Abstract
The respiratory epithelium plays a central role in innate immunity by secreting networks of inflammatory mediators in response to respiratory syncytial virus (RSV) infection. Previous proteomic studies focusing on the host cellular response to RSV indicated the existence of a nuclear heat shock response and cytoplasmic depletion of antioxidant proteins in model type II-like airway epithelial cells. Here, we increased the depth of nuclear proteomic interrogation by using fluorescence difference labeling followed by liquid isoelectric focusing prefractionation/two-dimensional gel electrophoresis (2-DE) to identify an additional 41 proteins affected by RSV infection. Surprisingly, we found inducible oligomers and shifts in isoelectric points for peroxiredoxin 1 (Prdx-1), Prdx-3, and Prdx-4 isoforms without changes in their total abundance, indicating that Prdxs were being oxidized in response to RSV. To address the role of Prdx-1 and Prdx-4 in RSV infection, isoforms were selectively knocked down by small interfering RNA (siRNA) transfection. Cells lacking Prdx-1, Prdx-4, or both showed increased levels of reactive oxygen species formation and a higher level of protein carbonylation in response to RSV infection. Using a novel saturation fluorescence labeling 2-DE analysis, we showed that 15 unique proteins had enhanced oxidative modifications of at least >1.2-fold in the Prdx knockdowns in response to RSV, including annexin A2 and desmoplakin. Our results suggest that Prdx-1 and Prdx-4 are essential for preventing RSV-induced oxidative damage in a subset of nuclear intermediate filament and actin binding proteins in epithelial cells.
The paramyxovirus respiratory syncytial virus (RSV) is a significant human pathogen that accounts for 100,000 hospitalizations in the United States annually (26). In immunocompetent individuals, RSV replicates in the airway mucosa, where it may produce uncomplicated upper respiratory infection or spread distally to the lower airways, producing more severe lower respiratory tract infection (LRTI). Although RSV infection can lead to LRTI in any age group, childhood LRTI is associated with enhanced morbidity when it is etiologically linked to recurrent episodic wheezing and asthma (21). In fatal cases of LRTI, RSV produces epithelial eosinophilic inclusions, necrosis mucous plugging, and apoptotic crisis leading to airway obstruction (29).
The respiratory epithelium plays an important regulatory role in initiating and coordinating innate immune responses by virtue of its ability to synthesize and secrete defensins, antiviral interferons (IFNs), and potent inflammatory chemokines of the C, CC, and CXC classes (8, 28, 32). In LRTI, RSV-infected alveolar type II epithelial cells produce distinct quantitative and qualitative patterns of chemokine expression different from those produced by nasal and upper airway cells, indicating that host responses are cell type dependent and linked to disease pathogenesis (22). For these reasons, understanding the type II epithelial host genomic and proteomic cellular response to RSV infection has been intensively investigated. Previously, using a high-resolution two-dimensional gel electrophoresis (2-DE), we have shown that RSV infection induces changes in cellular protein expression and have identified 24 different proteins, some of which include nuclear heat shock response proteins, with induction of 70- and 60-kDa heat shock protein (hsp) isoforms and rearrangement of nuclear dot 10 (ND10) structures (3). Moreover, RSV replication induces an oxidative stress response in vitro (5) and in vivo (6, 13), a phenomenon that we have found is linked to activation of transcription factors important in the innate inflammatory response (5, 14, 20). Interestingly, RSV infection increases reactive oxygen species (ROS) production in epithelial cells by activating NADPH oxidase while downregulating cytoplasmic catalase and superoxide dismutase (13). In fact, antioxidant treatment significantly ameliorates RSV-induced clinical disease and pulmonary inflammation (6). Together, RSV-induced ROS produce profound proteomic and genomic changes in infected cells in a manner linked to disease severity.
Despite the significant understanding of the host cell genomic responses to RSV infection, the coordinated proteomic response has been incompletely characterized. To gain greater insight into the nuclear response to viral replication, we sought to increase the depth of proteomic coverage. Here, we report our experience using isoelectric focusing (IEF) as a prefractionation step by multicompartment electrolyzer (MCE) followed by fluorescence difference gel electrophoresis (DIGE). Using this approach, we identified 41 proteins differentially affected by RSV infection, including nuclear antioxidant peroxiredoxins (Prdxs). The biological functions of Prdxs were investigated by small interfering RNA (siRNA) knockdown, where enhanced ROS stress and perturbations in inflammatory signaling were observed. We further applied a novel proteomics labeling technique that covalently links free cysteine side chains with fluorescent dyes to systematically identify proteins sensitive to RSV-induced oxidation in the presence or absence of Prdx-1, Prdx-4, or both. Prdx-1 and Prdx-4 protect a subset of nuclear proteins, including Lam A, annexin, and centromere E-associated protein. Together, these studies indicate that Prdxs perform important nuclear antioxidant responses to virus-induced ROS stress, despite the concurrent depletion of cytoplasmic antioxidases.
MATERIALS AND METHODS
Cell culture.
Human A549 pulmonary epithelial cells (American Type Culture Collection, Manassas, VA) were grown in F12K medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a 5% CO2 incubator.
Virus preparation and infection.
The human RSV A2 strain was propagated in Hep2 cells and purified on a sucrose cushion gradient (11). A549 cells were infected with RSV at a multiplicity of infection (MOI) of 1.0 for the times indicated on the figures.
Preparation of cytoplasmic and nuclear extracts.
A549 cells were scraped from plates and washed in phosphate-buffered saline (PBS) twice by centrifugation. The cell pellet was suspended in cold hypotonic buffer A (50 mM HEPES [pH 7.9], 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF], and 10 μl/ml of protease inhibitor cocktail [Sigma-Aldrich Chemical Co., St. Louis, MO]) and 0.1% Igepal CA-630 and incubated on ice for 10 min (9). The lysates were centrifuged at 6,000 rpm (4,000 × g) for 30 s at 4°C. The supernatant (cytoplasmic fraction) was saved, and the pellet (containing the nuclei) was resuspended in buffer B (buffer A containing 1.0 M sucrose) and centrifuged at 12,000 rpm (15,000 × g) for 10 min at 4°C. The supernatant was discarded, and the pellet was incubated in buffer C (50 mM HEPES [pH 7.9], 10% glycerol, 400 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1 mM PMSF, and protease inhibitor cocktail) with frequent shaking for 30 min at 4°C. After centrifugation at 15,000 × g for 10 min at 4°C, the supernatant (nuclear extract) was saved at −80°C. The protein content of both cytoplasmic and nuclear extracts was measured by Coomassie brilliant blue staining using bovine serum albumin (BSA) as a standard (Bio-Rad, Hercules, CA). For whole nuclear extracts, the purified nuclei were lysed in reagent 2 (catalog no. 163-2103; Bio-Rad) and tributylphosphine (TBP; catalog no. 163-2101; Bio-Rad) and vortexed for 5 min at room temperature. Nucleic acid was removed by adding 300 IU/ml of endonuclease (Sigma-Aldrich), sonicated, and incubated for 30 min at 22°C. After spinning at 15,000 × g at 4°C for 30 min, the supernatant was desalted using a MicroSpin G-25 column (GE Healthcare, Piscataway, NJ) (9). The extract was then normalized for protein amounts by Coomassie brilliant blue staining (Bio-Rad).
siRNA-mediated peroxiredoxin knockdown.
Scrambled Prdx-1- or Prdx-4-directed siRNA pools were commercially obtained (ON-TARGETplus smart pool siRNAs [Dharmacon Research, Inc., Lafayette, CO]) and transfected into A549 cells using TransIT-siQuest transfection reagent (Mirus Bio Corp., Madison, WI) with 5 nM of the indicated siRNA at a density of 0.25 × 106 cells/well for 48 h. Downregulation was determined by Western immunoblotting.
2-D DIGE.
A549 nuclear extracts were prepared as described above using a modified buffer C (containing 50 mM HEPES [pH 7.9], 10% glycerol, 400 mM NH4HCO3, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1 mM PMSF, and protease inhibitor cocktail), followed by removal of 400 mM NH4HCO3 by evaporation until the volume reached 50 μl. Buffer A (450 μl) was added to the tube and evaporated to 50 μl. This process was repeated three times. The final desalted sample was dialyzed versus buffer A with 2 mM tri-2(2-carboxyethyl) phosphine (TCEP; Geno Technology Inc., St. Louis, MO). Approximately 150 μg nuclear protein from uninfected or RSV-infected cells were labeled at a 15:1 molar excess to thiol with either Cy3 or Cy5, respectively, for 30 min at 22°C in the dark, according to the manufacturer's recommendation. DTT was added to stop the reaction, and excess dye was removed by centrifugation through a G-25 desalting column. The labeled sample was reduced and alkylated, pooled, and subjected to IEF using an MCE (Invitrogen Corp., Carlsbad, CA). The sample was diluted with MCE buffer (6 M urea, 2 M thiourea, 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 100 mM DTT, and 1% ampholyte [pH 3 to 10]) to a final concentration of 0.6 mg/ml. Protein (670 μl) was loaded into chambers spanning pH ranges 3 to 5.4, 5.4 to 7, and 7 to 10 and focused using constant voltage (100 V) for 20 min followed by constant power (1 W) for 160 min. The proteins within each chamber were individually collected, and protein concentration was determined. The sample volume was reduced to 50 μl using a YM-10 Millipore spin column.
IEF/2-DE.
IEF of the DIGE samples fractionated by the MCE was performed with 11-cm-long precast immobilized pH gradient (IPG) strips (pH 3 to 5.6, 5 to 8, and 6 to 11) (GE Healthcare) as appropriate, and for other samples, pH 3 to 10 IPG strips were used (3). Two hundred microliters of 1-mg/ml protein samples were loaded onto an IPG strip and rehydrated overnight. IEF was performed at 20°C with the following parameters: 50 V for 11 h, 250 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, 8,000 V for 2 h, and 8,000 V for 6 h, with a total 48,000 V hour. After IEF, the IPG strips were stored at −80°C until 2-D SDS-PAGE was performed. For the 2-D SDS-PAGE, the IPG strips were incubated in 4 ml of equilibration buffer (6 M urea, 2% SDS, 50 mM Tris-HCl [pH 8.8], 20% glycerol) containing 10 μl of 0.5 M TCEP [Tris(2-carboxyethyl)phosphine] for 15 min at 22°C with shaking. The strips were then incubated in another 4 ml of equilibration buffer with 25 mg of iodoacetamide/ml for 15 min at 22°C with shaking. Electrophoresis was performed at 150 V for 2.25 h at 4°C with 8 to 16% precast polyacrylamide gels in Tris-glycine buffer (25 mM Tris-HCl, 192 mM glycine, 0.1% SDS [pH 8.3]). After 2-DE, the gels were fixed (10% methanol [MeOH], 7% acetic acid in double-distilled water [ddH2O]), stained with Sypro ruby (Bio-Rad), and destained (10% ethanol in ddH2O).
Gel imaging.
The destained gels were scanned at a 100-μm resolution using the ProXPRESS 2-D proteomic imaging system (Perkin-Elmer, Boston, MA) with 480-nm excitation and 620-nm emission filters for Sypro ruby and 480-nm excitation and 530-nm emission filters for BODIPY [4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene N-(2-aminoethyl)maleimide] staining. The exposure time was adjusted to achieve a value of 55,000- to 63,000-pixel intensity on the most intense protein spots on the gel. The 2-D gel images were subsequently analyzed using Progenesis discovery software version 2003.03 (Nonlinear Dynamics, Ltd., Newcastle Upon Tyne, United Kingdom). Eight gels stained with BODIPY were analyzed, two from each of the following groups: group 1, scrambled siRNA; group 2, Prdx-1 siRNA; group 3, Prdx-4 siRNA; and group 4, Prdx-1 and Prdx-4 siRNA with or without RSV infection. The software automatically selected the gel with the highest number of spots (1 out of the 8 gels) as the reference gel. The spot volumes were normalized based on total spot volume for each gel, and the control and RSV-infected nuclei were compared. A fold change greater than ±2.0 in the normalized spot volume was considered significantly changed; these spots were subsequently robotically picked and trypsin digested, and peptide masses were identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF)/TOF.
Protein identification and pathway analyses.
Protein gel spots were excised and prepared for MALDI-TOF mass spectrometry (MS) analysis using DigiLab's (Holliston, MO) ProPic and ProPrep robotic instruments by following the manufacturer's protocol. Briefly, gel pieces were incubated with trypsin (20 μg/ml in 25 mM ammonium bicarbonate, pH 8.0) (Promega Corp., Madison, WI) at 37°C for 4 h. MALDI-TOF/TOF was performed using an Applied Biosystems Voyager model DE STR for peptide mass fingerprinting and sequencing. Protein identification was performed using a Bayesian algorithm (31), in which high-probability matches are indicated by an expectation score, an estimate of the number of matches that would be expected in that database if the matches were completely random. Biological networks were inferred by pathway analyses using the Ingenuity knowledge base database (Ingenuity Systems).
Detection of oxidized proteins.
Changes in oxidized protein levels were determined by using an OxyBlot kit (Chemicon/Millipore, Temecula, CA) according to the manufacturer's recommendations and protocols published previously (1). Briefly, proteins were derivatized with 4-dinitrophenylhydrazine (DNPH) for 15 min, followed by incubation at 22°C with neutralization buffer (Chemicon/Millipore, Temecula, CA). DNPH-derivatized proteins were electrophoresed on a 10% SDS-polyacrylamide gel and blotted on Hybond polyvinylidene difluoride (PVDF) membranes (GE Healthcare). Blots were blocked with 5% nonfat dry milk (blocking buffer) in Dulbecco's phosphate-buffered saline containing 0.05% Tween 20 (PBS-T) for 3 h and incubated with rabbit anti-DNP (2,4-dinitrophenol) primary antibody (Ab) (1:150) overnight at 4°C. After three washes with PBS-T, membranes were incubated for 1 h at room temperature with horseradish peroxidase-conjugated mouse anti-rabbit secondary antibody (GE Healthcare) dissolved in blocking buffer (1:300). Immune complexes were visualized by enhanced chemiluminescence (ECL; GE Healthcare) according to the manufacturer's instructions.
Saturation fluorescence labeling (FL) for detection of cysteinyl-oxidized proteins.
A549 nuclear protein was labeled with BODIPY (Invitrogen) before IEF/2-DE according to our previously published protocol (23). To label proteins, 150 to 200 μg protein were denatured in 200 μl of reaction buffer (50 mM Tris-HCl [pH 7.5], 7 M urea, 2 M thiourea, 2% CHAPS) and incubated with 60-fold excess BODIPY over protein thiols for 2 h at 22°C. The reaction was stopped with the addition of 2-mercaptoethanol (2-ME; 150-fold molar excess over dye) to the mixture and incubated for 30 min at 22°C.
Assessment of cellular redox state.
Changes in intracellular ROS levels were determined by 2′,7′-dichlorodihydro-fluorescein diacetate (H2DCF-DA; Invitrogen) as we described previously (15). Briefly, cells at ∼70% confluence were infected with RSV (MOI, 1.0) or mock infected. At 0, 1, 2, 4, 6, 9, 12, 15, 18, and 24 h, mock- and RSV-infected cells were loaded with 50 μM (final concentration) H2DCF-DA for 15 min at 37°C. Changes in dichlorofluorescein (DCF) fluorescence intensities were determined with an Flx-800 microplate fluorescence reader (BioTek Instruments, Inc., Winooski, VT) at excitation (485 nm) and emission (528 nm) wavelengths. Each data point represents the mean fluorescence for 12,000 cells from three or more independent experiments, and data are expressed as ±standard errors of the mean (SEM). In control experiments, we show that cellular (RSV-infected or control) uptake of H2DCF-DA reached a plateau at about 15 min. At 1 h postloading of 0.4% DCF, the oxidized form of H2DCF released in the medium was measured in a manner similar to that described previously (2).
Western blot.
Proteins were fractionated by 12% or 8 to 16% gradient SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane by electroblotting. Membranes were blocked in 5% nonfat dry milk in Tris-buffered saline-0.1% Tween (TBS-T) and probed with the primary Ab indicated in the figure legends. Membranes were washed and incubated with IRDye 700-conjugated anti-mouse Ab or IRDye 800-conjugated anti-rabbit Ab (Rockland Inc., Gilbertsville, PA). In some cases, membranes were probed with rabbit anti-goat IgG Ab conjugated with horseradish peroxidase. Finally, membranes were washed three times with TBS-T and imaged by an Odyssey infrared scanner or subjected to chemiluminescence reaction and exposed to X-ray film (Kodak, Rochester, NY). Sources of primary Ab were anti-Prdx-1 and anti-Prdx-4 polyclonal Abs (Abcam, Cambridge, MA), anti-α-tubulin Ab (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and anti-Lam B Ab (Calbiochem, San Diego, CA).
RESULTS
Prefractionation using membrane isoelectric focusing to extend the depth of proteome interrogation.
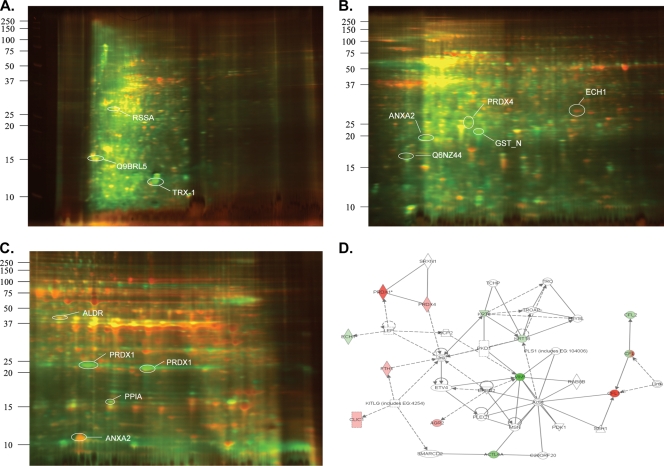
The transformed human A549 epithelial cell grows as a polarized monolayer that retains alveolar epithelial cell-like morphology, expressing surfactant-containing lamellar bodies, and shows inducible genomic responses similar to those of primary airway epithelial cells and is therefore used as a model type II alveolar cell in investigation of cellular responses to RSV infection (3, 10, 11). Previously, we investigated RSV-induced changes in the nuclear proteome of A549 cells by quantitative 2-DE, identifying 24 differentially expressed proteins and defining a nuclear heat shock response and ND10 reorganization (3). This work was further extended using high-pressure liquid chromatography (HPLC) prefractionation/2-DE of uninfected nuclear proteins from uninfected cells which significantly extended the depth of proteome coverage (9); however, HPLC fractionation resulted in significant adsorptive protein loss. To circumvent this problem, we applied a liquid IEF step using an efficient multicompartment analyzer (MCE; Invitrogen Corp.) following CyDye labeling for difference gel electrophoresis for more reliable detection of differentially expressed proteins. After MCE focusing in three different pH ranges, 3 to 5.4, 5.4 to 7, and 7 to 10, the protein pools were separated by 2-DE with three different pH ranges, from pH 3 to 5.6 (Fig. 1 A), pH 5 to 8 (Fig. 1B), and pH 6 to 11 (Fig. 1C). Comparison of normalized spot volumes of the two dyed samples identified spot volumes that were increased or decreased following RSV infection (Fig. 1A). Approximately 450 spots were detected in each gel, and differentially expressed proteins (<−2.0 or >2.0) were selected for protein identification by MS, resulting in 11, 17, and 18 protein identifications in gels 1 to 3, shown in Fig. 1A, 1B, and 1C, respectively.
FIG. 1.
RSV-induced changes of nuclear proteins of A549 cells by 2-D DIGE. Dual channel overlay of representative gel images in the pH ranges of 3.0 to 5.4 (A), 5.4 to 7.0 (B), and 7.0 to 10.0 (C). Uninfected and RSV-infected nuclear extracts were labeled with Cy3 (green) and Cy5 (red), respectively. For clarity, only a few representative spots are illustrated and found in Table 1. (D) Graphical representation of a pathway containing the largest number of focus proteins. Each node represents individual proteins, and relationships are represented by lines. Nodes colored red indicate a protein differentially upregulated by RSV, while nodes colored green indicate a downregulated protein. Uncolored nodes are inferred. Squares indicate cytokines, circles indicate chemokines, and ovals indicate transcription factors. Lines terminating with an arrow indicate “acts on.” Act6A, actin binding protein; CFL, cofilin; ECH, enoyl coenzyme A hydratase; KRT, keratin; Jnk, Jun N-terminal kinase; Prdx, peroxiredoxin; VMT, vimentin.
Identification of differentially expressed proteins.
Proteins identified by tryptic peptide mass fingerprinting are summarized in Table 1 and organized by gel number. Of the 11 proteins identified in gel 1, thioredoxin, ribosomal protein SA, and calmodulin 3 were reliably identified and increased in abundance following RSV infection. Conversely, 8 proteins, including heat shock protein 70, keratin 8, and vimentin, were decreased in abundance by RSV infection. In gels 2 and 3, we observed that RSV induced three isoforms of Prdx, Prdx-1, Prdx-2, and Prdx-3, which were not detected by our 2-DE analysis previously (3). Specifically, Prdx-1 and Prdx-4 isoforms were induced 5.3- and 2.6-fold, respectively, by RSV infection (Table 1). This finding was surprising, because RSV was shown to inhibit expression of cytoplasmic antioxidant proteins SOD-1, SOD-3, catalase, and glutathione S-transferase (13).
TABLE 1.
Classification of differentially expressed proteins in RSV-infected A549 nucleia
| Gel no. | Protein name | Common name | Accession no. | MM (Da) | pI | F/C | Expectation score |
|---|---|---|---|---|---|---|---|
| 1 | Vimentin | VME | P08670 | 52,406 | 4.99 | −17.43 | 2.40E−27 |
| Heat shock 70-kDa protein 5 | GRP78 | EAW87621* | 51,140 | 4.91 | −5.36 | 2.40E−56 | |
| Laminin-binding protein | RSSA | P08865 | 14,104 | 7.03 | −5.31 | 3.00E−19 | |
| Keratin 8 | K2C8 | P05787 | 53,671 | 5.52 | −5.10 | 6.10E−73 | |
| Vimentin | VME | P08670 | 52,406 | 4.99 | −4.95 | 7.60E−68 | |
| Vimentin | VME | P08670 | 53,653 | 5.06 | −4.40 | 1.20E−60 | |
| Heat shock 70-kDa protein 5 | GRP78 | P11021 | 51,140 | 4.91 | −4.24 | 1.50E−22 | |
| Thioredoxin | TRX-1 | Q09433 | 11,746 | 4.82 | 7.47 | 1.20E−05 | |
| Thioredoxin | TRX-1 | Q09433 | 11,746 | 4.82 | 5.13 | 2.40E−13 | |
| Ribosomal protein SA | RSSA | EAW64587* | 19,625 | 8.37 | 2.36 | 9.60E−08 | |
| Calmodulin 3 | Q9BRL5 | Q9BRL5 | 16,496 | 4.33 | 2.19 | 2.70E−05 | |
| 2 | Actin-like protein 6A | ACL6A | O96019 | 47,350 | 5.48 | −9.43 | 1.90E−15 |
| Stress-70 protein, mitochondrial | GRP75 | P38646 | 73,734 | 5.97 | −6.15 | 2.40E−50 | |
| Serum album | Q56G89 | Q56G89 | 69,039 | 5.85 | −5.80 | 1.50E−06 | |
| Aldehyde dehydrogenase 1 | AL1A1 | P00352 | 54,800 | 6.30 | −4.57 | 1.90E−63 | |
| Keratin 18 variant | K1C18 | P05783 | 47,957 | 5.40 | −4.50 | 1.20E−20 | |
| Enoyl coenzyme A hydratase 1 | ECH1 | Q13011 | 35,735 | 8.47 | −4.34 | 3.00E−37 | |
| Actin-related protein 3 | ARP3 | P61158 | 47,341 | 5.61 | −4.18 | 9.60E−22 | |
| Stress-70 protein, mitochondrial | HSPA9 | P38646 | 73,734 | 5.97 | −4.04 | 7.60E−47 | |
| Albumin | ALBU | P02768 | 65,993 | 5.69 | −3.73 | 1.20E−05 | |
| Peroxiredoxin 3 | Prdx3 | P30048 | 27,705 | 7.11 | 6.68 | 2.40E−22 | |
| Glutathione S-transferase | GST_N | 2PGTA* | 23,327 | 5.43 | 4.19 | 4.80E−35 | |
| Tubulin, alpha, isoform 10 | TUB_A | XP_001107503* | 46,284 | 4.96 | 3.82 | 4.80E−17 | |
| Glutathione S-transferase | GST_N | 2PGTA* | 23,327 | 5.43 | 2.72 | 3.00E−20 | |
| Peroxiredoxin 4 | Prdx-4 | Q13162 | 30,571 | 5.86 | 2.66 | 3.00E−21 | |
| Chloride intracellular channel 1 (nuclear) | CLIC1 | O00299 | 26,906 | 5.09 | 2.36 | 1.20E−15 | |
| Ferritin heavy subunit | Q6NZ44 | Q6NZ44 | 21,228 | 5.12 | 2.30 | 2.10E−03 | |
| Annexin A2 | ANXA2 | P07355 | 40,503 | 8.41 | 2.29 | 1.50E−15 | |
| 3 | Phosphoglycerate kinase 1 | PGK | EAW98602* | 28,434 | 9.21 | −9.11 | 6.10E−07 |
| Septin 7 | SEPT_7 | Q16181 | 48,628 | 8.76 | −7.40 | 3.00E−07 | |
| Cofilin 2 | COF2 | Q9Y281 | 18,725 | 7.66 | −5.91 | 7.60E−10 | |
| Enolase 1 variant | ENOA | P06733 | 47,111 | 7.01 | −5.38 | 9.60E−51 | |
| Voltage-dependent anion channel 1 | VDAC1 | P21796 | 30,754 | 8.62 | −5.35 | 1.20E−35 | |
| Aldehyde dehydrogenase 1 | AL1A1 | P00352 | 54,800 | 6.30 | −4.79 | 4.80E−26 | |
| Cleavage and polyadenylation factor | CPSF5 | O43809 | 26,153 | 9.06 | −4.30 | 6.10E−47 | |
| Cofilin 1 actin modulating protein | COF1 | P23528 | 18,491 | 8.22 | −3.15 | 4.80E−33 | |
| UDP-glucose dehydrogenase | UDPG | XP_001142450* | 47,572 | 5.64 | −3.18 | 3.80E−53 | |
| Ligand of annexin II | S100a1 | 1A4PA* | 11,065 | 7.30 | 7.06 | 3.00E−20 | |
| Cofilin 1 | COF1 | XP_866236* | 16,801 | 8.54 | 6.50 | 1.50E−12 | |
| hCG2017792 | CRA_B | EAW8645* | 32,661 | 8.50 | 6.45 | 2.40E−02 | |
| Peroxiredoxin 1 | Prdx-1 | Q06830 | 18,964 | 6.41 | 5.32 | 2.40E−50 | |
| Anterior gradient 2 homolog | Q53G64 | Q53G64 | 19,951 | 9.03 | 2.86 | 1.20E−36 | |
| Peroxiredoxin 1 | Prdx-1 | Q06830 | 18,964 | 6.41 | 2.59 | 6.10E−40 | |
| Cyclophilin A | PPIA | 5CYH_A* | 17,870 | 7.82 | 2.12 | 7.60E−39 | |
| Annexin A2 | ANXA2 | P07355 | 38,594 | 7.57 | 5.92 | 1.50E−07 | |
| Aldose reductase | ALDR | P15121 | 35,645 | 6.56 | 2.45 | 6.10E−17 |
Shown are high-probability identifications from MCE prefractionation DIGE 2-DE. For each protein, the gel used for identification, common name, Swiss Prot accession number, molecular mass (MM), isoelectric point (pI), fold change relative to uninfected cells (F/C), and expectation score are given. The expectation score is the number of matches expected if the matches were completely random. An expectation value of 1Ε−3 means the chance was 1 in 1,000 that the match was random. *, Swiss Prot ID not available; GenBank number provided for identification.
Network analysis of differentially expressed nuclear proteins.
To identify meaningful biological processes affected by the differentially expressed proteins, we subjected them to network analysis by the Ingenuity knowledge base. This analysis compares proteins in the input group to a curated database of publicly available pathways and relationships and displays a rank-ordered list of networks whose activities are most likely to be affected. The top-ranked network, its members, and their relationships (functional and physical) are displayed graphically in Fig. 1D. This network showed Prdx-1 and Prdx-4 isoforms acting on Jun N-terminal kinase and a cytoskeletal complex of actin, keratin, and vimentin proteins. Because of their important role in redox regulation, we were interested in further validating, characterizing, and determining the biological role of Prdxs in RSV-infected epithelial cells.
Validation of the peroxiredoxin upregulation in nuclear extracts.
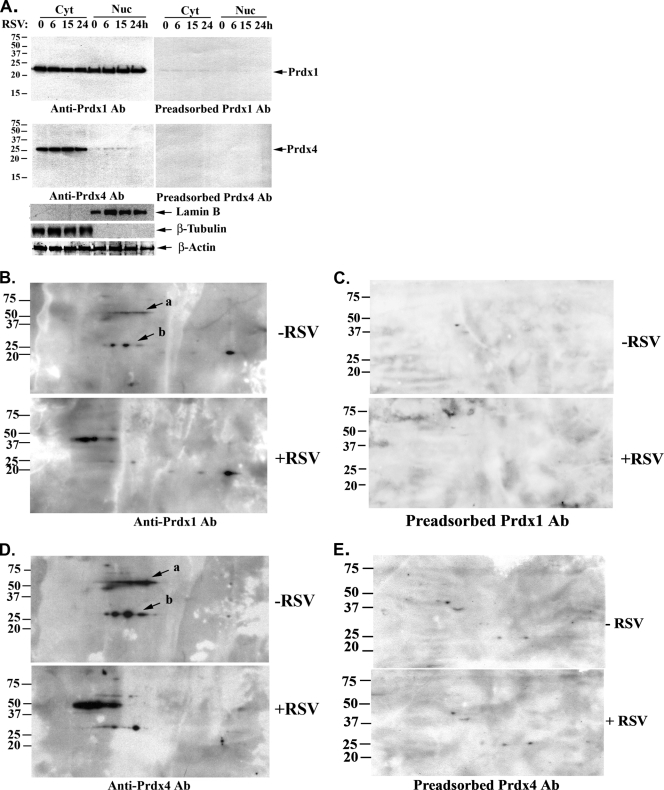
Validation of the RSV effect on nuclear Prdx expression was performed by Western immunoblotting of cytoplasmic and nuclear extracts from a time series of RSV-infected A549 cells fractionated by one-dimensional (1-D) SDS-PAGE (1-DE) (Fig. 2A, top). Under these conditions, specific Prdx-1 staining was seen as a single ∼24-kDa band that was abolished by peptide preadsorption of the anti-Prdx-1 Ab (Fig. 2A, top right). We observed that specific Prdx-1 staining was equally distributed in both cytoplasmic and nuclear fractions, and its total abundance was not affected by RSV infection. In contrast, specific Prdx-4 staining was detected as a 27-kDa band that was abolished by peptide preadsorption (Fig. 2A, middle right). We noted that Prdx-4 was predominately cytoplasmic, but RSV infection also did not change its expression (Fig. 2A, middle panel). These findings indicated that despite the appearance of Prdx-1 and Prdx-4 in the 2-DE DIGE, the total abundance of these proteins was not affected by RSV infection.
FIG. 2.
Validation of peroxiredoxin (Prdx) 1 and Prdx-4 expression in A549 cells by Western immunoblotting. (A) Effect of RSV infection on cytoplasmic-nuclear localization of Prdx-1 and Prdx-4 in A549 cells by Western blotting. Cells were infected with RSV for 0 to 24 h, and 50 μg of cytoplasmic (Cyt) or nuclear (Nuc) extracts were separated on a 12% SDS-PAGE gel and transferred to PVDF membranes. Membranes were probed with either Prdx-1 antibody (anti-Prdx-1 Ab) or Prdx-4 Ab (anti-Prdx-4 Ab) (left) or immunogen peptide-preadsorbed Prdx-1 and Prdx-4 Abs (right). After the membranes were stripped, the membranes were probed with either anti-Lam B, anti-β-tubulin, or anti-β-actin Abs. Molecular weight markers are shown on the left. (B) Induction of Prdx-1 by RSV infection. Nuclear extracts (150 μg) from uninfected (−RSV) or RSV-infected (+RSV) A549 cells were separated on a 2-D gel and transferred to PVDF membranes. Membranes were probed with Prdx-1 Ab (anti-Prdx-1 Ab). Arrows labeled “a” and “b” indicate dimeric and monomeric forms of Prdx-1, respectively. Molecular weight markers are shown on the left. (C) Prdx-1 Ab specificity. Identical amounts of proteins were separated by 2-DE and transferred to Immobilon membranes. Prdx-1 Ab was preadsorbed by mixing with blocking peptides before exposure to membranes. (D) RSV-induced changes of Prdx-4 by 2-DE Western blotting. Prdx-4 was detected by Western blotting using Prdx-4 Ab (anti-Prdx-4 Ab). The arrow labeled “a” designates dimer; the arrow labeled “b” designates the monomer of Prdx-4. (E) Prdx-4 Ab was preadsorbed prior to blotting, as described for panel C.
Prdx proteins form covalent oligomeric adducts with oxidized sulfenic-containing proteins to recycle them to their reduced cysteine state. These oligomeric structures would be lost by the typical 1-DE SDS-PAGE conditions, where proteins are denatured, reduced, and heated before electrophoresis. To determine if posttranslationally modified Prdx isoforms were produced by RSV infection, 2-DE Western immunoblot assays were performed on control and RSV-infected A549 nuclear extracts. Here, multiple Prdx-1 isoforms were detected, and their levels were dramatically increased by RSV infection (Fig. 2B). Interestingly, a pair of Prdx-1 spots (∼25 kDa), present in uninfected cells at high levels, were reduced in intensity and shifted in apparent pI. Concomitantly, the levels of an ∼50-kDa Prdx-1 isoform, whose levels were low in uninfected cells, increased sharply upon RSV infection (Fig. 2B). The identity of these isoforms was verified by peptide immunoadsorption (Fig. 2C). Similarly, Prdx-4 was detected in multiple forms by 2-DE Western blots, and their pI and apparent sizes changed upon RSV infection in a similar manner (Fig. 2D). In an overexposed 1-DE Western blot, we observed multiple forms of Prdx-1 and Prdx-4 isoforms (not shown). Together, these data indicate that although Prdx-1 and Prdx-4 isoforms were detected by MCE-DIGE as differentially expressed, these spots corresponded to oligomeric Prdx isoforms and do not reflect a change in the steady-state levels of Prdx-1 or Prdx-4. These data suggest that Prdx-1 and Prdx-4 may participate in protecting nuclear proteins against RSV-induced cysteinyl oxidation. Prdxs are also known to modulate NF-κB signaling in response to oxidative stress (12). We therefore sought to determine the role of Prdxs in signaling and protection against nuclear protein oxidation.
Prdx-1 and Prdx-4 modulate RSV-induced NF-κB signaling.
RSV-induced ROS is an important second messenger signaling in controlling NF-κB activation via a pathway converging on an activating phosphorylation of RelA at serine 276 (14, 15). To determine whether Prdx-1 or Prdx-4 modulated NF-κB signaling in response to RSV infection, endogenous Prdx-1 and Prdx-4 were downregulated by siRNA transfection. We noted >70% reduction of Prdx-1 and >90% reduction of Prdx-4 abundance measured by Western immunoblotting (Fig. 3A). We next examined the effect of Prdx silencing on RSV-induced expression of three NF-κB-dependent genes, interleukin-6 (IL-6), IL-8, and Gro-β, in A549 cells after RSV infection (Fig. 3B). Our results showed that neither Prdx-1 nor Prdx-4 knockdown affects basal or RSV-inducible NF-κB-dependent gene expression. However, in cells where both Prdx-1 and Prdx-4 were simultaneously downregulated, there was an increase in the basal expression of all three genes without RSV. Upon RSV infection, expression of all three genes was inhibited significantly, indicating that in the absence of both antioxidant enzymes, ROS negatively affect NF-κB-dependent gene expression.
FIG. 3.
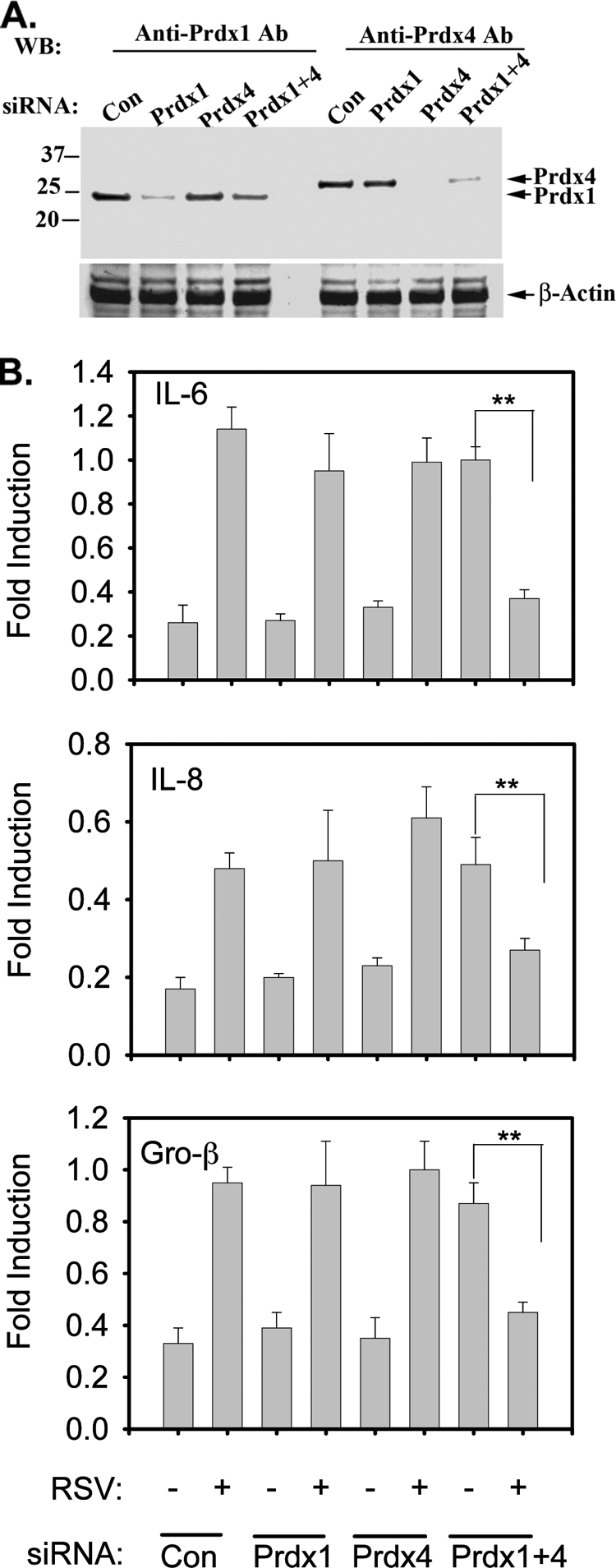
siRNA-mediated knockdown of Prdx-1 and Prdx-4 in RSV-treated A549 cells. Cells were transfected with 100 nM scrambled (Con), Prdx-1, Prdx-4, or Prdx-1 plus Prdx-4 siRNA. (A) After 72 h, cells were harvested and Prdx-1 and Prdx-4 were detected in whole-cell extract (WCE) by Western immunoblotting. Τhe same membrane was probed with β-actin Ab as a loading control. (B) A549 cells were transfected with either scrambled, Prdx-1, Prdx-4, or Prdx-1 plus Prdx-4 siRNA for 72 h, followed by no infection or RSV infection for 15 h. Total RNA was extracted and subjected to quantitative reverse transcription-PCR (qRT-PCR) for IL-6, Gro-β, and IL-8. Data are normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and expressed as fold changes relative to the uninfected control. Each bar is the mean ± standard deviation from triplicate determinations. **, P < 0.001.
Prdx-1 and Prdx-4 reduce virus-inducible ROS.
Peroxiredoxins efficiently catalyze the breakdown of H2O2 and alkyl peroxides to eliminate intracellular ROS produced in response to epidermal growth factor and tumor necrosis factor alpha (TNF-α) stimulation (17). We therefore examined the effect of Prdx-1 and Prdx-4 knockdown on the RSV-induced intracellular ROS in A549 cells. For this purpose, RSV-induced intracellular ROS production was measured by DCF-DA oxidation in cells transfected with scrambled or Prdx isoform-specific siRNA (15). As a positive control, H2O2 (100 μM) treatment resulted in ROS production 4- to 5-fold over mock treatment (Fig. 4A). Compared to scrambled siRNA treatment, there was a slight increase in basal ROS production in Prdx-1 and Prdx-4 siRNA-transfected cells. RSV infection alone induced ROS production at the same magnitude as H2O2. However, in the Prdx-1 or Prdx-4 siRNA transfectants, RSV infection induced a much higher level of DCF-DA oxidation than that produced by RSV alone. The oxidation was further increased by RSV in cells where both Prdx-1 and Prdx-4 were downregulated (Fig. 4A). To confirm this finding independently, we measured the effect of RSV infection on protein carbonylation in Prdx-1 or Prdx-4 siRNA-transfected A549 cells by Western immunoblotting. Compared to the effect of exogenous H2O2, RSV infection dramatically increased protein carbonylation (Fig. 4B), and the magnitude of protein carbonylation was further increased in both Prdx-1 and Prdx-4 knockdowns. These data indicate that Prdx-1 and Prdx-4 both contribute to cellular antioxidant defense in response to RSV infection.
FIG. 4.
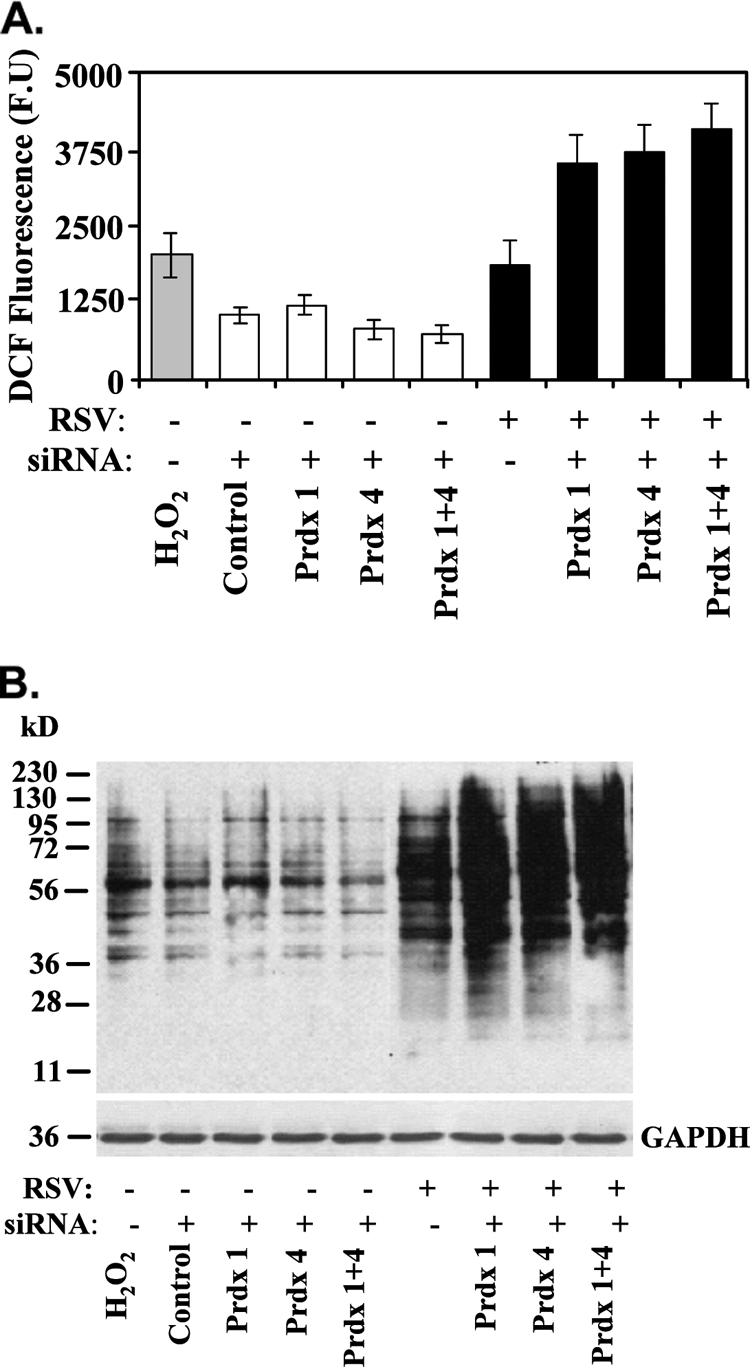
ROS production in A549 cells lacking Prdx-1, Prdx-4, or both. (A) ROS generation was measured by monitoring changes in oxidized DCF fluorescence in A549 cells transfected with control, Prdx-1, Prdx-4, or both Prdx-1 and Prdx-4 siRNA, after which the cells were infected with RSV for 15 h. Bars represent the means ± standard errors of two independent measurements in duplicate. H2O2 was used as a positive control. (B) Enhanced carbonylation of proteins by RSV infection in Prdx-1, Prdx-4, or Prdx-1 plus Prdx-4 knockdown cells by Western blotting. Cellular extracts from an experiment identical to that described for panel A were subjected to SDS-PAGE and transferred to a PVDF membrane. The membrane was probed with anti-DNP antibody. GAPDH is shown as a loading control.
Identification of proteins protected by Prdx-1 and Prdx-4 from RSV-induced cysteinyl oxidation.
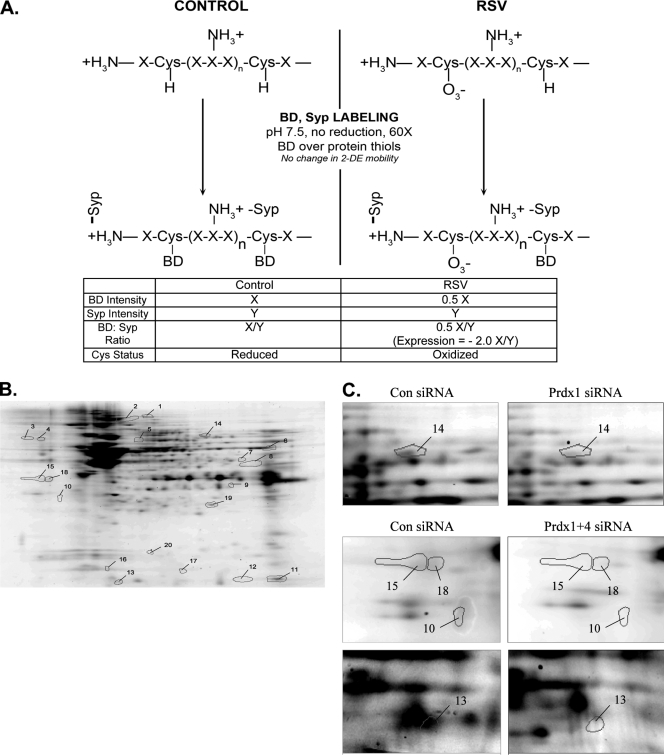
Our data indicate that Prdxs play an important role in reduced RSV-induced oxidative stress. However, the nuclear proteins that are protected are unknown. To determine their identities, we adapted a sensitive cysteinyl-reactive saturation fluorescence labeling technique to identify proteins that are oxidized in response to RSV in the presence or absence of Prdx knockdown (23). This technique uses BODIPY to covalently label protein cysteines when they are reduced. Briefly, nuclear extracts from mock- and RSV-infected cells were prepared and saturation fluorescence labeled with BODIPY at 60-fold excess of dye over protein thiols. The proteins were then separated by IEF/2-DE and spot intensities compared. Proteins containing oxidized forms of cysteines are unreactive with BODIPY, and this is reflected in the protein's lower fluorescence signal (Fig. 5A), because modified cysteines will not be labeled. Because the siRNA knockdown treatments are likely to change protein abundances, to differentiate between changes in cysteine oxidation due to the siRNA and adaptive changes in protein abundance, we stained the gels with Sypro ruby, which is a nonspecific protein stain. Changes in cysteine modifications result in increases or decreases in the ratio of BODIPY fluorescence signal to Sypro ruby fluorescence signal. Changes in BODIPY intensity indicate the presence of S-oxidized thiols.
FIG. 5.
Saturation fluorescence labeling with BODIPY-FL. (A) Schematic diagram of an example of the saturation fluorescence labeling assay for detection of cysteine oxidation. Protein extracts were assayed for cysteine content by amino acid analysis. Enough BODIPY-FL-maleimide was added at pH 7.5 to the unreduced protein mixture to yield a ratio of 60-fold BODIPY to protein thiols. BODIPY will react with reduced cysteine thiols but will not label oxidized thiols. Sypro ruby stain is directed to free amino groups and not affected by cysteine oxidation. The BODIPY/Sypro ruby ratio reflects the oxidation status of cysteine relative to the total protein. The example shows only the partial oxidation of the protein; complete cysteinyl oxidation would result in a very low BODIPY/Sypro ratio, indicating extreme oxidation. BD, BODIPY; Syp, Sypro ruby. (B) A representative image of a 2-D gel of proteins labeled with BODIPY. Nuclear extracts from A549 cells transfected with Prdx-1 siRNA and infected with RSV were labeled with BODIPY and separated by 2-DE. The numbers on the gel indicate individual protein spots whose abundance has changed >1.2-fold. These were identified by electrospray tandem MS on the OrbiTrap Velo. Their identities are listed in Table 2. (C) Expanded view of a few exemplary nuclear protein spots with notable differences in abundances from cells transfected with control, Prdx-1, or Prdx-1 plus Prdx-4 siRNAs.
To identify proteins protected by Prdx-1 from RSV-induced cysteinyl oxidation, A549 cells transfected with control or Prdx-1-specific siRNA for 48 h were subsequently mock or RSV infected. Proteins were labeled in the absence or presence of ascorbate and compared by IEF/2-DE. We observed that there were no major differences in staining intensities of high-abundance proteins between Sypro and BODIPY. However, there was a clear difference in staining of low-abundance proteins between uninfected and RSV-infected cells (Fig. 5B). The same experiment was repeated for Prdx-4-silenced cells and Prdx-1 and Prdx-4 cells. The identity of the 20 proteins whose oxidation was affected by Prdx-1 or Prdx-4 silencing is shown in Fig. 5B and Table 2. Examples of specific proteins in RSV-infected NE in control or Prdx-1 knockdowns are shown in Fig. 5C for enoyl-coenzyme A (CoA) hydratase (spot 14), desmoplakin (spot 15), annexin A2 (spot 18), and centromere-associated protein E (spots 10, 13), where the BODIPY staining is reduced. This can be ascribed to the nature of the dye binding. Sypro ruby is an ionic dye that binds by virtue of its electrostatic attraction to protonated amines in the protein at low pHs. The amount of dye that binds is related to this attraction and results in highly sensitive detection. However, it is at best semiquantitative, similar to other postseparation adsorptive dyes. On the other hand, the BODIPY modification is covalent and is strictly related to the number of cysteinyl residues in the protein under the conditions of labeling. However, whereas Sypro may therefore result in more sensitive detection, the BODIPY labeling is more accurate in estimating relative abundance of proteins and is more reproducible, both due to the saturating excess of dye over protein thiols and its specificity for cysteines (24).
TABLE 2.
Identification of proteins whose cysteine residues were protected from oxidation by either Prdx-1, Prdx-4, or both in RSV-infected A549 cellsa
| Protein name | Common name | Accession no. | Expectation score | Protection index |
||||
|---|---|---|---|---|---|---|---|---|
| Control | Prdx-1 siRNA | Prdx-4 siRNA | Prdx-1 + Prdx-4 siRNA | Prdx-1, Prdx-1 + Prdx-4 or Prdx 4, Prdx-1 + Prdx-4 <−1.25 | ||||
| Carbamoyl-phosphate synthase | CPSM | P31327 | 3.20E−41 | 1.29 | −1.16 | −1.50 | −3.07 | −3.07 |
| Leucine-rich PPR motif-containing protein | LPPRC | P42704 | 8.00E−84 | −1.08 | −2.59 | −1.29 | −1.27 | −2.59 |
| Stress-70 protein | GRP75 | P38646 | 5.00E−12 | −1.26 | 1.37 | −1.69 | −1.39 | −1.69 |
| Desmoplakin | DESP | P15924 | 1.00E−19 | −2.87 | 1.13 | −1.87 | −1.84 | −1.87 |
| 70-kDa lamin | LMNA | P02545 | 6.30E−16 | 1.50 | 1.13 | −2.70 | −1.64 | −2.70 |
| ATP synthase subunit alpha | ATPA | P25705 | 8.00E−43 | −1.29 | 1.05 | −2.97 | −1.43 | −2.97 |
| Sulfide:quinone oxidoreductase | SQRD | Q9Y6N5 | 6.30E−35 | 1.34 | −1.01 | −1.58 | −2.73 | −2.73 |
| Aspartate aminotransferase | AATM | P04259 | 1.60E−17 | 1.33 | −1.37 | 1.76 | −1.72 | −1.72 |
| Annexin A2 | ANXA2 | P07355 | 2.00E−22 | 18.90 | −1.72 | 1.98 | −1.33 | −1.72 |
| Centromere-associated protein E | CENPE | Q02224 | 8.00E−07 | 2.06 | 1.90 | −1.86 | −1.35 | −1.86 |
| Periplakin | PEPL | O60437 | 3.80E−06 | −1.05 | 1.11 | −3.92 | −1.50 | −3.92 |
| Centromere-associated protein E | CENPE | Q02224 | 8.00E−09 | 4.36 | −1.75 | 2.00 | −1.25 | −1.75 |
| Centromere-associated protein E | CENPE | Q02224 | 3.90E−05 | 1.35 | 1.28 | −2.29 | −1.36 | −2.29 |
| Enoyl-CoA hydratase | ECHA | P40939 | 8.00E−48 | 1.60 | −1.34 | −1.03 | −2.20 | −2.20 |
| Desmoplakin | DESP | P15924 | 3.20E−64 | −2.23 | 1.44 | −3.52 | −2.17 | −3.52 |
| Glial fibrillary acidic protein | GFAP | P14136 | 1.30E−08 | −1.78 | −2.14 | 1.23 | −1.32 | −2.14 |
| Glial fibrillary acidic protein | GFAP | P14136 | 3.60E−05 | −1.32 | −1.23 | −1.29 | −5.57 | −5.57 |
| Annexin A2 | ANXA2 | P04259 | 3.20E−18 | −8.91 | −2.61 | −4.61 | −1.61 | −4.61 |
| Centrosomal protein (290 kDa) | CE290 | O15078 | 2.10E−05 | −1.27 | −1.40 | −3.60 | −1.51 | −3.60 |
| Peroxiredoxin-5 | Prdx5 | P30044 | 5.00E−14 | −2.48 | −1.05 | −1.35 | −10.95 | −10.95 |
Protection was determined by the ratio of spot volume of a protein labeled with BODIPY-FL-maleimide to Sypro ruby. A protection index is presented in the far right section of the table by number (a positive value indicates protection, while a negative value indicates Cys oxidation).
Prdx-1 and Prdx-4 protect proteins involved in nuclear assembly from RSV-induced ROS.
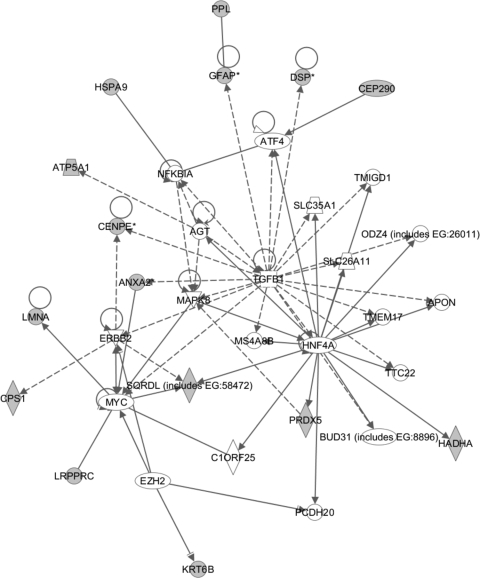
To determine the biological functions of the proteins protected from RSV-induced oxidation, the proteins were subjected to pathway analysis by the Ingenuity pathways knowledge base. The top-scoring network is shown in Fig. 6. Here, we observe that all 15 unique proteins are represented, representing a highly significant enrichment of this biological processes (−log; P value of 43). These proteins are identified based on published literature to interact with hepatocyte nuclear factor (HNF), Myc, and transforming growth factor β1 (TGF-β1) proteins. The data indicate that Prdx-1 and Prdx-4 play a major antioxidant role in the protection of a network of proteins important in nuclear assembly and organization from RSV-induced oxidation.
FIG. 6.
Network analysis of nuclear proteins which have been oxidized by RSV infection in Prdx-1- or Prdx-4-silenced A549 cells. Shown is the molecular interaction network program using the 15 unique proteins found to be most oxidized by RSV infection as the input. Note that all 15 unique proteins occur in the network (gray). ALDH1B1, aldehyde dehydrogenase 1 family, member B1; ANXA2, annexin A2; ATF4, activating transcription factor 4 (tax-responsive enhancer element B67); ATP5A1, ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle; CDC42SE1, CDC42 small effector 1; CENPE, centromere protein E, 312 kDa; CEP290, centrosomal protein, 290 kDa; CPS1, carbamoyl-phosphate synthetase 1, mitochondrial; DSP, desmoplakin; ERBB2, v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma-derived oncogene homolog (avian); GFAP, glial fibrillary acidic protein; GOT2, glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2); HADHA, hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase (trifunctional protein), alpha subunit; HSPA9, heat shock 70-kDa protein 9 (mortalin); IMPACT, impact homolog (mouse); LEP, leptin; LMNA, Lam A/C; LRPPRC, leucine-rich PPR-motif containing; MAPK8, mitogen-activated protein kinase 8; MYC, v-myc myelocytomatosis viral oncogene homolog (avian); NFKB2, nuclear factor of kappa light polypeptide gene enhancer in B cells 2 (p49/p100); NNMT, nicotinamide N-methyltransferase; PAK1IP1, PAK1 interacting protein 1; PPL, periplakin; Prdx5, peroxiredoxin 5; PRL2C3, prolactin family 2, subfamily c, member 3; RPL17, ribosomal protein L17; SLC11A1, solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1; SOLH, small optic lobes homolog (Drosophila); SP1, Sp1 transcription factor; SPAG4, sperm-associated antigen 4; SQRDL, sulfide quinone reductase-like (yeast); STMN3, stathmin-like 3; TGFB1, transforming growth factor, beta 1.
DISCUSSION
RSV is a significant human pathogen responsible for pediatric LRTI linked to chronic airway hyperreactivity (21). In this setting, RSV is trophic for respiratory epithelial cells, where its replication initiates a panoply of cellular responses that induce host cellular stress response, innate immunity, and apoptosis pathways. There is increasing evidence that inducible ROS responses play critical second-messenger roles in signaling and disease pathogenesis by modulating the activity of NF-κB and IRF3, central transcription factors that mediate innate immunity (5, 13, 14). In this study, we employed differential proteomics analysis to understand the complex homeostatic responses occurring in RSV-infected nuclei. To this end, we identified an additional 41 proteins differentially expressed by RSV infection using an all-liquid IEF preseparation 2-DE DIGE approach. Although nuclear Prdx-1, Prdx-3, and Prdx-4 isoforms were identified by this approach, their total abundances are not changed by RSV infection, but rather they exhibit shifts in isoelectric points, suggesting likely oxidative cysteine modifications in response to RSV infection. These conclusions are supported by decreased reactivity of cysteinyl thiols of these proteins with BODIPY-FL-maleimide and increased levels of carbonylation. Examination of the role of Prdx-1 and Prdx-4 indicates that these proteins have redundant functions as essential components of cellular antioxidant pathways controlling nuclear assembly and signaling.
Proteomic approaches have illuminated the complex host cellular response to RSV infection. In earlier studies, we demonstrated application of subcellular fractionation prior to 2-DE to identify organelle responses to RSV infection. These studies indicated that RSV infection produces a nuclear heat shock response and cytoplasmic translocation of SP100, a major structural protein important in ND10 function and whose cytoplasmic translocation may contribute to the innate immune response (3). Subsequent analyses indicated that nuclear HPLC fractionation improved the identification of proteins, producing a comprehensive functional proteomics map of uninfected A549 cells (9). However, HPLC fractionation produced irreversible losses of up to 50% of the nuclear protein on the immobilized surface, reducing the general application of this method (9). As an alternative approach, here we used a liquid isoelectric focusing method by MCE prior to 2-D DIGE that substantially reduces the amount of protein loss during fractionation. Subsequent 2-DE separations allow us to identify proteins undergoing stimulus-dependent posttranslational modification. The spots containing Prdxs identified here were, in fact, oxidized oligomeric forms induced by RSV infection, thereby suggesting a functional role of Prdxs in the antioxidant response to RSV replication.
Prdx-1 and Prdx-4 are members of a highly conserved 2-Cys peroxiredoxin family that catalyze reduction of H2O2 and alkyl hydroperoxides in the presence of thioredoxin (TRX), TRX reductase, and NADPH (24, 30). Under normal oxidative stress (e.g., in unstimulated cells), Prdx-1 and Prdx-4 catalyze the destruction of peroxides through a sulfenic acid intermediate (Cys-SOH), immediately followed by intermolecular homodimerization to a covalent disulfide form. The active enzymes are then regenerated when the disulfides are reduced by thioredoxin. The disulfide formation is relatively slow, so that in the presence of elevated peroxide, the sulfenic acid intermediate closer to the NH2 terminus of the enzyme (Cys-52) may undergo further oxidation to a thiosulfinic (Cys-SO2) or thiosulfonic acid (Cys-SO3), neither of which is reducible by thioredoxin, and thereby inactivates the enzymes. This hyperoxidation produces the observed shift in pI observed in our 2-DE analysis (7). After removal of the oxidative stress, regeneration of the active Prdxs is mediated by sulfiredoxin and ATP. Previously, we have shown that RSV replication is a potent inducer of ROS formation, and enhanced ROS has been measured by multiple independent techniques, including oxidation of DCF-DA, formation of prostaglandin F2-8 isoprostanes, and depletion of reduced cytoplasmic glutathione (6, 13). Here, we extend our understanding to show that RSV replication also induces cellular protein carbonylation. Carbonylation is an irreversible oxidation of Lys, Arg, and Pro residues that targets proteins for proteasomal degradation (18). Although it is well established that Prdx-1 and Prdx-4 prevent cysteine oxidation of substrates, we observed that the level of oxidation-induced protein carbonylation is dramatically increased upon Prdx silencing. Together, the shift in pI, inducible oligomer formation, and enhanced protein carbonylation indicate that the 2-Cys Prdx-1 and Prdx-4 are actively participating in redox regulation in response to RSV infection.
Our earlier understanding of the antioxidant response of airway epithelial cells to RSV replication indicated that RSV infection depletes a primary group of cytoplasmic antioxidant enzymes, including glutathione S-transferase, catalase, and superoxide dismutases 1 and 3 (13). This event renders the infected epithelial cell sensitive to oxidative stress, where H2O2 formation is important in intracellular signaling of the innate immune pathway (14, 20). Upon depletion of these antioxidant cellular pathways, it is not known if ancillary antioxidant pathways operate to partially maintain cellular homeostasis. Our experiments using siRNA knockdown indicate that both Prdx-1 and Prdx-4 play an important role in reducing virus-induced ROS formation, because enhanced DCF-DA oxidation and protein carbonylation are seen in cells lacking either Prdx-1 or Prdx-4. Together, these data indicate that Prdx-1 and Prdx-4 play an important antioxidative role in viral infection.
Emerging literature indicates that, in response to receptor stimulation by TNF-α, platelet-derived growth factor (PDGF), or oxidized low-density lipoprotein (LDL) (7, 16), Prdxs modulate second-messenger intracellular H2O2 signaling. Our earlier studies have shown that virus-induced ROS mediate key steps in the phosphorylation control of IRF3, STAT, and NF-κB transcription factors, whose functions are important in mediating inflammation and disease pathogenesis (5, 13, 14). The relationship between Prdxs and NF-κB signaling has been suggested, where cytoplasmic Prdx-1 targeting inhibits inducible NF-κB activation through a mechanism that involves cysteine oxidation of the NF-κB1/p50 DNA-binding subunit (12). Our siRNA inhibition experiment shows that only in the absence of both Prdx-1 and Prdx-4 isoforms is a paradoxical regulation of NF-κB-dependent genes observed. We infer from these data that constitutive Prdx-1 and Prdx-4 expression plays redundant, inhibitory actions on RSV-induced NF-κB-dependent gene expression. The mechanism for this paradoxical regulation will require further investigation.
The Prdxs comprise an important antioxidative pathway in response to virus-induced ROS. However, little is known about the cellular proteins that are protected by their antioxidant effects. Here, we have adapted a covalent cysteine-reactive labeling assay using BODIPY-FL-maleimide to monitor oxidized cysteine residues in RSV-infected proteins. Coupled with Prdx-1 and Prdx-4 knockdown manipulations, we can identify proteins whose oxidation state is increased in the presence of virus and Prdx inhibition. Staining the 2-DE with a secondary stain (Sypro ruby) that is not thiol reactive controls for changes in total protein abundance, allowing the identification of only those proteins whose oxidative state is changed. Fundamental to this analysis is the uncharged nature of the BODIPY dye, so that the fluorescence-labeled cysteine of the proteins does not play a role in their electrophoretic mobility, and spot intensities can be directly compared.
Our assay indicates that Prdxs protect at least 15 unique proteins that map to a protein interaction network controlling cellular assembly and organization. To this end, it is notable that RSV induces significant cytoskeletal changes in infected epithelial cells, including cytopathic effects, reduction in cellular motility, and the formation of multinucleated syncytia (19). The Prdx-sensitive proteins that we identified include annexin A2, centromere-associated protein E, and desmoplakin. Annexin A2 is a calcium-regulated membrane-bound F-actin binding protein important in signal transduction. In the membrane, annexin A2 forms a heterotetrameric complex with S100A10. Here, the annexin A2-S100A10 complex formation is known to be regulated by the redox status of its sulfhydryl groups, where oxidation of Cys residues 8 and 132 inhibits its ability to interact with phospholipid liposomes and F-actin (4). Another essential cytostructural protein sensitive to ROS-induced oxidation in the absence of Prdx-1 or Prdx-4 is desmoplakin, a member of the plakin family of cytolinker proteins that binds intermediate filaments and actin. Importantly, nitrosylation of 4 highly conserved cysteine residues in the COOH-terminal intermediate fiber binding domain of this protein family results in intermediate fiber collapse (27). Finally, we identified centromere-associated protein E in multiple spots, which is a kinesin motor responsible for chromosome movement and/or spindle elongation. The NH2-terminal motor domain contains four cysteine residues essential for function (25). The network analysis indicates that these cytoskeletal proteins interact with nodes composed of HNF, Myc, and TGF-β1 proteins, suggesting that future studies on the effects of RSV on cellular proliferation and growth will be informative. Together, these data indicate that Prdxs play important homeostatic roles, protecting proteins important in cellular structure and chromosomal segregation from virus-induced cysteine oxidation. In future studies, it will be interesting to analyze the structure of intermediate filaments in RSV-infected cells after Prdx-1 or Prdx-4 silencing.
In summary, using a liquid isoelectric focusing/2-DE difference gel assay of isolated nuclear proteins, we identified Prdx-1 and Prdx-4 isoforms as targets of RSV-induced oxidation. Functional studies show that this pathway contributes to the cellular antioxidant response, despite depletion of cytoplasmic antioxidant enzymes, including glutathione S-transferase, catalase, and SOD-1 and SOD-3 isoforms. Prdx-1 and Prdx-4 function to modulate RSV-induced ROS signaling, functioning as nonredundant inhibitors of basal NF-κB-dependent gene expression. Finally, a novel cysteine-reactive fluorescent labeling assay was adapted to 2-DE, in which 15 unique reductive Prdx targets were identified. These studies further underscore the complexity of host cellular response to viral homeostatic insults and illustrate the role of inducible ROS as a mediator of innate immunity.
Acknowledgments
This work was supported, in part, by NIH grants 1U54RR02614 UTMB CTSA (A.R.B.), AI062885 (A.R.B.), NHLBI contract BAA-HL-02-04 (A.R.B.), and ES06676 (to K. Elferink, UTMB).
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Aguilera-Aguirre, L., A. Bacsi, A. Saavedra-Molina, A. Kurosky, S. Sur, and I. Boldogh. 2009. Mitochondrial dysfunction increases allergic airway inflammation. J. Immunol. 183:5379-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldogh, I., A. Basci, B. Choudhary, N. Dharajiya, R. Alam, T. Hazdra, S. Mitra, R. Goldblum, and S. Sur. 2005. ROS generated by pollen NADPH oxidase provide a signal that augments antigen-induced allergic airway inflammation. J. Clin. Invest. 115:2169-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brasier, A. R., H. Spratt, Z. Wu, I. Boldogh, Y. Zhang, R. P. Garofalo, A. Casola, J. Pashmi, A. Haag, B. Luxon, and A. Kurosky. 2004. Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected A549 cells identified by high-resolution two-dimensional gel electrophoresis. J. Virol. 78:11461-11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caplan, J. F., N. R. Filipenko, S. L. Fitzpatrick, and D. M. Waisman. 2004. Regulation of annexin A2 by reversible glutathionylation. J. Biol. Chem. 279:7740-7750. [DOI] [PubMed] [Google Scholar]
- 5.Casola, A., N. Burger, T. Liu, M. Jamaluddin, A. R. Brasier, and R. P. Garofalo. 2001. Oxidant tone regulates RANTES gene expression in airway epithelial cells infected with respiratory syncytial virus. Role in viral-induced interferon regulatory factor activation. J. Biol. Chem. 276:19715-19722. [DOI] [PubMed] [Google Scholar]
- 6.Castro, S. M., A. Guerrero-Plata, G. Suarez-Real, P. A. Adegboyega, G. N. Colasurdo, A. M. Khan, R. P. Garofalo, and A. Casola. 2006. Antioxidant treatment ameliorates respiratory syncytial virus-induced disease and lung inflammation. Am. J. Respir. Crit. Care Med. 174:1361-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conway, J. P., and M. Kinter. 2006. Dual role of peroxiredoxin I in macrophage-derived foam cells. J. Biol. Chem. 281:27991-28001. [DOI] [PubMed] [Google Scholar]
- 8.Easton, A. J., J. B. Domachowske, and H. F. Rosenberg. 2004. Animal pneumoviruses: molecular genetics and pathogenesis. Clin. Microbiol. Rev. 17:390-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbus, J., H. Spratt, J. Wiktorowicz, Z. Wu, I. Boldogh, L. Denner, A. Kurosky, R. C. Brasier, B. Luxon, and A. R. Brasier. 2006. Functional analysis of the nuclear proteome of human A549 alveolar epithelial cells by HPLC-high resolution 2-D gel electrophoresis. Proteomics 6:2656-2672. [DOI] [PubMed] [Google Scholar]
- 10.Foster, K. A., C. G. Oster, M. M. Mayer, M. L. Avery, and K. L. Audus. 1998. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 243:359-366. [DOI] [PubMed] [Google Scholar]
- 11.Garofalo, R., M. Sabry, M. Jamaluddin, R. K. Yu, A. Casola, P. L. Ogra, and A. R. Brasier. 1996. Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J. Virol. 70:8773-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen, J. M., S. Moriarty-Craige, and D. P. Jones. 2007. Nuclear and cytoplasmic peroxiredoxin-1 differentially regulate NF-kappaB activities. Free Radic. Biol. Med. 43:282-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosakote, Y. M., T. Liu, S. M. Castro, R. P. Garofalo, and A. Casola. 2009. Respiratory syncytial virus induces oxidative stress by modulating antioxidant enzymes. Am. J. Respir. Cell Mol. Biol. 41:348-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamaluddin, M., B. Tian, I. Boldogh, R. Garofalo, and A. R. Brasier. 2009. Respiratory syncytial virus infection induces a ROS-MSK1-phospho-Ser-276 RelA pathway required for cytokine expression. J. Virol. 83:10605-10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamaluddin, M., S. Wang, I. Boldogh, B. Tian, and A. R. Brasier. 2007. TNF-α-induced NF-κB/RelA Ser 276 phosphorylation and enhanceosome formation on the IL-8 promoter is mediated by a reactive oxygen species (ROS)-dependent pathway. Cell. Signal. 9:1419-1433. [DOI] [PubMed] [Google Scholar]
- 16.Kang, S. W., H. Z. Chae, M. S. Seo, K. Kim, I. C. Baines, and S. G. Rhee. 1998. Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-alpha. J. Biol. Chem. 273:6297-6302. [DOI] [PubMed] [Google Scholar]
- 17.Kang, S. W., S. G. Rhee, T. S. Chang, W. Jeong, and M. H. Choi. 2005. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol. Med. 11:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine, R. L., D. Garland, C. N. Oliver, A. Amici, I. Climent, A. G. Lenz, B. W. Ahn, S. Shaltiel, and E. R. Stadtman. 1990. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 186:464-478. [DOI] [PubMed] [Google Scholar]
- 19.Levine, S., and R. Hamilton. 1969. Kinetics of the respiratory syncytial virus growth cycle in HeLa cells. Arch. Ges. Virusforsch. 28:122-132. [DOI] [PubMed] [Google Scholar]
- 20.Liu, T., S. Castro, A. R. Brasier, M. Jamaluddin, R. P. Garofalo, and A. Casola. 2004. Reactive oxygen species mediate virus-induced STAT activation: role of tyrosine phosphatases. J. Biol. Chem. 279:2461-2469. [DOI] [PubMed] [Google Scholar]
- 21.Mohapatra, S. S., and S. Boyapalle. 2008. Epidemiologic, experimental, and clinical links between respiratory syncytial virus infection and asthma. Clin. Microbiol. Rev. 21:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olszewska-Pazdrak, B., A. Casola, T. Saito, R. Alam, S. Crowe, F. Mei, P. L. Ogra, and R. Garofalo. 1998. Cell-specific expression of RANTES, MCP-1, and MIP-1α by lower airway epithelial cells and eosinophils infected with respiratory syncytial virus. J. Virol. 72:4756-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pretzer, E. P., and J. E. Wiktorowicz. 2008. Saturation fluorescence labeling of proteins for proteomic analyses. Anal. Biochem. 374:250-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhee, S. G., H. Z. Chae, and K. Kim. 2005. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 38:1543-1552. [DOI] [PubMed] [Google Scholar]
- 25.Rosenfeld, S. S., M. van Duffelen, W. M. Behnke-Parks, C. Beadle, J. Correia, and J. Xing. 2009. The ATPase cycle of the mitotic motor CENP-E. J. Biol. Chem. 284:32858-32868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shay, D. K., R. C. Holman, G. E. Roosevelt, M. J. Clarke, and L. J. Anderson. 2001. Bronchiolitis-associated mortality and estimates of respiratory syncytial virus-associated deaths among US children, 1979-1997. J. Infect. Dis. 183:16-22. [DOI] [PubMed] [Google Scholar]
- 27.Spurny, R., K. Abdoulrahman, L. Janda, D. Runzler, G. Kohler, M. J. Castanon, and G. Wiche. 2007. Oxidation and nitrosylation of cysteines proximal to the intermediate filament (IF)-binding site of plectin. J. Biol. Chem. 282:8175-8187. [DOI] [PubMed] [Google Scholar]
- 28.Tian, B., Y. Zhang, B. A. Luxon, R. P. Garofalo, A. Casola, M. Sinha, and A. R. Brasier. 2002. Identification of NF-κB-dependent gene networks in respiratory syncytial virus-infected cells. J. Virol. 76:6800-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welliver, T. P., R. P. Garofalo, Y. Hosakote, K. H. Hintz, L. Avendano, K. Sanchez, L. Velozo, H. Jafri, S. Chavez-Bueno, P. L. Ogra, L. McKinney, J. L. Reed, and R. C. Welliver, Sr. 2007. Severe human lower respiratory tract illness caused by respiratory syncytial virus and influenza virus is characterized by the absence of pulmonary cytotoxic lymphocyte responses. J. Infect. Dis. 195:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood, Z. A., E. Schroder, J. R. Harris, and L. B. Poole. 2003. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28:32-40. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, W., and B. T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72:2482-2489. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y., B. A. Luxon, A. Casola, R. P. Garofalo, M. Jamaluddin, and A. R. Brasier. 2001. Expression of RSV-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J. Virol. 75:9044-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]