech, Inc., were used to express and purify recombinant proteins with or without a glutathione S-transferase (GST) tag. For P20 and its mutants, the DNA fragments were cloned into the EcoRI and XhoI sites of the vector. For NC (N36-L6-C34) polypeptide, the DNA sequences of N36 and C34 were amplified by PCR, ligated by the BamHI and BglII fusion sites, and then cloned into the BamHI and XhoI sites of the pGEX-6p-1 vector. For 3NC, three N peptide segments and three C peptide segments are alternately linked (N-C-N-C-N-C) using short Gly/Ser peptide linkers, with the exception of the last N-to-C linker, GGRGG (35, 41), so that the expressed protein consists of three copies of sequentially linked N36-L6-C34 that can automatically form a 6-HB in a manner similar to that of the 5-helix protein (44). The purified proteins were identified with SDS-PAGE, and the protein concentrations were calculated with absorbance at a UV wavelength of 280 nm.
Enzyme-linked immunosorbent assay (ELISA).
To determine the activity of P20 and its mutants binding to the gp41 fusion core domains, wells of microtiter plates were coated with 50 μl P20 and its mutant (10 μg/ml) in 0.1 M NaHCO3 buffer (pH 8.6) overnight. The wells then were blocked with phosphate-buffered saline (PBS, pH 7.5) containing 0.25% gelatin. After three washes with PBS containing 0.1% Tween 20 (PBS-T), NC polypeptide diluted in PBS at a 4-fold dilution (starting from 10 μg/ml) was added, followed by incubation at room temperature for 1 h. After extensive washes, the amount of bound 6-HB was detected by the addition of monoclonal antibody (MAb) NC-1 (21), peroxidase-conjugated anti-mouse antibody, and the substrate o-phenylendiamine sequentially. The absorbance at 450 nm (A450) was recorded. To determine whether P20 could block gp41 6-HB formation, a microplate was coated with 100 μl rabbit anti-N36/C34 IgG (4 μg/ml in 0.1 M Tris, pH 8.8) at 4°C overnight. After being blocked with 1% nonfat milk at 37°C for 1 h, 50 μl P20 at 4-fold serial dilutions was mixed with 25 μl N36 (4 μM). After incubation at 37°C for 30 min, 25 μl C34 (4 μM) was added and incubated at 37°C for an additional 30 min. ADS-J1, a small-molecule HIV-1 fusion inhibitor that blocks gp41 6-HB formation (24, 49), was included as a control. The mixture was added to the coated wells, followed by incubation at 37°C for 1 h. After extensive washes, the 6-HB formed by N36 and C34 was detected by sequentially adding 100 μl MAb NC-1 (5 μg/ml), peroxidase-conjugated anti-mouse IgG, and the substrate o-phenylendiamine.
SPR assay.
The affinities of the interaction of P20 and its mutant with the gp41 6-HB were determined using a surface plasmon resonance (SPR) assay on a Biacore X instrument (Biacore AB, Uppsala, Sweden) essentially as described previously (11, 42). In short, the 6-HB (3NC) was immobilized by amine coupling to the surface of a CM5 sensor chip (Biacore). Immobilization and interaction studies were conducted at 25°C in 10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.005% of the surfactant polyoxyethylenesorbitan (HBS-EP buffer; Biacore) as a running buffer. P20 and its variants were diluted in the running buffer and injected as 35-μl solutions at a flow rate of 5 μl/min. The dissociation reaction was done by washing with running buffer for at least 2 min. The responses all were analyzed by the fit of a 1:1 interaction model using the BIAevaluation program (Biacore).
FN-PAGE.
Fluorescence native polyacrylamide gel electrophoresis (FN-PAGE) was performed to detect the 6-HB formed by mixing N36 (100 μM) with an equimolar concentration of C34-fluorescein isothiocyanate (FITC) as described previously (34). Briefly, a test peptide was preincubated with an equal amount of N36 at 37°C for 30 min, followed by the addition of C34-FITC at 37°C for 30 min. The mixtures then were diluted in Tris-glycine native sample buffer (Invitrogen, Carlsbad, CA), and the samples (20 μl) were loaded onto Tris-glycine gels (18%; Invitrogen, Carlsbad, CA), which were run at a constant voltage of 120 V at room temperature for 1 h. The gels were visualized with the FluorChem 8800 imaging system (Alpha Innotech Corp., San Leandro, CA) using a transillumination UV light source with an excitation wavelength of 302 nm and a fluorescence filter with an emission wavelength of 520 nm. The same gels then were stained with Coomassie blue and imaged with the FluorChem 8800 imaging system using a visible light source.
Assay for HIV-1 Env-mediated syncytium formation.
Target (3T3.T4.CXCR4) cells (5 × 104) resuspended in Dulbecco's modified Eagle medium containing 10% FBS were plated in wells of 48-well plates and incubated overnight at 37°C, followed by washes with glutamine-deficient minimal essential medium. A total of 3 × 104 effector (CHO-WT) cells prestimulated with 6.5 mM sodium butyrate for about 20 h were added in the absence or presence of an inhibitor at graded concentrations. After being cocultured for 24 h at 37°C, the syncytia, defined as giant cells with diameters more than four times larger than those of single cells, were counted under a microscope. The percent inhibition of syncytium formation was calculated using the following formula: % inhibition = [1 − (number of syncytia in a well containing an inhibitor)/(number of syncytia in a well containing no inhibitor)] × 100. To determine the biological functions, P20 peptide at a serial 2-fold dilution (starting from 37.5 μg/ml) was preincubated with CHO-WT cells at 37°C for 60 min, followed by the addition of the mixture to the target cells. The GST protein was used as a negative control. Other steps were the same as those described above. The concentration for 50% inhibition (IC50) was calculated using the CalcuSyn software (6).
Flow cytometry.
Cells were detached and washed with wash buffer (PBS containing 1% FBS) three times. They were incubated with the GST-P20 fusion protein or GST protein alone for 1 h at 4°C. After three washes, FITC-conjugated GST antibodies were added and incubated for 1 h at 4°C. After at least three washes, the cells were examined by flow cytometry and the fluorescence intensity was recorded by a FACSCalibur (Becton Dickinson).
Inhibition of HIV-1 infection.
Inhibitory activities of P20 and its mutants on infection by HIV-1 X4 strains IIIB and NL4-3 as well as the T-20-resistant variants (43) were determined as previously described (40). Briefly, 1 × 104/ml MT-2 cells in RPMI 1640 medium containing 10% FBS were infected with HIV-1 isolates at 100 TCID50 (50% tissue culture infective doses) in 200 μl culture medium in the presence or absence of the test peptide overnight. The culture supernatants were removed, and fresh media were added the next day. On the fourth day postinfection, 100 μl of culture supernatants were collected from each well, mixed with equal volumes of 5% Triton X-100, and assayed for p24 antigen by ELISA. The inhibition of P20 and its variants on infection by the HIV-1 R5 strain Bal, the primary HIV-1 isolates, and the VSV-G pseudovirus was determined as described before (31). Briefly, 100 μl of TZM-bl cells (1 × 105/ml) was precultured overnight and infected with a virus at 100 TCID50 in the presence or absence of the test peptide overnight. 293T cells, instead of TZM-bl cells, were used for assessing the infectivity of the IAV HA pseudovirus. The cells were harvested and lysed on the fourth day postinfection with 50 μl of lysing reagent. The luciferase activity was analyzed using a luciferase kit (Promega, Madison, WI) and a luminometer (Ultra 386; Tecan, Durham, NC) according to the manufacturer's instructions. The percent inhibition of luciferase activity and the IC50s were calculated using the CalcuSyn software (6).
Analysis of cytotoxicity.
The potential cytotoxicity of P20 on the MT-2 and TZM-bl cells was measured by using the colorimetric XTT assay as previously described (25). Briefly, 100 μl of a compound at graded concentrations was added to equal volumes of cells (105 cells/ml) in wells of 96-well plates. After incubation at 37°C for 4 days, 50 μl of XTT solution (1 mg/ml) containing 0.02 μM phenazine methosulfate was added. After 4 h, the absorbance at 450 nm was measured with an ELISA reader and the percentage of cytotoxicity was calculated.
RESULTS
Identification of a gp41-binding molecule with sequence homology to human TNNI3K-like protein.
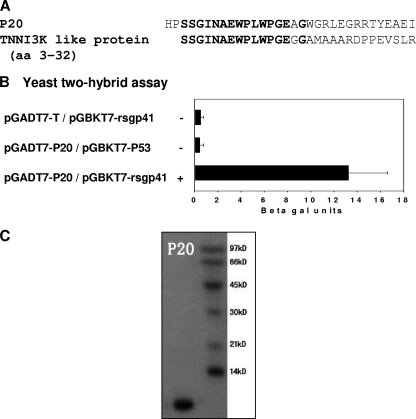
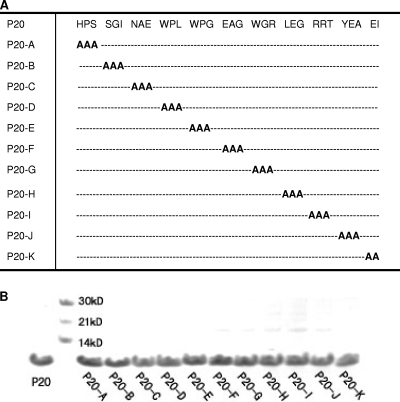
To identify a gp41-binding molecule with sequence homology to a human protein, we conducted a yeast two-hybrid screening on a human bone marrow cDNA library using rsgp41e as the bait. Out of more than 1 × 106 clones screened, a blue clone on a −Try/−Leu/−His/−Ade/X-Gal plate was identified, and the corresponding plasmid was recovered and sequenced. Since it was the 20th protein on the plate, it was named P20; it consists of 32 amino acids (Fig. 1A). Blasting the human protein database at NCBI showed that it had similarity to human TNNI3K-like protein (GenBank accession no. CAE45949.1) (Fig. 1A), confirming that P20 is partly derived from a human protein-coding clone in the human bone marrow cDNA library. To demonstrate the specificity of the interaction, pGADT7-P20 and the control plasmid pGADT7-T were transformed into yeast strain AH109, which contains the bait plasmid pGBKT7-rsgp41e or the control bait plasmid pGBKT7-P53. Only the yeasts containing pGADT7-P20/pGBKT7-rsgp41e and pGADT7-T/pGBKT7-P53 displayed the His+/Ade+/Mel+ phenotype (Fig. 1B), suggesting that P20 can specifically interact with rsgp41e, as the control T protein specifically interacts with P53 (29). The recombinant P20 peptide was expressed in Escherichia coli and purified with affinity chromatography. We found that P20 was highly soluble and easily purified, with an estimated molecular mass of 3.5 kDa (Fig. 1C), making it easier to pursue further research and applications.
FIG. 1.
Identification of a gp41-binding molecule, P20. (A) The amino acid sequence of P20 with homology to human TNNI3K-like protein. (B) Specific interaction between rsg41e and P20 as determined by yeast two-hybrid assay. (C) SDS-PAGE analysis of the purified P20. The estimated molecular mass of P20 is 3.5 kDa.
P20 could not significantly interact with the individual NHR and CHR peptides but strongly bound to the gp41 6-HB core.
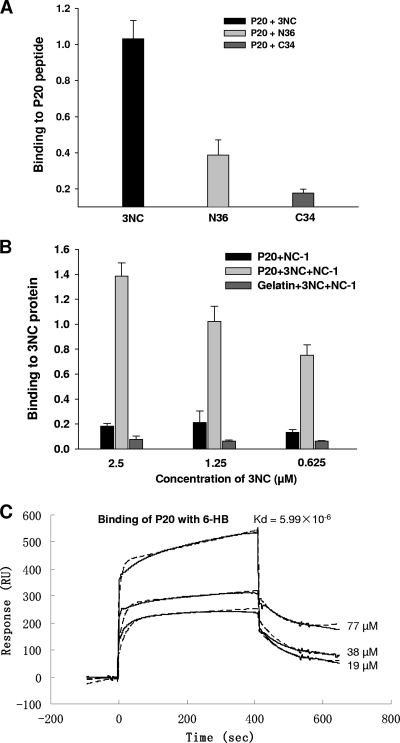
Since the gp41 NHR and CHR regions are the critical functional domains involved in the fusion core formation and the viral fusion process and serve as important targets for HIV fusion inhibitors, we tested the binding activity of P20 with the NHR and CHR peptides and the 6-HB core formed by the NHR and CHR peptides in different assays. As shown in Fig. 2A and B, P20 exhibited weak binding activity with gp41 NHR and CHR peptides (e.g., N36 and C34, respectively), but it strongly bound to 3NC polypeptide, an in vitro model of the gp41 6-HB core that was formed by the NHR and CHR peptides (37), in a dose-dependent manner. The interaction between P20 and 3NC was further confirmed by the SPR analysis (Fig. 2C). Experimental curves (solid line) were shown overlaid with fitted curves (dashed line) obtained with the 1:1 binding-with-drifting baseline model, suggesting a 1:1 stoichiometry of P20 binding to the gp41 6-HB. These results indicate that P20 specifically binds to the conformational gp41 core structure formed by the gp41 NHR and CHR domains but cannot interact with the linear NHR and CHR sequences.
FIG. 2.
Binding of P20 to the HIV-1 gp41 6-HB core. (A) Binding of P20 to the N peptide N36, the C peptide C34, and the 6-HB 3NC, as determined by direct ELISA. (B) Binding of P20 to 6-HB, as detected by sandwich ELISA using the 6-HB-specific MAb NC-1. Gelatin was used as the negative control. (C) Kinetic analyses of P20 and 6-HB interaction by SPR. The y axis of the sensorgram is denoted as the RU signal, whereas the time (in seconds) is represented on the x axis. A series of concentrations of P20 were injected at a flow rate of 5 μl/min. Experimental curves (solid line) are shown overlaid with fitted curves (dash line) obtained with the 1:1 binding-with-drifting baseline model.
P20 was ineffective in blocking gp41 6-HB core formation between the NHR and CHR peptides.
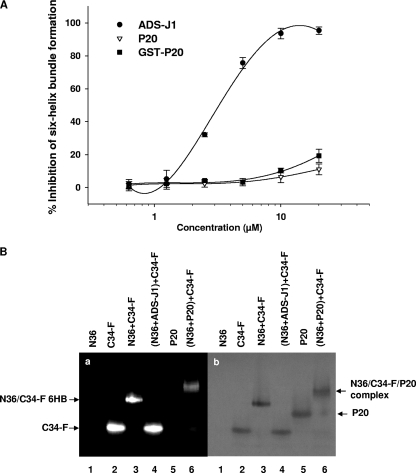
We subsequently tested whether P20 could inhibit the formation of 6-HB formed by the peptides N36 and C34 by ELISA and FN-PAGE. ADS-J1, a small-molecule HIV-1 fusion inhibitor that was proven to block gp41 core formation (24, 49), was used as a control. In ELISA, ADS-J1 strongly blocked the gp41 6-HB formation with an IC50 of 3 μM, while P20 peptide at concentrations of up to 20 μM exhibited no significant inhibition on 6-HB formation (Fig. 3 A). In FN-PAGE (Fig. 3B), C34-FITC alone showed a clear fluorescence band at the lower position in the gel (lane 2). The mixture of N36 and C34-FITC displayed a fluorescence band at the upper position corresponding to the gp41 6-HB (34) and, at the same time, a lower band corresponding to the C34-FITC disappeared (lane 3), suggesting that N36 and C34-FITC associate to form a 6-HB. The addition of ADS-J1 to the mixture of N36 and C34-FITC resulted in the disappearance of the upper band and the reappearance of the lower band (line 4), confirming that ADS-J1 is able to block 6-HB formation. However, after the addition of P20 to the N36/C34-FITC mixture, the lower band did not reappear, but instead a new band at a position higher than that of the 6-HB band appeared. These results indicate that P20 does not block 6-HB formation but rather binds to the N36/C34-FITC 6-HB to form a complex with sizes different from that of 6-HB.
FIG. 3.
Effect of P20 on the gp41 6-HB formation between N36 and C34 by ELISA (A) and FN-PAGE (B). ADS-J1 was used as a positive control. (a) After native polyacrylamide gel electrophoresis, the gel was visualized using a transillumination UV light source to reveal the bands containing C34-FITC, including C34-FITC alone (line 2), N36/C34-FITC complex (line 3), and N36/C34-FITC/P20 complex (line 5). (b) The same gel then was stained with Coomassie blue to show all of the protein bands.
P20 inhibited HIV-1 Env-mediated membrane fusion.
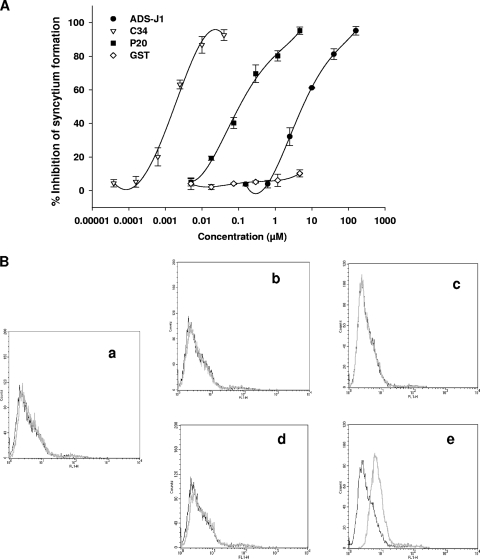
We then investigated whether the P20 peptide could inhibit HIV-1 Env-mediated syncytium formation. Like the C peptide C34 and small-molecule HIV-1 fusion inhibitor ADS-J1, P20 potently inhibited syncytium formation in a dose-dependent manner with an IC50 of 0.108 μM (Fig. 4A). To further confirm whether P20 interacts with a fusion-active conformation of HIV-1 Env, we used flow cytometry to determine the binding activity of P20 to the effector and target cells in the fusion and nonfusion states. Our previous studies have shown that only the sodium butyrate-treated CHO-WT cells (7) are able to fuse with 3T3.T4.CXCR4 cells, resulting in syncytium formation. Here, we found that P20 bound with neither the 3T3.T4.CXCR4 cells (Fig. 4B, graph a) nor the CHO-WT cells (Fig. 4B, graph b). P20 also could not bind to the cocultured 3T3.T4.CXCR4 cells and CHO-WT cells not treated with sodium butyrate (with no syncytium formation) (Fig. 4B, graph c), nor could it bind to the sodium butyrate-treated CHO-WT cells (Fig. 4B, graph d). However, it significantly bound to the cocultured 3T3.T4.CXCR4 cells and sodium butyrate-treated CHO-WT cells (with syncytium formation) (Fig. 4B, graph e). These results suggest that P20 does not bind to the native HIV-1 Env expressed on the effector cells but is effective in binding to the Env in the fusion-active state, possibly to the gp41 fusion core at the late stage of membrane fusion.
FIG. 4.
Effect of P20 on HIV-1 Env-mediated membrane fusion. (A) Inhibition of P20 on HIV-1 Env-mediated syncytium formation. GST protein was used as the negative control. (B) Binding of P20 (light line) or GST protein (dark line) with 3T3.T4.CXCR4 (target) cells and/or CHO-WT (effector) cells as analyzed by flow cytometry: graph a, 3T3.T4.CXCR4 cells only; b, CHO-WT cells only; c, coculture of 3T3.T4.CXCR4 and CHO-WT cells; d, CHO-WT cells treated with sodium butyrate for 20 h; and e, coculture of 3T3.T4.CXCR4 and sodium butyrate-treated CHO-WT cells.
P20 inhibited infection by a broad spectrum of HIV-1 strains.
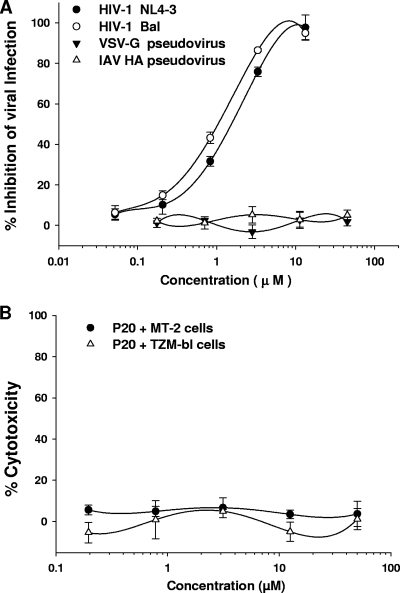
The antiviral activity of P20 against infection by laboratory-adapted HIV-1 strains was determined using different assays. We found that P20 significantly inhibited infection by the laboratory-adapted HIV-1 X4 (NL4-3 and IIIB) and R5 (Bal) strains with IC50s at low-μM levels (Table 1), while it exhibited no inhibition on VSV-G and IAV HA pseudovirus infection (Fig. 5A). P20 showed no cytotoxicity to MT-2 and TZM-bl cells at concentrations as high as 50 μM (Fig. 5B). These results suggest that P20-mediated antiviral activity is specific for HIV-1 and that P20 has no significant toxic effect on CD4+ cells that were used for testing the antiviral activity of P20. We compared the antiviral activity of T-20 to that of T-20-sensitive and -resistant strains that bear mutations in the principal determinant responsible for T-20 resistance (aa 36 to 45; GIVQQQNNLL) in the gp41 NHR domain (10, 15, 28, 36). Unlike T-20, which is much less effective against T-20-resistant strains than against T-20-sensitive strains, P20 showed similar potency against both T-20-sensitive and -resistant variants (Table 2). This result indicates that because P20 has a different target site in gp41 from that for T-20, P20 is able to inhibit infection by T-20-resistant viruses. Subsequently, we tested the inhibitory activities of P20 against a panel of 14 representative primary HIV-1 isolates with distinct subtypes (A to G and group O) and coreceptor tropism (R5 and X4R5), using T-20 as a control. As shown in Table 3, P20 could significantly inhibit infection by all of the primary HIV-1 isolates tested, with IC50s at low-μM levels. T-20, the highly potent HIV fusion inhibitor targeting gp41, inhibited infection by these primary HIV-1 isolates at the submicromolar level. These results suggest that like T-20, P20 is effective against a broad spectrum of HIV-1 strains with different genotype and phenotypes.
TABLE 1.
Inhibitory activity of P20 and its mutants on infection by laboratory-adapted HIV-1 strainsa
| Peptide | IC50 (μM) for inhibiting infection by HIV-1 |
||
|---|---|---|---|
| NL4-3 (X4) | IIIB (X4) | Bal (R5) | |
| P20 | 1.52 ± 0.82 | 5.31 ± 3.24 | 1.03 ± 0.45 |
| P20-A | 0.91 ± 0.72 | 2.31 ± 1.51 | 0.68 ± 0.37 |
| P20-B | 1.98 ± 0.87 | 6.75 ± 3.55 | 1.66 ± 2.18 |
| P20-C | 1.36 ± 1.25 | 5.73 ± 2.55 | 1.22 ± 0.87 |
| P20-D | 2.35 ± 1.72 | 6.32 ± 0.96 | 2.01 ± 1.05 |
| P20-E | 15.4 ± 6.14 | >25 | 10.2 ± 5.17 |
| P20-F | 4.56 ± 1.13 | 12.4 ± 5.23 | 3.49 ± 1.91 |
| P20-G | >25 | >25 | >25 |
| P20-H | 13.4 ± 8.12 | 20.1 ± 5.98 | 8.24 ± 8.27 |
| P20-I | 18.3 ± 6.92 | >25 | 13.8 ± 7.15 |
| P20-J | 2.56 ± 1.55 | 5.38 ± 3.57 | 1.52 ± 0.85 |
| P20-K | 1.73 ± 0.51 | 5.99 ± 3.29 | 1.32 ± 1.19 |
The experiment was performed in triplicate, and the data are presented as means ± standard deviations.
FIG. 5.
Antiviral activities of P20 and its cytotoxicity. (A) Inhibition of P20 on infection by HIV-1 NL4-3 (X4) and Bal (R5) strains as well as VSV-G and IAV HA pseudoviruses. The infectivity of the HIV-1 NL4-3 was detected in MT-2 cells using ELISA p24 antigen, while the infectivity of HIV-1 Bal and VSV-G pseudovirus was determined in TZM-bl cells using a luciferase assay. The infectivity of IAV HA pseudovirus was assessed in 293T cells using a luciferase assay. (B) Cytotoxicity of P20. The potential toxic effect of P20 on the CD4+ TZM-bl and MT-2 cells, which were used for testing anti-HIV-1 activity, was determined by a colorimetric XTT assay.
TABLE 2.
Inhibition of infection by T-20-sensitive and -resistant HIV-1 strainsa
| HIV-1 strain | T-20 phenotype | IC50 (μM) |
|||
|---|---|---|---|---|---|
| T-20 | P20 | P20-A | P20-G | ||
| HIV-1NL4-3(36G) N42S | Sensitive | 0.02 ± 0.01 | 1.32 ± 0.80 | 0.81 ± 0.08 | >15 |
| HIV-1NL4-3(36G) V38A/N42D | Resistant | 0.32 ± 0.05 | 1.11 ± 0.30 | 0.73 ± 0.09 | >15 |
| HIV-1NL4-3(36G) V38E/N42S | Resistant | 9.62 ± 2.60 | 2.31 ± 0.70 | 0.95 ± 0.05 | >15 |
The experiment was performed in triplicate, and the data are presented as means ± standard deviations.
TABLE 3.
Inhibition of P20 and its variants on infection by primary HIV-1 isolatesa
| HIV-1 isolate (subtype, coreceptor tropism) | IC50 (μM) |
|||
|---|---|---|---|---|
| P20 | P20-A | P20-G | T-20 | |
| 94UG103 (A, X4R5) | 5.24 ± 0.05 | 2.19 ± 0.08 | >45 | 0.027 ± 0.007 |
| 92RW008 (A, R5) | 9.33 ± 1.51 | 4.96 ± 0.35 | >45 | 0.021 ± 0.01 |
| 92US657 (B, R5) | 4.20 ± 0.83 | 1.89 ± 0.16 | >45 | 0.105 ± 0.01 |
| 92BR014 (B, X4R5) | 3.79 ± 0.53 | 1.63 ± 0.12 | >45 | 0.058 ± 0.12 |
| 93MW959 (C, R5) | 2.99 ± 0.07 | 1.57 ± 0.04 | >45 | 0.009 ± 0.002 |
| 92IN101 (C, R5) | 4.79 ± 0.19 | 2.94 ± 0.11 | >45 | 0.022 ± 0.005 |
| 92BR025 (C, R5) | 4.86 ± 0.85 | 1.72 ± 0.17 | >45 | 0.011 ± 0.007 |
| 94UG118 (D, R5) | 32.9 ± 17.6 | 13.1 ± 0.52 | >45 | 0.927 ± 0.061 |
| 92UG001 (D,X4R5) | 10.5 ± 1.73 | 4.43 ± 0.28 | >45 | 0.123 ± 0.072 |
| 92TH009 (A/E, R5) | 4.69 ± 0.48 | 2.79 ± 0.09 | >45 | 0.087 ± 0.012 |
| 92TH023 (E, R5) | 41.5 ± 10.9 | 23.2 ± 0.33 | >45 | 0.487 ± 0.078 |
| 93BR020 (F, X4R5) | 13.6 ± 2.34 | 7.25 ± 1.69 | >45 | 0.093 ± 0.025 |
| RU570 (G, R5) | 16.2 ± 4.10 | 7.87 ± 2.01 | >45 | 0.116 ± 0.021 |
| BCF02 (O, R5) | 10.2 ± 1.12 | 4.52 ± 0.14 | >45 | 0.063 ± 0.015 |
The samples were tested in triplicate, and the data are presented as means ± standard deviations. T-20 was included as a control.
The critical functional domain is localized in a region adjacent to the C terminus of P20.
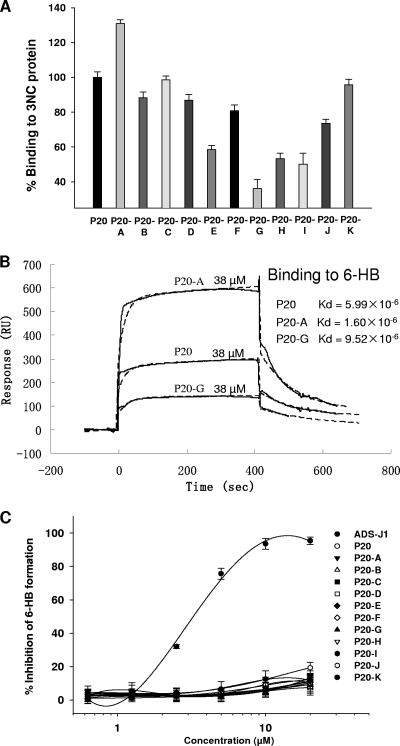
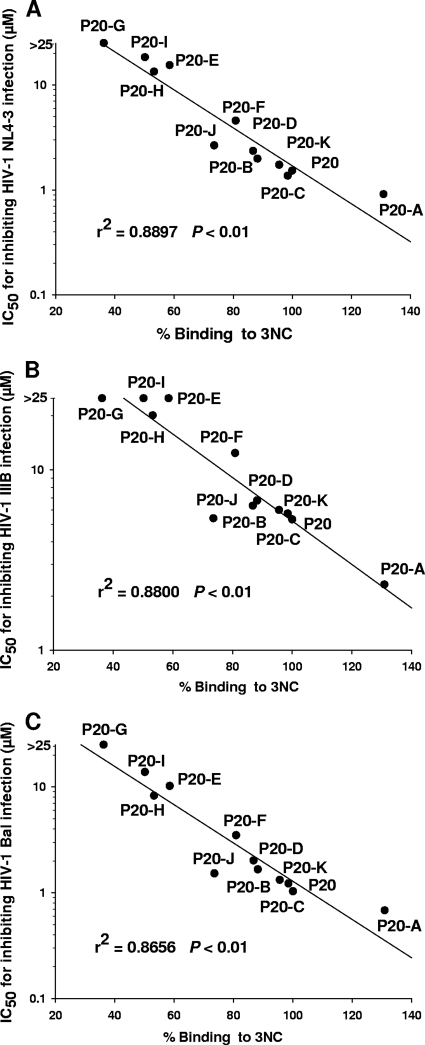
To identify the critical functional domain in the P20 peptide, we designed 11 triple-alanine mutants overlapping the entire sequence of P20 (Fig. 6A). All of these mutants were well expressed and purified in a manner similar to that of P20 (Fig. 6B). Results showed that the site-specific binding capacities of these P20 mutations to the 6-HB were severely affected. For example, while mutant P20-A showed enhanced affinity to the 6-HB, the mutants P20-E, P20-G, P20-H, and P20-I exhibited decreased affinity to the 6-HB (Fig. 7A). As determined by SPR analysis, the binding affinities of P20 with the 6-HB (3NC) were lower and higher than those of P20-A and P20-G, respectively (Fig. 7B). Unlike ADS-J1 but like P20, not all P20 mutants significantly inhibited gp41 6-HB formation at concentrations up to 20 μM (Fig. 7C). By analyzing the anti-HIV-1 activity of P20 and its mutants, we found that those mutants with decreased affinity of binding to 6-HB (e.g., P20-E, P20-G, P20-H, and P20-I) also were less effective than P20 and P20-A in inhibiting HIV-1 infection (Table 1 to 3). Interestingly, the 6-HB core-binding activity of P20 and its mutants is well correlated with their anti-HIV-1 activity (Fig. 8). Since those mutants with decreased biological activity share a motif (WGRLEGRRT) located in the region adjacent to the C terminus of P20, we suspect that this region is its critical functional domain. It is expected that the optimization of sequence in this region generates analog peptides with improved anti-HIV-1 activity.
FIG. 6.
P20 and its mutants. (A) The sequences of P20 and its mutants. (B) SDS-PAGE analysis of P20 and its mutants.
FIG. 7.
Biological analysis of P20 and its mutants. (A) Binding activity of P20 and its mutants to the gp41 6-HB core as measured by direct ELISA. (B) SPR analyses of P20 and its variants binding to the gp41 6-HB (3NC). The same amounts (38 μM) of P20 and its mutants were injected at a flow rate of 5 μl/min. Experimental curves (solid line) are overlaid with the fitted curves (dashed line) obtained with the 1:1 binding-with-drifting baseline model. (C) Assessment of inhibitory activities of P20 and its mutants on gp41 6-HB formation detected by sandwich ELISA.
FIG. 8.
Correlation between the binding activities of P20 and its mutants to 6-HB and their antiviral activity against HIV-1 NL4-3 (A), IIIB (B), and Bal (C). The relative binding activities of P20 and its mutants (x axis) were plotted against their IC50s for inhibiting HIV-1 infection (y axis).
DISCUSSION
Recently, a number of human proteins that are required for HIV infection have been identified through genomewide screenings (2, 27, 56). Some human proteins, such as apolipoprotein B mRNA-editing enzyme 3G (APOBEC3G) and tripartite motif protein 5 alpha (Trim5α), serve as restriction factors targeting HIV replication (12, 48). The sequences in these proteins responsible for binding with the HIV-1 molecules, e.g., gp41, may be used for designing novel anti-HIV therapeutics.
Using a yeast two-hybrid system with rsgp41e as the bait to screen a human bone marrow cDNA library, we have identified a novel gp41 core-binding molecule, P20, which is effective in blocking HIV-1 Env-mediated cell fusion and inhibiting infection by a broad spectrum of HIV-1 strains with distinct subtypes and coreceptor tropism, but it exhibits no inhibitory activity against other enveloped viruses, such as VSV and influenza virus. P20 peptide has a sequence partly homologous to human TNNI3K-like protein or some other unknown human protein, although the potential interaction of this human protein with HIV-1 gp41 has not been defined yet. In addition to its low molecular mass, P20 may have advantages for drug development over the HIV-1 gp41-based HIV-1 fusion inhibitors like T-20, since it may not induce strong antibody responses when it is used in humans, thus avoiding rapid clearance by specific human antibodies. Furthermore, it was highly soluble for easy expression and purification via the prokaryotic expression system.
Unlike the current HIV fusion inhibitors derived from the HIV-1 gp41 NHR and CHR domains, such as T-20, which target the corresponding CHR and NHR regions, respectively, P20 may inhibit HIV entry by targeting the gp41 core conformation, since it bound neither the N peptide N36 nor the C peptide C34 but interacted with the 6-HB core formed by N36 and C34. Furthermore, P20 could not block 6-HB formation, while most of the N and C peptides are effective in inhibiting the formation of the gp41 6-HB core. It was reported that some gp41 6-HB-specfic human MAbs (167-D and 1281), mouse MAbs (9F2 and 2G8), and some protein inhibitors also could inhibit HIV-1 Env-mediated cell-cell fusion at the suboptimal temperature 31.5 or 37°C (13, 14, 17, 19, 30, 57). In contrast, the human MAbs that recognize the cluster I epitopes in gp41, such as 240-D and 181-D (54), did not block HIV-1 Env-mediated membrane fusion at both 31.5 and 37°C (13). Most recently, Roymans et al. (45) demonstrated that TMC353121, a potent respiratory syncytial virus (RSV) fusion inhibitor, bound with the amino acid residues in both NHR and CHR domains, resulting in the stabilization of an alternate 6-HB conformation and causing a local disturbance of the natural 6-HB conformation rather than completely blocking 6-HB formation. These findings provide clues for unraveling the mechanism of action of P20 and other 6-HB-binding molecules in inhibiting viral fusion.
Thus far, HIV Env-mediated membrane fusion is a largely unknown process, especially whether or not membranes fuse immediately after 6-HB formation. Miyauchi et al. provided compelling evidence to show that HIV enters cells primarily by endocytosis (39), which proves that the virus and target cell membranes do not start to fuse immediately after gp41 changes conformation. Instead, a series of complex molecular events, which facilitate viral entry by endocytosis or other means, occur during this short period. Therefore, it is possible that cell fusion takes place inside the target cell only moments later. Our previous study demonstrated that gp41 6-HB could interact with epsin (18), which is an essential accessory factor of endocytosis and docks to the plasma membrane by interacting with the lipid. Here, we found that P20 interacted with the gp41 6-HB and blocked HIV-1-mediated membrane fusion. It bound neither the effector cells nor the target cells, but it interacted with the syncytium formed by the fusion between the effector and target cells (Fig. 5).
To dissect the specific role of each amino acid in P20, we have designed 11 mutants and tested their biological and biochemical functions. Interestingly, the anti-HIV-1 activity of P20 and its mutants is well correlated with their binding affinities to the gp41 6-HB (Fig. 8). Mutations of the first three amino acids both increase the binding ability of P20 to the 6-HB and its antiviral efficacy, while the mutations in P20-E, P20-G, P20-H, and P20-I peptides, which share the motif WGRLEGRRT, show a significant decrease of gp41 6-HB-binding affinity and antiviral efficacy. This suggests that these amino acids compose the active site of P20 that plays a vital role in the inhibition of viral fusion. Further research should focus on these important sites to further optimize the antiviral efficacy of P20.
T-20 is a first-generation fusion inhibitor antiviral drug that has been successfully used in the last few years. However, T-20 is increasingly found to be ineffective against resistant viruses. Thus, it is necessary to replace it with a next-generation drug with an improved resistance profile. Both P20 and P20-A inhibited the T-20-resistant strain, suggesting that their mechanism of action and target are different from those of T-20.
In summary, by yeast two-hybrid screening on a human bone marrow cDNA library using rsgp41e as the bait, we have successfully identified a novel HIV-1 fusion inhibitor, P20, which has sequence homologous to that of a human protein and is able to bind to the gp41 6-HB core. P20 can be further developed as a novel anti-HIV-1 therapeutic and used as a probe for studying the possible role of the gp41 6-HB core in the late stages of membrane fusion.
Acknowledgments
We thank Lanying Du for providing IAV HA pseudovirus and Hong Lu for the propagation and titration of primary HIV-1 isolates.
This work was supported by 973-2007CB914402 and 2008zx10001-007 to Y.-H.C. and an NIH grant (AI46221) to S.J.
Footnotes
Published ahead of print on 30 June 2010.
REFERENCES
- 1.Blumenthal, R., and D. S. Dimitrov. 2007. Targeting the sticky fingers of HIV-1. Cell 129:243-245. [DOI] [PubMed] [Google Scholar]
- 2.Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921-926. [DOI] [PubMed] [Google Scholar]
- 3.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 4.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 5.Chang, L. J., V. Urlacher, T. Iwakuma, Y. Cui, and J. Zucali. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 6:715-728. [DOI] [PubMed] [Google Scholar]
- 6.Chou, T. C., and P. Talalay. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22:27-55. [DOI] [PubMed] [Google Scholar]
- 7.D'Anna, J. A., R. A. Tobey, and L. R. Gurley. 1980. Concentration-dependent effects of sodium-butyrate in Chinese-hamster cells-cell-cycle progression, inner-histone acetylation, histone H-1 dephosphorylation, and induction of an H1-like protein. Biochemistry 19:2656-2671. [DOI] [PubMed] [Google Scholar]
- 8.Dimitrov, D. S. 1997. How do viruses enter cells? The HIV coreceptors teach us a lesson of complexity. Cell 91:721-730. [DOI] [PubMed] [Google Scholar]
- 9.Dwyer, J. J., K. L. Wilson, K. Martin, J. E. Seedorff, A. Hasan, R. J. Medinas, D. K. Davison, M. D. Feese, H. T. Richter, H. Kim, T. J. Matthews, and M. K. Delmedico. 2008. Design of an engineered N-terminal HIV-1 gp41 trimer with enhanced stability and potency. Protein Sci. 17:633-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggink, D., C. E. Baldwin, Y. Deng, J. P. Langedijk, M. Lu, R. W. Sanders, and B. Berkhout. 2008. Selection of T1249-resistant human immunodeficiency virus type 1 variants. J. Virol. 82:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geitmann, M., T. Unge, and U. H. Danielson. 2006. Interaction kinetic characterization of HIV-1 reverse transcriptase non-nucleoside inhibitor resistance. J. Med. Chem. 49:2375-2387. [DOI] [PubMed] [Google Scholar]
- 12.Goila-Gaur, R., and K. Strebel. 2008. HIV-I Vif, APOBEC, and intrinsic immunity. Retrovirology 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golding, H., M. Zaitseva, E. de Rosny, L. R. King, J. Manischewitz, I. Sidorov, M. K. Gorny, S. Zolla-Pazner, D. S. Dimitrov, and C. D. Weiss. 2002. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J. Virol. 76:6780-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorny, M. K., and S. Zolla-Pazner. 2000. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 74:6186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg, M. L., and N. Cammack. 2004. Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 54:333-340. [DOI] [PubMed] [Google Scholar]
- 16.Guo, Y., E. Rumschlag-Booms, J. Wang, H. Xiao, J. Yu, L. Guo, G. F. Gao, Y. Cao, M. Caffrey, and L. Rong. 2009. Analysis of hemagglutinin-mediated entry tropism of H5N1 avian influenza. Virol. J. 6:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, J. H., Z. Q. Liu, S. Liu, S. Jiang, and Y. H. Chen. 2006. Identification of the HIV-1 gp41 core-binding motif-HXXNPF. FEBS Lett. 580:4807-4814. [DOI] [PubMed] [Google Scholar]
- 18.Huang, J. H., Z. Qi, F. Wu, L. Kotula, S. B. Jiang, and Y. H. Chen. 2008. Interaction of HIV-1 gp41 core with NPF motif in Epsin-implication in endocytosis of HIV. J. Biol. Chem. 283:14994-15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, J. H., H. W. Yang, S. Liu, J. Li, S. Jiang, and Y. H. Chen. 2007. The mechanism by which molecules containing the HIV gp41 core-binding motif HXXNPF inhibit HIV-1 envelope glycoprotein-mediated syncytium formation. Biochem. J. 403:565-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, S., and A. K. Debnath. 2000. Development of HIV entry inhibitors targeted to the coiled-coil regions of gp41. Biochem. Biophys. Res. Commun. 269:641-646. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. Inhibition of HIV-1 infection by a fusion domain binding peptide from the HIV-1 envelope glycoprotein GP41. Biochem. Biophys. Res. Commun. 195:533-538. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, S., K. Lin, L. Zhang, and A. K. Debnath. 1999. A screening assay for antiviral compounds targeted to the HIV-1 gp41 core structure using a conformation-specific monoclonal antibody. J. Virol. Methods 80:85-96. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, S. B., K. Lin, and A. R. Neurath. 1991. Enhancement of human immunodeficiency virus type 1 infection by antisera to peptides from the envelope glycoproteins gp120/gp41. J. Exp. Med. 174:1557-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilby, J. M., and J. J. Eron. 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 348:2228-2238. [DOI] [PubMed] [Google Scholar]
- 27.König, R., Y. Y. Zhou, D. Elleder, T. L. Diamond, G. M. C. Bonamy, J. T. Irelan, C. Y. Chiang, B. P. Tu, P. D. De Jesus, C. E. Lilley, S. Seidel, A. M. Opaluch, J. S. Caldwell, M. D. Weitzman, K. L. Kuhen, S. Bandyopadhyay, T. Ideker, A. P. Orth, L. J. Miraglia, F. D. Bushman, J. A. Young, and S. K. Chanda. 2008. Global analysis of host-pathogen interactions that regulate early stage HIV-1 replication. Cell 135:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labrosse, B., L. Morand-Joubert, A. Goubard, S. Rochas, J. L. Labernardiere, J. Pacanowski, J. L. Meynard, A. J. Hance, F. Clavel, and F. Mammano. 2006. Role of the envelope genetic context in the development of enfuvirtide resistance in human immunodeficiency virus type 1-infected patients. J. Virol. 80:8807-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, B., and S. Fields. 1993. Identification of mutations in P53 that affect its binding to SV40 large T-antigen by using the yeast 2-hybrid system. FASEB J. 7:957-963. [DOI] [PubMed] [Google Scholar]
- 30.Li, J., X. Chen, J. Huang, S. Jiang, and Y. H. Chen. 2009. Identification of critical antibody-binding sites in the HIV-1 gp41 six-helix bundle core as potential targets for HIV-1 fusion inhibitors. Immunobiology 214:51-60. [DOI] [PubMed] [Google Scholar]
- 31.Li, L., L. He, S. Tan, X. Guo, H. Lu, Z. Qi, C. Pan, X. An, S. Jiang, and S. Liu. 2010. 3-Hydroxyphthalic anhydride-modified chicken ovalbumin exhibits potent and broad anti-HIV-1 activity: a potential microbicide for preventing sexual transmission of HIV-1. Antimicrob. Agents Chemother. 54:1700-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, S., S. Wu, and S. Jiang. 2007. HIV entry inhibitors targeting gp41: from polypeptides to small-molecule compounds. Curr. Pharm. Des. 13:143-162. [DOI] [PubMed] [Google Scholar]
- 33.Liu, S. W., H. Lu, J. Niu, Y. J. Xu, S. G. Wu, and S. B. Jiang. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 280:11259-11273. [DOI] [PubMed] [Google Scholar]
- 34.Liu, S. W., Q. Zhao, and S. B. Jiang. 2003. Determination of the HIV-1 gp41 fusogenic core conformation modeled by synthetic peptides: applicable for identification of HIV-1 fusion inhibitors. Peptides 24:1303-1313. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Z., and Y. H. Chen. 2004. Design and construction of a recombinant epitope-peptide gene as a universal epitope-vaccine strategy. J. Immunol. Methods 285:93-97. [DOI] [PubMed] [Google Scholar]
- 36.Lu, J., S. G. Deeks, R. Hoh, G. Beatty, B. A. Kuritzkes, J. N. Martin, and D. R. Kuritzkes. 2006. Rapid emergence of enfuvirtide resistance in HIV-1-infected patients: results of a clonal analysis. J. Acquir. Immune Defic. Syndr. 43:60-64. [DOI] [PubMed] [Google Scholar]
- 37.Lu, L., Y. Zhu, J. H. Huang, X. Chen, H. W. Yang, S. B. Jiang, and Y. H. Chen. 2008. Surface exposure of the HIV-1 Env cytoplasmic tail LLP2 domain during the membrane fusion process-interaction with gp41 fusion core. J. Biol. Chem. 283:16723-16731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 39.Miyauchi, K., Y. Kim, O. Latinovic, V. Morozov, and G. B. Melikyan. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137:433-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neurath, A. R., N. Strick, S. Jiang, Y. Y. Li, and A. K. Debnath. 2002. Anti-HIV-1 activity of cellulose acetate phthalate: synergy with soluble CD4 and induction of “dead-end” gp41 six-helix bundles. BMC Infect. Dis. 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiao, Z. S., M. Kim, B. Reinhold, D. Montefiori, J. H. Wang, and E. L. Reinherz. 2005. Design, expression, and immunogenicity of a soluble HIV trimeric envelope fragment adopting a prefusion gp41 configuration. J. Biol. Chem. 280:23138-23146. [DOI] [PubMed] [Google Scholar]
- 42.Rich, R. L., and D. G. Myszka. 2007. Survey of the year 2006 commercial optical biosensor literature. J. Mol. Recognit. 20:300-366. [DOI] [PubMed] [Google Scholar]
- 43.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 45.Roymans, D., H. L. De Bondt, E. Arnoult, P. Geluykens, T. Gevers, M. Van Ginderen, N. Verheyen, H. Kim, R. Willebrords, J. F. Bonfanti, W. Bruinzeel, M. D. Cummings, H. van Vlijmen, and K. Andries. 2010. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc. Natl. Acad. Sci. U. S. A. 107:308-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sista, P. R., T. Melby, D. Davison, L. Jin, S. Mosier, M. Mink, E. L. Nelson, R. DeMasi, N. Cammack, M. P. Salgo, T. J. Matthews, and M. L. Greenberg. 2004. Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. AIDS 18:1787-1794. [DOI] [PubMed] [Google Scholar]
- 47.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Towers, G. J. 2007. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, H. T., Z. Qi, A. G. Guo, Q. C. Mao, H. Lu, X. L. An, C. L. Xia, X. J. Li, A. K. Debnath, S. G. Wu, S. W. Liu, and S. B. Jiang. 2009. ADS-J1 inhibits human immunodeficiency virus type 1 entry by interacting with the gp41 pocket region and blocking fusion-active gp41 core formation. Antimicrob. Agents Chemother. 53:4987-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 52.Wild, C., T. Greenwell, and T. Matthews. 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retrovir. 9:1051-1053. [DOI] [PubMed] [Google Scholar]
- 53.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U. S. A. 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, J. Y., M. K. Gorny, T. Palker, S. Karwowska, and S. Zolla-Pazner. 1991. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 65:4832-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, L., S. Hue, S. Taylor, D. Ratcliffe, J. A. Workman, S. Jackson, P. A. Cane, and D. Pillay. 2002. Minimal variation in T-20 binding domain of different HIV-1 subtypes from antiretroviral-naive and -experienced patients. AIDS 16:1684-1686. [DOI] [PubMed] [Google Scholar]
- 56.Yeung, M. L., L. Houzet, V. S. R. K. Yedavalli, and K. T. Jeang. 2009. A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem. 284:19463-19473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, M. Y., V. Choudhry, I. A. Sidorov, V. Tenev, B. K. Vu, A. Choudhary, H. Lu, G. M. Stiegler, H. W. Katinger, S. Jiang, C. C. Broder, and D. S. Dimitrov. 2006. Selection of a novel gp41-specific HIV-1 neutralizing human antibody by competitive antigen panning. J. Immunol. Methods 317:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]