Abstract
Foamy viruses (FVs), or spumaviruses, are integrating retroviruses that have been developed as vectors. Here we generated nonintegrating foamy virus (NIFV) vectors by introducing point mutations into the highly conserved DD35E catalytic core motif of the foamy virus integrase sequence. NIFV vectors produced high-titer stocks, transduced dividing cells, and did not integrate. Cells infected with NIFV vectors contained episomal vector genomes that consisted of linear, 1-long-terminal-repeat (1-LTR), and 2-LTR circular DNAs. These episomes expressed transgenes, were stable, and became progressively diluted in the dividing cell population. 1-LTR circles but not 2-LTR circles were found in all vector stocks prior to infection. Residual integration of NIFV vectors occurred at a frequency 4 logs lower than that of integrase-proficient FV vectors. Cre recombinase expressed from a NIFV vector mediated excision of both an integrated, floxed FV vector and a gene-targeted neo expression cassette, demonstrating the utility of these episomal vectors. The broad host range and large packaging capacity of NIFV vectors should make them useful for a variety of applications requiring transient gene expression.
Several nonintegrating retrovirus vectors have been produced previously in order to provide transient transgene expression and/or avoid the consequences of insertional mutagenesis. These nonintegrating vectors are rendered integration deficient by mutations in the viral integrase or att sites and can express transgenes (1, 17, 41, 42, 59). After infection, nonintegrating vector genomes are found as linear, 1-long-terminal-repeat (1-LTR), or 2-LTR episomal DNA molecules and are stable, but because these episomes do not replicate, they are diluted over time in proliferating cells (1, 5, 7, 41). Most nonintegrating retroviral vectors have been derived from the lentivirus (LV) system, but they have also been produced from Moloney leukemia virus (MLV) and feline immunodeficiency virus (48, 63). These vectors have been used to transduce both dividing and nondividing cells in vitro and in vivo (4, 17, 41, 48, 57, 59, 60). Nonintegrating LV vectors expressing zinc finger endonucleases or I-SceI endonuclease have also been used to introduce double strand breaks and provide substrates for gene targeting (8, 34). These studies have shown the utility of nonintegrating retroviral vectors.
Foamy virus (FV) vectors have been developed as gene transfer vehicles because of their many unique properties. Wild-type (wt) FVs, members of the spumaretrovirus subfamily of retroviruses, are endemic to a number of animal species, including nonhuman primates, cats, cows, and horses, but not to humans (13, 22, 45). Humans have been infected with FVs after exposure to nonhuman primates, but no resulting pathology or human-to-human transmission has been reported (13, 22, 45). FVs have a DNA genome created by reverse transcription in the virus-producing cell, and they are the largest of all retroviruses (>13 kb), with a packaging capacity of more than 9 kb. They can be produced at high titers without contaminating replication-competent retrovirus (54). Also, FV vectors can transduce many cell types from a variety of species (25, 30, 45, 54, 58), have a favorable integration profile (2), and form stable preintegration complexes in nondividing cells (47, 55). FV vectors have shown promise in hematopoietic stem cell gene therapy and in the treatment of genetic diseases (2, 24).
As with other retroviruses, FV vector integration is mediated by the integrase protein encoded at the 3′ end of the pol gene. The highly conserved integrase protein is comprised of three distinct regions, the HHCC zinc-binding domain, the DNA-binding domain, and the catalytic core domain (see Fig. 1). The catalytic core contains the DD35E motif, a universal feature of integrases and transposases that participates in catalyzing the phosphotransfer reaction during strand transfer (7). Class I mutations in the catalytic site result in vectors that are defective for integration, and these mutations have been used to generate nonintegrating LV (NILV) and MLV vectors (59, 63). FV vectors with class I mutations have also been produced and have been shown to generate viral particles that infected cells and did not integrate into the host genome (9, 12, 19, 20, 35). However, transgene expression was minimal or nonexistent in these studies, for unclear reasons (12, 19, 20). Here, we show that nonintegrating FV (NIFV) vectors can be produced that efficiently transduce cells and express transgenes from their episomal genomes.
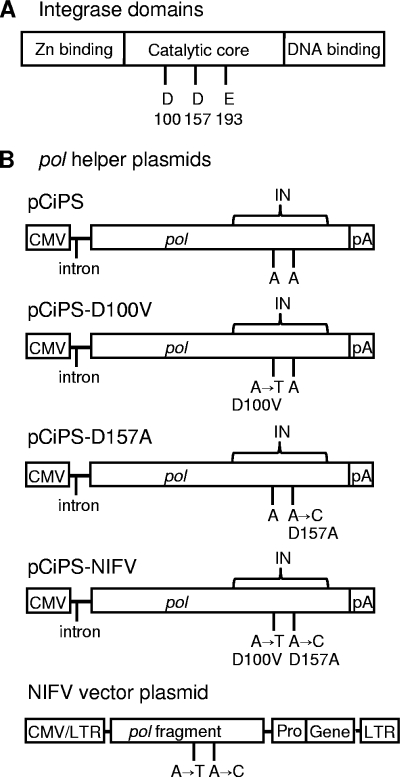
FIG. 1.
Schematic representation of mutations introduced into integrase, the pol helper plasmid, and the NIFV vector plasmid. (A) Domain structure of the integrase protein. The highly conserved DDE residues are shown in the catalytic core. (B) Mutations producing the indicated amino acid changes were introduced into the integrase sequence (IN) of the pol helper plasmid, pCiPS, and the pol fragment of FV vector plasmids to generate the NIFV pol helper and vector plasmids shown. Locations of the CMV promoter, simian virus 40 (SV40) intron (intron), SV40 polyadenylation site (pA), internal promoter (Pro), transgenes (Gene), LTRs, and pol open reading frame (pol) are shown.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney 293D cells (16), HT-1080 human fibrosarcoma (HT-1080) cells (44), and human fibroblasts (Coriell Institute for Medical Research, Camden, NJ, repository number GM05387) were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT), 100 U/ml of penicillin, and 100 μg/ml streptomycin. Osteogenesis imperfecta (OI) mesenchymal stem cells (OI12 MSCs) were established from the vertebral fragments from a patient with type III OI that were obtained during surgery under an Institutional Review Board-approved protocol as described previously (6). OI12 MSCs were grown in low-glucose DMEM with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml streptomycin and supplemented with 2 mM l-glutamine (Invitrogen).
Plasmids.
The NIFV pol helper plasmids were based on the plasmid pCiPS (54). Site-directed mutagenesis (QuikChange; Stratagene, La Jolla, CA) was used to introduce point mutations at nucleotides 2635 and 2806 of the pol open reading frame (accession number U21247). Single mutant pol helper plasmids were generated using the primers D100V Forward (5′-GATAAATTCTTTATTGTCTATATTGGACCTTTGCC-3′) and D100V Reverse (5′-GGCAAAGGTCCAATATAGACAATAAAGAATTTATC-3′) for pCiPS-D100V and D157A Forward (5′-CAAAGGTGATTCACTCTGCTCAAGGTGCAGCATTCAC-3′) and D157A Reverse (5′-GTGAATGCTGCACCTTGAGCAGAGTGAATCACCTTTG-3′) for pCiPS-D157A. pCiPS-NIFV was generated by sequential rounds of mutagenesis to include both these mutations. All mutations were verified by sequencing.
FV vector plasmids were derived from the plasmid pΔφ (54). pNIFV was created by inserting a PstI-EcoRI restriction fragment from pCiPS-NIFV into the cis-acting pol region of pΔφ. pNIFV-MscvF contains the murine stem cell virus (MSCV) long terminal repeat promoter (18) expressing green fluorescent protein (GFP) and was derived from pΔΦMscvF (54) by incorporating both integrase mutations in pNIFV. pNIFV-MscvCre is similar to pNIFV-MscvF but contains the mammal-optimized Cre gene (a gift from Rolf Sprengel, Max-Planck-Institute, Heidelberg, Germany), in place of GFP. pNIFV-CnZPNO was constructed by inserting an AflII fragment isolated from pNIFV-MscvF into pΔφ-CnZPNO (36). pΔφMscvTKNloxP is similar to pNIFV-MscvF but contains a truncated thymidine kinase-neomycin phosphotransferase fusion gene (50, 51) instead of GFP and has a loxP site inserted into the XbaI site of the 3′ LTR. All plasmid sequences are available by request.
Vector production.
FV and NIFV vector stocks were produced by transient cotransfection as previously described (14). Pseudostocks were generated similarly to vector stocks except that no pol helper plasmid was included in the transfection. Seventy-two hours after transfection, the culture medium was collected, filtered through a 45-μM filter, concentrated by ultracentrifugation, resuspended in DMEM with 5% dimethyl sulfoxide, and stored at −80°C.
DNA isolation and DNA analysis.
Genomic DNA was isolated by using the Puregene DNA purification system (Gentra Systems, Minneapolis, MN). Southern blotting was performed by standard techniques. Radiolabeled probes were synthesized by random priming using a Ready Prime kit (Amersham). Quantitation of Southern blots was performed on a PhosphorImager (Storm 820; Amersham Biosciences, Piscataway, NJ). To identify HT-1080 clones containing a single integrant of ΔφMscvTKNloxP and to document removal of the floxed FV provirus, a 640-bp HindIII-SalI 3′ FV LTR probe fragment from pΔφMscvTKNloxP was used. The COL1A2 probe corresponds to COL1A2 genomic DNA chr7:94,026,171-94,027,234 (February 2009 assembly). Low-molecular-weight DNA was isolated from cells by Hirt extraction (23).
Vector genomes were isolated from concentrated FV and NIFV vector stocks by extracting twice with phenol, twice with phenol-chloroform, and once with chloroform and then precipitating with 2 volumes of 100% ethanol. Extracted DNA was digested with DpnI to fragment contaminating residual plasmid DNA from the transfection and analyzed by Southern blotting using a GFP probe (735-bp AgeI-NotI fragment from pΔφMscvF), a neo probe (686-bp EagI-BssSI fragment from ΔφMscvTKNloxP), or a Cre probe (1,069-bp AgeI-NotI fragment from pΔφMscvCre).
Transduction conditions.
To generate data for Table 1, HT-1080 cells were seeded at 105 cells/well in 6-well plates on day 1 and then infected with serial dilutions of vector stocks on day 2, and GFP expression was determined by flow cytometry (Becton Dickinson, San Jose, CA) on day 4. The mean fluorescence intensity was determined by infecting HT-1080 cells with all vectors at a multiplicity of infection (MOI) of 5 genome-containing particles per cell.
TABLE 1.
Titers of NIFV vectors in HT-1080 cells
| Vector | No. of vector DNA genomes/ml | No. of transducing units/ml | No. of genomes/transducing unit | MFIa |
|||
|---|---|---|---|---|---|---|---|
| Day 1 |
Day 2 |
||||||
| Raw | Relative | Raw | Relative | ||||
| ΔφMscvF | 3.77 × 108 | 5.97 × 106 | 63 | 113 | 1.00 | 475 | 1.00 |
| NIFV-MscvF | 6.40 × 108 | 1.35 × 107 | 47 | 65 | 0.58 | 116 | 0.24 |
| D100V-MscvF | 3.72 × 108 | 8.45 × 106 | 44 | 73 | 0.65 | 125 | 0.26 |
| D157A-MscvF | 3.95 × 108 | 6.28 × 106 | 63 | 61 | 0.54 | 107 | 0.23 |
MFI determined by gating on GFP+ cells with a window containing <0.1% GFP+ cells in uninfected cultures.
The GFP expression time course results shown in Fig. 3 were obtained by seeding human fibroblasts at 105 cells/well in 6-well plates on day 1 and then infecting with vector stocks at an MOI of 50 genome-containing particles per cell on day 2. At designated time points, 10 to 20% of the cells were harvested and analyzed by flow cytometry.
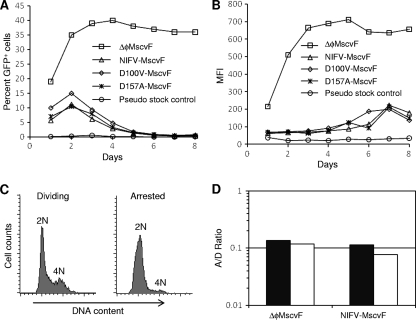
FIG. 3.
GFP expression profile from cells infected with NIFV vectors. Human fibroblasts were infected with ΔφMscvF, NIFV-MscvF, D100V-MscvF, or D157A-MscvF or with a pseudostock control (stock prepared without pol expression plasmid) at an MOI of 50 genome-containing particles per cell. The percentage of GFP+ cells (A) or the MFI (B) was monitored over time by flow cytometry. (C) Cell cycle analysis of the DNA content in dividing and arrested cell populations by propidium iodide staining. (D) Dividing and arrested human fibroblast cultures were infected with ΔφMscvF and NIFV-MscvF at an MOI of 15 genome-containing particles per cell. The percentage of GFP+ cells in the arrested culture divided by that in dividing cultures (A/D ratio) is shown for both infections, with solid and open columns representing separate experiments.
To determine if NIFV vectors are expressed in quiescent cells, human fibroblasts were seeded at 1.5 × 105 cells/well in a 12-well plate on day 1 and grown for 3 days until confluent. On day 4, the serum was reduced to 0.5%, and the cells were grown for an additional 4 days prior to infection. To obtain dividing cells, human fibroblast cultures were grown in DMEM with 10% FBS and plated at 1.5 × 105 cells/well 24 h prior to transduction. On day 8, the NIFV-MscvF and ΔφMscvF vectors were added to dividing and nondividing cell cultures at an MOI of 15 genomes/cell, and on day 10, cells were harvested and analyzed by flow cytometry.
Neomycin resistance titers were determined by seeding HT-1080 cells at 105 cells/well in 6-well plates on day 1, followed by infection with serial dilutions of vector stocks on day 2. Each well was replated into 10-cm dishes at serial dilutions on day 3, and G418 selection (700 μg/ml) was started on day 4. After 10 days of selection, G418-resistant colonies were stained with Coomassie blue and counted.
To analyze vector DNA in infected cells (see Fig. 4), 60-mm dishes were seeded with 2.2 × 105 human fibroblasts on day 1 and then infected with either ΔφMscvF or NIFV-MscvF at an MOI of 1,000 genome-containing particles per cell (or an equivalent volume of pseudostock as a control) on day 2. The cells were expanded continuously, and portions were harvested for low-molecular-weight DNA isolation 4, 7, and 10 days after infection.
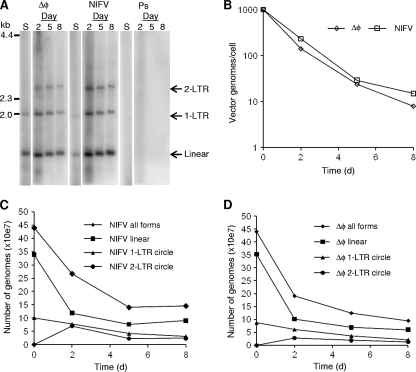
FIG. 4.
Analysis of episomal vector genomes. Human fibroblasts were transduced with ΔφMscvF (Δφ) and NIFV-MscvF (NIFV) at a multiplicity of infection of 1,000 genome-containing particles per cell or a pseudostock control (Ps). (A) Low-molecular-weight DNA was isolated on days 2, 5, and 8 after infection, cut with DpnI, PstI, and AgeI, and then analyzed by Southern blotting. DNA purified from each vector stock was also included (S). See Fig. 2 for diagrams of the predicted sizes. (B) The number of vector genomes present per cell over time. (C and D) The total number of vector genomes and those of individual species of linear, 1-LTR circular, and 2-LTR circular genome forms are shown for each time point after infection with NIFV (C) or Δφ (D).
Quantifying NIFV integration.
Integration frequencies were determined by infecting 1 × 105 human fibroblasts with ΔφCnZPNO or NIFV-CnZPNO at an MOI of 100 genome-containing particles per cell and then replating and selecting with G418 (700 μg/ml) starting 2 days after infection. After 10 days of selection, single integrant clones from cells infected with NIFV-CnZPNO were picked and expanded. Colony counts in the ΔφCnZPNO-infected cells were determined by staining with Coomassie blue.
Five μg of genomic DNA from each NIFV-CnZPNO integrant clone was digested with PmeI and PshAI for 4 h, extracted with phenol-chloroform, and precipitated with ethanol. Five μg of digested DNA was circularized with 20 U of T4 DNA ligase overnight at room temperature in 20 μl and then used to transform Escherichia coli strain DH10B by electroporation. Kanamycin-resistant colonies were picked, and plasmid DNA containing rescued proviruses was purified and sequenced using the primers FV3′LTR (5′-AAGTAACGAGGAGAGGGT-3′), FV5′PGK (5′-ATGCTCCAGACTGCCTTGGG-3′), and FV5′CMV (5′-CCATTTACCGTCATTGACG-3′).
Cre recombinase-mediated excision.
Floxed FV proviral clones were generated by infecting HT-1080 cells with ΔφMscvTKNloxP at an MOI of 0.05 transducing units per cell and selecting in G418. Genomic DNA was isolated from G418-resistant colonies and analyzed by Southern blotting using the 3′ FV LTR probe. To assess Cre expression from NIFV-MscvCre, HT-1080 cells harboring a single ΔφMscvTKNloxP provirus were transduced with either NIFV-MscvCre at an MOI of 100 genome-containing particles per cell or soluble Cre protein (16 μM) as previously described (6). Five days after Cre treatment, the cells were selected with ganciclovir (50 μM) for 14 days, and single colonies were expanded and analyzed by Southern blotting. Colony counts were determined by staining with Coomassie blue.
To measure Cre-mediated excision at a defined chromosomal locus, OI12 MSCs targeted at exon 2 of COL1A2 with a floxed internal ribosome entry site (IRES)-neo cassette (6) were transduced at MOIs of 0, 5, 50, and 500 or 0, 5, 50, 500, and 1,000 genome-containing particles per cell with ΔφMscvCre and NIFV-MscvCre, respectively (or an equivalent volume of pseudostock as a control) and expanded for 14 days, and then genomic DNA was isolated for Southern blot analysis.
RESULTS
NIFV vector design and stock production.
To produce NIFV vectors, we introduced class I integrase mutations into the FV pol gene in the helper plasmid pCiPS (Fig. 1). These mutations resulted in an aspartic acid-to-valine substitution at residue 100 (D100V) or an aspartic acid-to-alanine substitution at residue 157 (D157A) of the FV integrase protein (Fig. 1B). The D100V mutation is similar to the D64V mutation used previously in NILV vectors (1, 39, 41, 60), and the D157A mutation has been shown to produce integrase-deficient FV vectors (9, 12). These mutations do not interfere with reverse transcription, nuclear localization of the preintegration complex, or circularization of the viral genome (9, 12, 29). We also introduced both mutations into the cis-acting pol fragment present in vector plasmids to avoid possible recombination with the pol expression plasmid and mutation reversion during vector production. Three different vector plasmids were produced that expressed either the green fluorescent protein (GFP), the neomycin resistance gene (neo), or Cre recombinase (Cre) under the control of an internal MSCV promoter. Each of these vector plasmids was cotransfected with gag- and env-expressing plasmids and either the wt pol helper plasmid pCiPS or an integrase-defective helper plasmid (pCiPS-NIFV, pCiPS-D100V, or pCiPS-D157A) to create vector stocks. A pseudostock control was produced by omission of the pol helper plasmid.
To determine the impact of integrase mutations on vector production, we compared the vector genome titers of our GFP-expressing, nonintegrating FV vectors NIFV-MscvF, D100V-MscvF, D157A-MscvF and integrase-proficient deleted FV vector ΔφMscvF. DNA vector genomes were purified directly from vector stocks by phenol-chloroform extraction, digested with DpnI to remove contaminating transfected plasmids, and analyzed by Southern blotting (Fig. 2). The titers were similar for both the integrase-proficient vector and the nonintegrating vectors, indicating that the class I integrase mutations did not affect vector production (Table 1).
FIG. 2.
Southern blot analysis of NIFV vector stocks. Purified DNAs from vector stocks of ΔφMscvF (Δφ), NIFV-MscvF (NIFV), D100V-MscvF (100), D157A-MscvF (157), and a pseudostock control (Ps) were cut with PstI and AgeI and analyzed by Southern blotting. DNAs were also digested with DpnI to remove transfected plasmid DNA. The positions of the linear DNA genome (linear), replication intermediates (R1 and R2), and 1-LTR circles (1-LTR) are shown. Diagrams of the predicted sizes (not to scale) of LTR-containing fragments are below the Southern blot. The position of the probe is denoted by the black bar.
We noted two additional bands present in undigested vector DNA preparations on the Southern blot (Fig. 2) that did not migrate at the position of full-length linear vector genomes. These bands may represent vector replication intermediates (labeled R1 and R2). After digestion with restriction enzymes and probing for an LTR-containing fragment, 1-LTR circular vector forms could also be identified (Fig. 2). Our results showed that in both NIFV and integrase-proficient vector stocks, ∼80% of the vector genomes were linear and ∼20% were 1-LTR circles. Thus, 1-LTR circles are generated in vector-producing cells before the vectors infect cells. This agrees with previous reports noting the presence of 1-LTR circles in FV vector stocks (55). We did not detect 2-LTR circles in our vector stocks, although these have been observed in wt FV stocks (55).
Gene expression from NIFV vectors.
HT-1080 human sarcoma cells were transduced with NIFV-MscvF, D100V-MscvF, D157A-MscvF, ΔφMscvF, and the pseudostock control, and 2 days after infection, GFP expression was monitored by flow cytometry. We observed gene expression from all the vectors, and their titers were similar when measured as transducing units (Table 1). The ratio of vector genomes to transducing units ranged from 44 to 63, demonstrating that most input vector virions do not produce cells that express the transgene at detectable levels. This was the case for both integrase-proficient and NIFV vectors. We also compared the level of transgene expression by infecting cells at an MOI of 5 DNA genome-containing particles/cell and measuring the mean fluorescence intensity (MFI) of GFP-expressing cells 1 and 2 days after infection. The MFIs were 1.5- to 1.9-fold lower for integrase-deficient vectors than for the integrase-proficient vector on day 1 and 3.8- to 4.3-fold lower on day 2, consistent with results for other nonintegrating retroviral vectors. For example, in one study, a 10-fold-higher dose of NILV vector was required to equal the expression from the integrase-proficient LV vector (8).
Next, we evaluated transgene expression from NIFV vectors over time in a dividing population of human fibroblasts (Fig. 3A and B). The integrase-proficient vector produced the maximal percentage of GFP+ cells on day 2, and this did not decrease over time, consistent with provirus integration. In contrast, the integrase-deficient vectors reached a maximum percentage of GFP+ cells by day 2, which decreased to background levels over the next 8 days. This is the expected result from unintegrated vector genomes that do not replicate and become diluted over time. The relative MFI from NIFV vectors was lower in human fibroblasts than in HT-1080 cells (14% versus 24% on day 2), most likely due to variation in NIFV expression between cell lines. Surprisingly, the MFI of cells infected with NIFV vectors increased over time, suggesting that the intracellular GFP protein level may take days to reach a steady state in the subpopulation of cells that retain and express the NIFV vector genome.
Although FV vectors do not transduce quiescent cells as efficiently as dividing cells (55), it has been shown that FV genomes enter the nuclei of stationary cells (33, 49). To assess expression from NIFV vectors in nondividing cells, we infected arrested and dividing cultures of human fibroblasts with NIFV-MscvF and ΔφMscvF and measured the percentage of GFP+ cells 2 days after transduction. Propidium iodide staining showed that only 3.6% of the arrested cells contained an intermediate amount of DNA, consistent with S phase (Fig. 3C). Determination of the arrested-cell-to-dividing-cell transduction ratio (A/D ratio) (percent GFP+ cells in the arrested culture/percent GFP+ cells in the dividing culture) showed that both NIFV vectors and integrase-proficient vectors require cell division for efficient gene expression (Fig. 3D).
Status of vector genomes in infected cells.
The distribution of different episomal vector genome forms was followed over time by Southern blot analysis of low-molecular-weight DNA isolated from infected human fibroblasts (Fig. 4A). The total number of episomal genomes per cell decreased as the cell population expanded, at a similar rate for both integrase-proficient FV and NIFV vectors (Fig. 4B). Two days after infection, we recovered 60.7 and 43.2% of the input viral genomes from NIFV-MscvF and ΔφMscvF, respectively. Since free vector virions should have been washed away during the medium changes and cell pelleting, these presumably represent intracellular molecules, and the high genome-containing particle/transducing unit ratios we observed (Table 1) were not due to inefficient cellular entry. When the total amounts of each form of the NIFV genome were examined (Fig. 4C), there was a decrease in the linear and 1-LTR circle forms over time, with an appearance of 2-LTR circle forms not present in the vector stock. Thus, the 2-LTR circle vector forms are produced after cellular entry, most likely due to circularization of linear molecules. Similar results were obtained with integrase-proficient vectors (Fig. 4D), except the number of 2-LTR circles was lower, consistent with integration of some linear vector forms rather than their conversion to 2-LTR circles.
Quantification of rare NIFV vector integration events.
Integration of other nonintegrating retroviral vectors has been shown to be 3 to 4 logs lower than that of their wt counterparts and not mediated by integrase (43, 59). To address this possibility in NIFV vectors, we compared the integration frequencies of an NIFV shuttle vector and its integrase-proficient counterpart. The CnZPNO cassette present in these vectors contains a neo gene under the control of both a cytomegalovirus (CMV) promoter and a prokaryotic promoter, as well as a bacterial replication origin, allowing us to expand integrated proviruses by selecting transduced cells in G418 and then recover the integrated proviruses and flanking DNA as bacterial plasmids (Fig. 5). We infected human fibroblasts with equal amounts of NIFV and integrase-proficient CnZPNO genome-containing particles, and the number of G418-resistant colonies obtained was more than 4 logs higher for the integrase-proficient vector (4.91 × 105 versus 43 CFU per 107 input vector genomes).
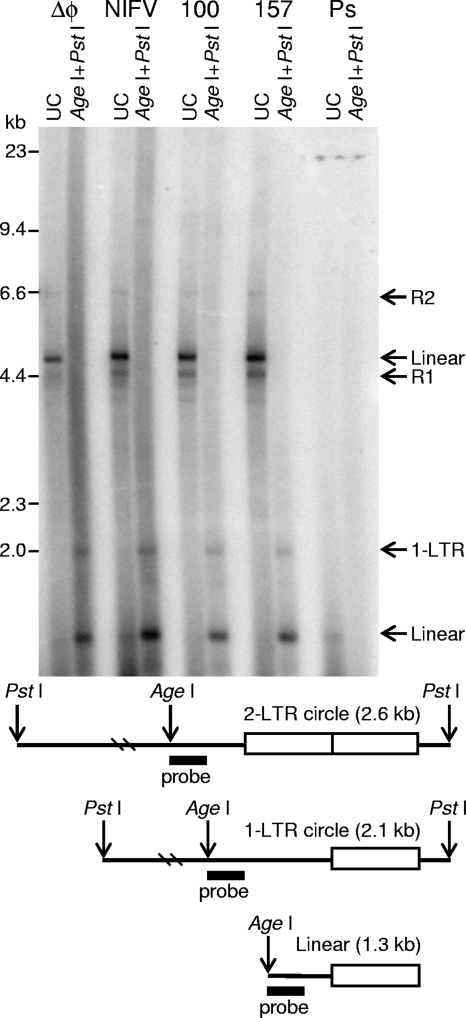
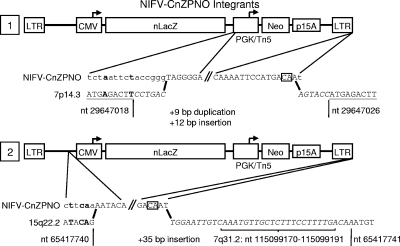
FIG. 5.
Sequence of vector-chromosome junctions from NIFV vector integrants. Two integrated proviruses were rescued as bacterial plasmids, and their junctions were sequenced. In each case, the junction-containing portions of the provirus are expanded below the vector map, and the predicted vector genome sequence is aligned above the chromosomal sequences obtained, with the junction points indicated by bold black lines. Sequences present in each provirus are in uppercase, and extensions of the vector sequences not seen in the rescued DNA are in lowercase. Insertions are denoted with italics, and duplications of the chromosomal DNA are underlined. Bold font indicates regions of microhomology between the vector and the chromosome, and the terminal CA dinucleotides are boxed. Specific nucleotide positions of the chromosomal sequences are indicated.
Integrated proviruses were rescued along with flanking chromosomal DNA from two G418-resistant clones transduced with NIFV-CnZPNO by restriction enzyme digestion of genomic DNA, circularization with ligase, electroporation of bacteria, and isolation of kanamycin-resistant colonies. Sequencing of the left and right junctions showed that integration was not mediated by integrase, which should have produced a 4-bp duplication of host DNA and a provirus with processed LTRs beginning with a TG dinucleotide at the 5′ junction and terminating with a CA dinucleotide at the 3′ junction (12, 40). One integrant was present on chromosome 7 and had a 7.1-kb deletion of vector sequences and a duplication of 9 bp of host DNA at the 5′ junction, distinct 6-bp insertions at both junctions, and an extra base at the 3′ LTR terminus (Fig. 5). The second integrant was present on chromosome 15 and had a 2.7-kb deletion of vector sequences at the 5′ junction and a 35-bp insertion at the 3′ junction (of which 22 bp were homologous to sequences on chromosome 7). These data demonstrate that rare NIFV integration events are not mediated by residual FV integrase activity but instead are consistent with ligation via nonhomologous end joining by host enzymes (32, 56). The 3′ LTRs also included one or two additional bases that are normally removed by FV integrase prior to integration.
Expression of Cre recombinase from a NIFV vector.
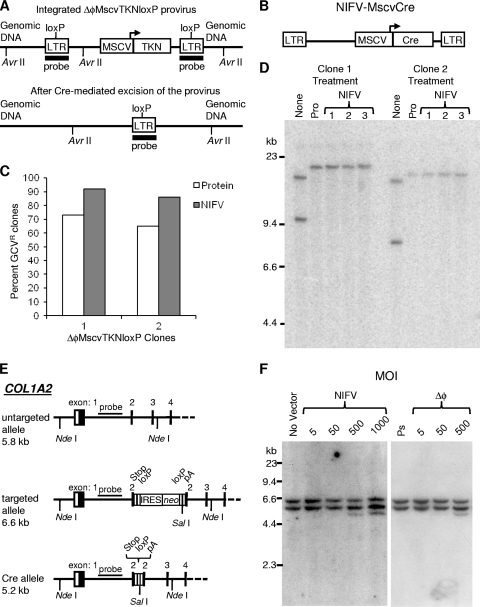
As a potential practical application, we tested whether Cre recombinase could be transiently expressed from a NIFV vector and permanently alter the host genome. We first created the floxed FV vector, ΔφMscvTKNloxP, containing a thymidine kinase-neo positive-negative selection cassette and a loxP site in the 3′ LTR that is copied to the 5′ LTR during replication (Fig. 6A). After HT-1080 cells were infected with ΔφMscvTKNloxP, followed by selection in G418, individual clones were isolated, expanded, and analyzed by Southern blotting. Two single integrant clones were identified and infected with NIFV-MscvCre, which expresses a mammalian codon-optimized Cre under the control of the MSCV promoter, and then selected with ganciclovir for loss of the thymidine kinase gene (Fig. 6B). Ninety-two and eighty-six percent of cells from clones 1 and 2, respectively, were ganciclovir resistant after infection with NIFV-MscvCre, compared to 73 and 65% of cells treated with the Cre protein (Fig. 6C). Southern blot analysis of isolated ganciclovir-resistant subclones revealed that the floxed FV vector had been removed as expected by recombination at the loxP sites, leaving behind a single deleted FV LTR. These results show that Cre can be expressed at functional levels from NIFV vectors and thereby used to accurately excise a floxed cassette from the host cell genome.
FIG. 6.
Cre recombinase is expressed from a NIFV vector. (A) Schematic representation of an integrated ΔφMscvTKNloxP provirus containing a thymidine kinase-neo fusion gene (TKN) driven by an internal MSCV promoter and the same genomic locus after Cre-mediated excision. The positions of loxP sites and AvrII restriction sites are indicated. (B) Illustration of the NIFV-MscvCre vector with its MSCV promoter and mammal-optimized Cre transgene (Cre). (C) Graph showing the percentage of ganciclovir-resistant (GCVR) colonies obtained after transduction of ΔφMscvTKNloxP-containing clones with the Cre protein (protein) or NIFV-MscvCre (NIFV). (D) Southern blot analysis of DNA isolated from clones 1 and 2 after no treatment with Cre (None), treatment with Cre protein (Pro), or infection with NIFV-MscvCre (NIFV). The DNAs were digested with AvrII and probed for LTR sequences. (E) Schematic representation of the COL1A2 locus showing the untargeted allele, the targeted allele with insertion of the IRES-neo cassette, and the targeted allele after excision of IRES-neo by Cre recombinase. (F) Southern blot of NdeI/SalI-digested genomic DNA isolated from the parental, OIMSC12-targeted clone (No Vector), cells infected with NIFV-MscvCre at MOIs of 5, 50, 500, and 1,000 genome-containing particles per cell, cells infected with ΔφMscvCre at MOIs of 5, 50, and 500 genome-containing particles per cell, or the highest volume equivalent of Cre pseudostock (Ps), as indicated. Expected fragment sizes are 5.8 kb (untargeted allele), 6.6 kb (targeted allele), and 5.2 kb (cre-excised locus) after probing for COL1A2 sequences as indicated in panel E.
We also tested if the NIFV-MscvCre vector could remove a floxed transgene in gene-targeted mesenchymal stem cells (MSCs). These MSCs were obtained from an individual with osteogenesis imperfecta (OI) due to a dominant-negative mutation in exon 34 of the COL1A2 gene. Previously we used an AAV gene-targeting vector to insert an internal ribosome entry site (IRES)-neo cassette into exon 2 of COL1A2 and eliminate abnormal collagen expression from the mutant allele in these MSCs (6). loxP sites flank the IRES-neo cassette, allowing us to remove the potentially antigenic neo gene before these cells would be transplanted in a clinical setting. MSCs targeted at exon 2 of COL1A2 were infected with ΔφMscvCre, NIFV-MscvCre, or a pseudostock control (prepared without the pol helper plasmid), expanded, and analyzed by Southern blotting. Cre-mediated excision of the IRES-neo cassette decreases the size of the 6.6-kb NdeI-SalI fragment to 5.2 kb (Fig. 6E). After transduction, removal of the IRES-neo cassette occurred in <1% of the targeted alleles treated with highest dose of Cre pseudostock and <1, 6, 24, and 36% of the targeted alleles treated with NIFV-MscvCre at MOIs of 5, 50, 500, and 1,000 genome-containing particles/cell, respectively (Fig. 6F). MSCs infected with ΔφMscvCre at MOIs of 5, 50, and 500 genome-containing particles/cell had excision of the IRES-neo cassette in 3, 12, and 36% of the targeted alleles, consistent with our prior finding that integrase-proficient FV vectors express their transgenes at higher levels than NIFV vectors (Fig. 3). Of course, the integrating vector produces permanent Cre expression, which is undesirable in most applications.
DISCUSSION
In this report, we have shown for the first time that NIFV vectors can efficiently transduce cells and express multiple transgenes. Consistent with results reported in previous articles, mutations in the FV integrase catalytic core specifically inhibited the integration of the FV provirus but did not alter vector production. The residual integration frequency of NIFV vectors was logs lower than that of integrase-proficient vectors and occurred by an integrase-independent mechanism. NIFV vectors expressing the Cre recombinase were able to mediate excision of floxed cassettes, including an integrated, floxed FV vector provirus that can be used for stable, removable transgene expression. NIFV vectors were not expressed in quiescent cells.
Previous studies did not detect transgene expression from a similar NIFV vector system (12, 19, 20). However, in our experiments, we used the same D157A mutation but detected robust GFP expression. There are two main differences between our vectors and the previously described NIFV system that may explain this discrepancy: the expression cassette and the stock production protocol. We employed a strong MSCV promoter in our vectors, and the previous group used the spleen focus-forming virus promoter (3). However, expression from these two promoters has been shown to be comparable (28). Instead, the most likely explanation is that the prior study used unconcentrated vector stocks with undetermined titers, while we concentrated our NIFV vectors more than 100-fold and infected with known MOIs of genome-containing particles. Thus, our experiments used significantly higher vector doses that enabled us to detect transgene expression.
Other retroviruses, such as LV and MLV, reverse transcribe their double-stranded RNA genomes into cDNA after entering the host cell (7), and these cDNAs can either integrate into the host genome or persist as linear, 1-LTR, or 2-LTR episomal molecules. 1-LTR circles are formed either by homologous recombination between the two LTRs (15, 52) or from a circular reverse transcription intermediate during second-strand transfer (11, 26, 37), while 2-LTR circles are thought to form by nonhomologous end joining (NHEJ) of the linear DNA (27, 31, 53). FV genomes also persist as linear, 1-LTR, and 2-LTR episomes in transduced cells, but unlike the case with other retroviruses, reverse transcription occurs in the virion before it is released from the virus-producing cell (38). Interestingly, we found that 1-LTR circular genomes were present in FV vector stocks before the virions entered host cells. Therefore, homologous recombination enzymes present in the infected cell nucleus are not required to produce these molecules, and our data are consistent with the production of 1-LTR circles from a reverse transcription intermediate (11, 26, 37). The same may be true for LV and MLV vectors, but this distinction cannot be made, since reverse transcription occurs after entry into the host cell. In contrast, 2-LTR circular forms were observed only in infected cells and not in vector stocks, indicating that cellular proteins are required to produce these forms, as expected for NHEJ, and suggesting that this form can be used as a specific marker of nuclear entry. Interestingly, this is not the case for wt FV stocks, which can contain 2-LTR circular forms (10, 55). This may be a unique feature of replication-competent FV, possibly resulting from viral replication during cellular infection.
In order to complete our studies, we needed to standardize our vector stocks. FV vector stocks contain a 5:1 ratio of RNA to DNA genomes, but only the DNA-containing particles have been shown to be infectious (38, 46, 61, 62). For this reason, we developed a Southern blot method that allows for a reliable determination of vector genome titers. This allowed us to show that the gene expression level of an NIFV vector was 1.5- to 4.3-fold lower than that of its integrase-proficient counterpart after infection at the same MOI. The reason for this gene expression difference is unknown, but this is an area where NIFV vectors may be improved in the future. The vector genome titer-determining method also allowed us to calculate particle-to-transducing-unit ratios for FV vectors. This could be a useful quality control analysis if FV vectors are used clinically.
NIFV vectors, with their broad tropism, large packing capacity, and lack of pathogenicity, have many useful applications. Interfering RNAs could be expressed from NIFV vectors to manipulate target cell gene expression transiently and then revert to a normal phenotype. The inclusion of a replication origin could produce replicating NIFV vectors, particularly the circular episomal forms. NIFV vectors could be used to deliver targeting constructs for homologous recombination either alone or in concert with nucleases, such as zinc finger nucleases. Certain applications may require stable transgene expression in proliferating cells for an extended but temporary period of time, such as generation of induced pluripotent stem cells. While NIFV vectors would not be appropriate for this purpose, the floxed FV vector we described would be ideal, and the integrated provirus could be removed at any time by infection with an NIFV-Cre vector. The residual, deleted LTR remnant produced by Cre-mediated excision would have minimal genotoxicity since it is transcriptionally silent (21).
Footnotes
Published ahead of print on 30 June 2010.
REFERENCES
- 1.Apolonia, L., S. N. Waddington, C. Fernandes, N. J. Ward, G. Bouma, M. P. Blundell, A. J. Thrasher, M. K. Collins, and N. J. Philpott. 2007. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol. Ther. 15:1947-1954. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, T. R., Jr., J. M. Allen, M. Hai, L. M. Tuschong, I. F. Khan, E. M. Olson, R. L. Adler, T. H. Burkholder, Y. C. Gu, D. W. Russell, and D. D. Hickstein. 2008. Successful treatment of canine leukocyte adhesion deficiency by foamy virus vectors. Nat. Med. 14:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baum, C., S. Hegewisch-Becker, H. Eckert, C. Stocking, and W. Ostertag. 1995. Novel retroviral vectors for efficient expression of the multidrug resistance (mdr-1) gene in early hematopoietic cells. J. Virol. 69:7541-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer, M., B. Kantor, A. Cockrell, H. Ma, B. Zeithaml, X. Li, T. McCown, and T. Kafri. 2008. A large U3 deletion causes increased in vivo expression from a nonintegrating lentiviral vector. Mol. Ther. 16:1968-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 6.Chamberlain, J. R., D. R. Deyle, U. Schwarze, P. Wang, R. K. Hirata, Y. Li, P. H. Byers, and D. W. Russell. 2008. Gene targeting of mutant COL1A2 alleles in mesenchymal stem cells from individuals with osteogenesis imperfecta. Mol. Ther. 16:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin, J. M., S. H. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 8.Cornu, T. I., and T. Cathomen. 2007. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 15:2107-2113. [DOI] [PubMed] [Google Scholar]
- 9.Delelis, O., C. Petit, H. Leh, G. Mbemba, J. F. Mouscadet, and P. Sonigo. 2005. A novel function for spumaretrovirus integrase: an early requirement for integrase-mediated cleavage of 2 LTR circles. Retrovirology 2:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delelis, O., A. Saib, and P. Sonigo. 2003. Biphasic DNA synthesis in spumaviruses. J. Virol. 77:8141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dina, D., and E. W. Benz, Jr. 1980. Structure of murine sarcoma virus DNA replicative intermediates synthesized in vitro. J. Virol. 33:377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enssle, J., A. Moebes, M. Heinkelein, M. Panhuysen, B. Mauer, M. Schweizer, D. Neumann-Haefelin, and A. Rethwilm. 1999. An active foamy virus integrase is required for virus replication. J. Gen. Virol. 80(Pt. 6):1445-1452. [DOI] [PubMed] [Google Scholar]
- 13.Falcone, V., M. Schweizer, and D. Neumann-Haefelin. 2003. Replication of primate foamy viruses in natural and experimental hosts. Curr. Top. Microbiol. Immunol. 277:161-180. [DOI] [PubMed] [Google Scholar]
- 14.Gharwan, H., R. K. Hirata, P. Wang, R. E. Richard, L. Wang, E. Olson, J. Allen, C. B. Ware, and D. W. Russell. 2007. Transduction of human embryonic stem cells by foamy virus vectors. Mol. Ther. 15:1827-1833. [DOI] [PubMed] [Google Scholar]
- 15.Gilboa, E., S. Goff, A. Shields, F. Yoshimura, S. Mitra, and D. Baltimore. 1979. In vitro synthesis of a 9 kbp terminally redundant DNA carrying the infectivity of Moloney murine leukemia virus. Cell 16:863-874. [DOI] [PubMed] [Google Scholar]
- 16.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-72. [DOI] [PubMed] [Google Scholar]
- 17.Haas, D. L., S. S. Case, G. M. Crooks, and D. B. Kohn. 2000. Critical factors influencing stable transduction of human CD34(+) cells with HIV-1-derived lentiviral vectors. Mol. Ther. 2:71-80. [DOI] [PubMed] [Google Scholar]
- 18.Hawley, R. G., F. H. Lieu, A. Z. Fong, and T. S. Hawley. 1994. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1:136-138. [PubMed] [Google Scholar]
- 19.Heinkelein, M., T. Pietschmann, G. Jarmy, M. Dressler, H. Imrich, J. Thurow, D. Lindemann, M. Bock, A. Moebes, J. Roy, O. Herchenroder, and A. Rethwilm. 2000. Efficient intracellular retrotransposition of an exogenous primate retrovirus genome. EMBO J. 19:3436-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinkelein, M., M. Rammling, T. Juretzek, D. Lindemann, and A. Rethwilm. 2003. Retrotransposition and cell-to-cell transfer of foamy viruses. J. Virol. 77:11855-11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrie, P. C., Y. Huo, R. B. Stolitenko, and D. W. Russell. 2008. A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol. Ther. 16:534-540. [DOI] [PubMed] [Google Scholar]
- 22.Heneine, W., M. Schweizer, P. Sandstrom, and T. Folks. 2003. Human infection with foamy viruses. Curr. Top. Microbiol. Immunol. 277:181-196. [DOI] [PubMed] [Google Scholar]
- 23.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 24.Josephson, N. C., G. Trobridge, and D. W. Russell. 2004. Transduction of long-term and mobilized peripheral blood-derived NOD/SCID repopulating cells by foamy virus vectors. Hum. Gene Ther. 15:87-92. [DOI] [PubMed] [Google Scholar]
- 25.Josephson, N. C., G. Vassilopoulos, G. D. Trobridge, G. V. Priestley, B. L. Wood, T. Papayannopoulou, and D. W. Russell. 2002. Transduction of human NOD/SCID-repopulating cells with both lymphoid and myeloid potential by foamy virus vectors. Proc. Natl. Acad. Sci. U. S. A. 99:8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Junghans, R. P., L. R. Boone, and A. M. Skalka. 1982. Products of reverse transcription in avian retrovirus analyzed by electron microscopy. J. Virol. 43:544-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz, R. A., C. A. Omer, J. H. Weis, S. A. Mitsialis, A. J. Faras, and R. V. Guntaka. 1982. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J. Virol. 42:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S., K. Lee, M.-D. Kim, S. Kang, C. W. Joo, J.-M. Kim, S. H. Kim, S. S. Yu, and S. Kim. 2006. Factors affecting the performance of different long terminal repeats in the retroviral vector. Biochem. Biophys. Res. Commun. 343:1017-1022. [DOI] [PubMed] [Google Scholar]
- 29.Leavitt, A., G. Robles, N. Alesandro, and H. Varmus. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leurs, C., M. Jansen, K. E. Pollok, M. Heinkelein, M. Schmidt, M. Wissler, D. Lindemann, C. von Kalle, A. Rethwilm, D. A. Williams, and H. Hanenberg. 2003. Comparison of three retroviral vector systems for transduction of nonobese diabetic/severe combined immunodeficiency mice repopulating human CD34+ cord blood cells. Hum. Gene Ther. 14:509-519. [DOI] [PubMed] [Google Scholar]
- 31.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, Y., and A. S. Waldman. 2001. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics 158:1665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo, Y. T., T. Tian, P. E. Nadeau, J. Park, and A. Mergia. 2010. The foamy virus genome remains unintegrated in the nuclei of G1/S phase-arrested cells, and integrase is critical for preintegration complex transport into the nucleus. J. Virol. 84:2832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lombardo, A., P. Genovese, C. M. Beausejour, S. Colleoni, Y. L. Lee, K. A. Kim, D. Ando, F. D. Urnov, C. Galli, P. D. Gregory, M. C. Holmes, and L. Naldini. 2007. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 25:1298-1306. [DOI] [PubMed] [Google Scholar]
- 35.Meiering, C. D., K. E. Comstock, and M. L. Linial. 2000. Multiple integrations of human foamy virus in persistently infected human erythroleukemia cells. J. Virol. 74:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, D. G., L. M. Petek, and D. W. Russell. 2003. Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol. Cell. Biol. 23:3550-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, M., B. Wang, and F. Bushman. 1995. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J. Virol. 69:3938-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moebes, A., J. Enssle, P. D. Bieniasz, M. Heinkelein, D. Lindemann, M. Bock, M. O. McClure, and A. Rethwilm. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 40.Neves, M., J. Peries, and A. Saib. 1998. Study of human foamy virus proviral integration in chronically infected murine cells. Res. Virol. 149:393-401. [DOI] [PubMed] [Google Scholar]
- 41.Nightingale, S. J., R. P. Hollis, K. A. Pepper, D. Petersen, X. J. Yu, C. Yang, I. Bahner, and D. B. Kohn. 2006. Transient gene expression by nonintegrating lentiviral vectors. Mol. Ther. 13:1121-1132. [DOI] [PubMed] [Google Scholar]
- 42.Philippe, S., C. Sarkis, M. Barkats, H. Mammeri, C. Ladroue, C. Petit, J. Mallet, and C. Serguera. 2006. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 103:17684-17689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philpott, N. J., and A. J. Thrasher. 2007. Use of nonintegrating lentiviral vectors for gene therapy. Hum. Gene Ther. 18:483-489. [DOI] [PubMed] [Google Scholar]
- 44.Rasheed, S., W. A. Nelson-Rees, E. M. Toth, P. Arnstein, and M. B. Gardner. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027-1033. [DOI] [PubMed] [Google Scholar]
- 45.Rethwilm, A. 2007. Foamy virus vectors: an awaited alternative to gammaretro- and lentiviral vectors. Curr. Gene Ther. 7:261-271. [DOI] [PubMed] [Google Scholar]
- 46.Roy, J., W. Rudolph, T. Juretzek, K. Gartner, M. Bock, O. Herchenroder, D. Lindemann, M. Heinkelein, and A. Rethwilm. 2003. Feline foamy virus genome and replication strategy. J. Virol. 77:11324-11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell, D., and A. Miller. 1996. Foamy virus vectors. J. Virol. 70:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saenz, D. T., N. Loewen, M. Peretz, T. Whitwam, R. Barraza, K. G. Howell, J. M. Holmes, M. Good, and E. M. Poeschla. 2004. Unintegrated lentivirus DNA persistence and accessibility to expression in nondividing cells: analysis with class I integrase mutants. J. Virol. 78:2906-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saib, A., F. Puvion-Dutilleul, M. Schmid, J. Peries, and H. de The. 1997. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J. Virol. 71:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salomon, B., S. Maury, L. Loubiere, M. Caruso, R. Onclercq, and D. Klatzmann. 1995. A truncated herpes simplex virus thymidine kinase phosphorylates thymidine and nucleoside analogs and does not cause sterility in transgenic mice. Mol. Cell. Biol. 15:5322-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz, F., N. Maeda, O. Smithies, R. Hickey, W. Edelmann, A. Skoultchi, and R. Kucherlapati. 1991. A dominant positive and negative selectable gene for use in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 88:10416-10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shank, P. R., S. H. Hughes, H. J. Kung, J. E. Majors, N. Quintrell, R. V. Guntaka, J. M. Bishop, and H. E. Varmus. 1978. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell 15:1383-1395. [DOI] [PubMed] [Google Scholar]
- 53.Swanstrom, R., W. J. DeLorbe, J. M. Bishop, and H. E. Varmus. 1981. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc. Natl. Acad. Sci. U. S. A. 78:124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trobridge, G., N. Josephson, G. Vassilopoulos, J. Mac, and D. W. Russell. 2002. Improved foamy virus vectors with minimal viral sequences. Mol. Ther. 6:321-328. [DOI] [PubMed] [Google Scholar]
- 55.Trobridge, G., and D. W. Russell. 2004. Cell cycle requirements for transduction by foamy virus vectors compared to those of oncovirus and lentivirus vectors. J. Virol. 78:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varga, T., and P. D. Aplan. 2005. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair 4:1038-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vargas, J., Jr., G. L. Gusella, V. Najfeld, M. E. Klotman, and A. Cara. 2004. Novel integrase-defective lentiviral episomal vectors for gene transfer. Hum. Gene Ther. 15:361-372. [DOI] [PubMed] [Google Scholar]
- 58.Vassilopoulos, G., G. Trobridge, N. C. Josephson, and D. W. Russell. 2001. Gene transfer into murine hematopoietic stem cells with helper-free foamy virus vectors. Blood 98:604-609. [DOI] [PubMed] [Google Scholar]
- 59.Wanisch, K., and R. J. Yanez-Munoz. 2009. Integration-deficient lentiviral vectors: a slow coming of age. Mol. Ther. 17:1316-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanez-Munoz, R. J., K. S. Balaggan, A. MacNeil, S. J. Howe, M. Schmidt, A. J. Smith, P. Buch, R. E. MacLaren, P. N. Anderson, S. E. Barker, Y. Duran, C. Bartholomae, C. von Kalle, J. R. Heckenlively, C. Kinnon, R. R. Ali, and A. J. Thrasher. 2006. Effective gene therapy with nonintegrating lentiviral vectors. Nat. Med. 12:348-353. [DOI] [PubMed] [Google Scholar]
- 61.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]
- 62.Yu, S. F., M. D. Sullivan, and M. L. Linial. 1999. Evidence that the human foamy virus genome is DNA. J. Virol. 73:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu, S. S., K. Dan, H. Chono, E. Chatani, J. Mineno, and I. Kato. 2008. Transient gene expression mediated by integrase-defective retroviral vectors. Biochem. Biophys. Res. Commun. 368:942-947. [DOI] [PubMed] [Google Scholar]