Abstract
HIV-1 R5 envelopes vary considerably in their capacities to exploit low CD4 levels on macrophages for infection and in their sensitivities to the CD4 binding site (CD4bs) monoclonal antibody (MAb) b12 and the glycan-specific MAb 2G12. Here, we show that nonglycan determinants flanking the CD4 binding loop, which affect exposure of the CD4bs, also modulate 2G12 neutralization. Our data indicate that such residues act via a mechanism that involves shifts in the orientation of proximal glycans, thus modulating the sensitivity of 2G12 neutralization and affecting the overall presentation and structure of the glycan shield.
The trimeric envelope (Env) spikes on HIV-1 virions are comprised of gp120 and gp41 heterodimers. gp120 is coated extensively with glycans (9, 11, 15) that are believed to protect the envelope from neutralizing antibodies. The extents and locations of glycosylation are variable and evolving (15). Thus, while some glycans are conserved, others appear or disappear in a host over the course of infection. Such changes may result in exposure or protection of functional envelope sites and can result from selection by different environmental pressures in vivo, including neutralizing antibodies.
We previously reported that HIV-1 R5 envelopes varied considerably in tropism and neutralization sensitivity (3, 4, 12-14). We showed that highly macrophage-tropic R5 envelopes were more frequently detected in brain than in semen, blood, and lymph node (LN) samples (12, 14). The capacity of R5 envelopes to infect macrophages correlated with their ability to exploit low levels of cell surface CD4 for infection (12, 14). Determinants within and proximal to the CD4 binding site (CD4bs) were shown to modulate macrophage infectivity (3, 4, 5, 12, 13) and presumably acted by altering the avidity of the trimer for cell surface CD4. These determinants include residues proximal to the CD4 binding loop, which is likely the first part of the CD4bs contacted by CD4 (1). We also observed that macrophage-tropic R5 envelopes were frequently more resistant to the glycan-specific monoclonal antibody (MAb) 2G12 than were non-macrophage-tropic R5 Envs (13).
Here, we investigated the envelope determinants of 2G12 sensitivity by using two HIV-1 envelopes that we used previously to map macrophage tropism determinants (4), B33 from brain and LN40 from lymph node tissue of an AIDS patient with neurological complications. While B33 imparts high levels of macrophage infectivity and is resistant to 2G12, LN40 Env confers very inefficient macrophage infection and is 2G12 sensitive (12-14).
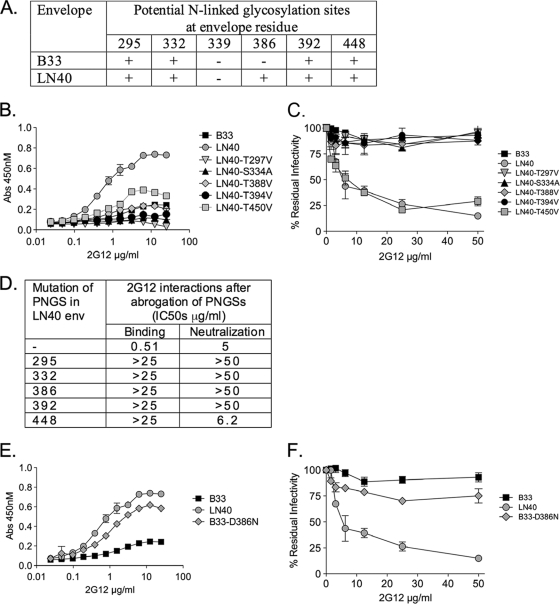
Mapping potential N-linked glycosylation sites implicated in 2G12 binding: role of the glycan at N386.
We first investigated the potential N-linked glycosylation sites (PNGSs) on LN40 that are targeted by 2G12. Previous studies showed that the most important PNGSs for binding 2G12 are at positions 295, 332, 339, and 392 (2, 7, 17-19), while PNGSs at positions 386 and 448 were reported to play an indirect role (7, 16, 17) (Fig. 1A). The LN40 envelope contains PNGSs at residues 295, 332, 386, 392, and 448 but lacks the one at residue 339. We constructed LN40 mutants to individually remove the implicated PNGSs (LN40-T297V, LN40-S334A, LN40-T388V, LN40-T394V, and LN40-T450V mutants, respectively) and evaluated their effect on 2G12 binding and neutralization. We used an LN40 construct that carried gp41 from B33 and used “single-round” pseudovirions (3, 4, 12-14). We employed an enzyme-linked immunosorbent assay (ELISA) where monomeric gp120 was captured from pseudovirions and 2G12 binding tested (3, 4). We found that LN40 lost binding to 2G12 when PNGSs at positions 295, 332, 386, and 392 were eliminated but retained low binding with the PNGS at position 448 removed (Fig. 1B). Similar results were obtained when these mutant Envs were tested for 2G12 neutralization, although surprisingly, efficient neutralization was observed for LN40 lacking the PNGS at position 448 (Fig. 1C). Of note, restoration of the “missing” PNGS at residue 339 did not confer 2G12 sensitivity to resistant wild-type B33 (B33wt) or LN40 lacking the PNGS at N386 (not shown). These results indicate that the glycans at N295, N332, and N392 are critical for 2G12 binding to the LN40 envelope and implicate a role for N386 (Fig. 1D).
FIG. 1.
Potential N-linked glycosylation sites involved in 2G12 binding and neutralization of Env+ pseudovirions. Presence of PNGSs implicated in 2G12 binding in LN40 and B33 envelopes (A). PNGSs were eliminated at N295 (T297V), N332 (S334A), N386 (T388V), N392 (T394V), and N448 (T450V). LN40 envelope mutants that lack each specific PNGS were tested for binding to 2G12 by ELISA (B) and for sensitivity to 2G12 neutralization by using HeLa TZM-bl cells to estimate residual infectivity (C). Removal of PNGSs at N295, N332, N386, and N392 led to a loss of 2G12 binding and neutralization. Removal of the PNGS at N448 conferred substantially reduced 2G12 binding, even though this mutant retained sensitivity to neutralization. These data are summarized in a table (D). Reconstitution of a PNGS at residue 386 in B33 confers 2G12 binding to monomeric gp120 in an ELISA (E) but only weak neutralization (F). The binding and neutralization assays shown were repeated at least once and usually multiple times. The ELISA results presented (B and E) are each from one representative experiment, with error bars representing standard deviations from triplicate readings. The neutralization results (C and F) represent data from two separate experiments, with error bars representing standard deviations. Abs, absorbance; IC50, 50% inhibitory concentration.
The glycan at N386 is absent in B33. To further test the role of N386 in 2G12 neutralization, we constructed a B33-D386N (B33-N386) mutant, which restores the PNGS at residue 386. This substitution conferred efficient 2G12 binding to the monomer (Fig. 1E) but only marginal neutralization (Fig. 1F). Nonetheless, these results confirm that the PNGS at N386 plays an important role in 2G12 binding and neutralization for both LN40 and B33 mutants carrying the N386 PNGS.
Mutations that affect the CD4bs impact the recognition of the 2G12 epitope.
Previously, we showed that residues within and proximal to the CD4bs modulated the capacity of B33 and LN40 envelopes to use low levels of CD4 for macrophage infection (3, 4). We next evaluated whether these residues also affected sensitivity to 2G12. The following LN40 mutants were tested for 2G12 sensitivity: LN40-T283N [LN40-N], LN40-Q363P/P364S (LN40-PS), and LN40-T283N/Q363P/P364S (LN40-N283-PS) mutants (Fig. 2A and B). These mutants carried substitutions that increased macrophage infectivity via low levels of CD4 (3, 4). Each of these mutants conferred decreases in 2G12 sensitivity (Fig. 2B). We also investigated the reverse substitutions using B33 carrying N386, which conferred 2G12 binding but not neutralization (Fig. 1E and F). Together, the B33 substitutions conferred increased sensitivities to 2G12 neutralization (Fig. 2C).
FIG. 2.
Residues that impact CD4 binding affect 2G12 neutralization. Substitutions within and proximal to the CD4 binding site were investigated for their effects on 2G12 neutralization. HeLa TZM-bl cells were used to estimate residual infectivity. (A) Alignment of LN40 and B33 from N283 to the start of the V4 loop. Residues investigated in experiments with results shown in panels B and C are boxed. The stars indicate CD4 contact residues. b. loop, binding loop. (B) LN40 substitutions in the C2 part of the CD4bs (T283N) and on the N-terminal flank of the CD4 binding loop (Q363P/P364S) conferred decreased sensitivity to 2G12. (C) The reverse substitutions in B33 conferred increased sensitivity to 2G12. (D) A complex B33 mutant that has low avidity for CD4 is substantially resistant to 2G12. The B33-T283-SHFE-INQP-RN386 mutant is non-macrophage-tropic and resistant to soluble CD4 (sCD4) yet nearly completely resistant to 2G12. In contrast, the B33-T283-HF-QP-RN386 mutant, which contains fewer substitutions than and has the same non-macrophage-tropic phenotype as the B33-T283-SHFE-INQP-RN386 mutant, is sensitive to 2G12. This result shows that CD4 avidity per se is not directly related to 2G12 sensitivity. Nonetheless, the data shown are consistent with the possibility that the overall orientation of the glycans present on the trimer influences 2G12 binding and sensitivity to neutralization. The table shown in panel E summarizes the properties of the envelopes tested in the experiments with results shown in panel D. Mac, macrophage; +++, highly mac-tropic; −, non-mac-tropic. The results from the neutralization assays represent data from two independent experiments, with error bars representing standard deviations.
These observations reveal that substitutions which affect CD4 binding also have a strong effect on 2G12 neutralization. Since these residues are not in PNGSs, they may affect 2G12 interactions by shifting the orientation of proximal glycans. However, it is also possible that envelopes with a higher avidity for CD4 could confer altered neutralization properties and may escape by efficiently binding sufficient CD4 molecules on the cell surface to displace bound 2G12 antibody.
B33 mutant with low CD4 avidity that is resistant to 2G12.
We next describe a B33 mutant with low CD4 avidity that is substantially resistant to 2G12 and thus counters the possibility that high CD4 avidity is required for 2G12 resistance. The B33-T283-SHFE-INQP-RN386 mutant carries 11 substitutions for residues present in LN40 (Fig. 2A). This mutant, along with the B33-T283-HF-QP-RN386 mutant, was prepared previously to study determinants of macrophage tropism (4). Neither of these mutants infects macrophages, and both have low avidity for CD4, requiring high levels for infection (4). However, while the B33-T283-HF-QP-RN386 mutant was sensitive to 2G12, the B33-T283-SHFE-INQP-RN386 mutant was substantially resistant (Fig. 2D and E). This result separates CD4 avidity from 2G12 sensitivity but remains consistent with the possibility that changes in glycan orientation affect 2G12 sensitivity.
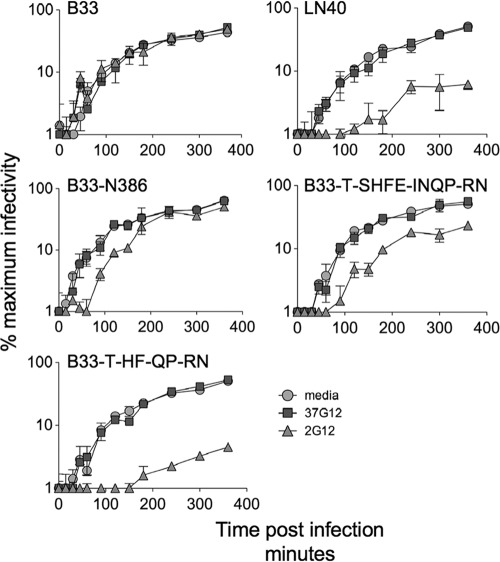
Entry kinetic assays provide evidence that 2G12 binds to Env+ pseudovirions that are substantially resistant to 2G12.
The 2G12-resistant B33-T283-SHFE-INQP-RN386 and B33-N386 envelopes carry all of the PNGSs implicated in 2G12 binding to LN40wt. We hypothesized that 2G12 resistance results from these glycans being inappropriately orientated. However, it was also possible that one or more of the PNGSs were not adequately glycosylated on these Envs to support 2G12 binding and neutralization. To investigate this, we reasoned that if 2G12 bound to trimers on virions, it might delay entry even if effective neutralization was not achieved. We tested whether 2G12 affected the entry of the B33-T283-SHFE-INQP-RN386 envelope (low CD4 avidity, 2G12 resistant) compared with that of the B33-N386 and B33wt envelopes (high CD4 avidity, 2G12 resistant) as well as that of the B33-T283-HF-QP-RN386 and LN40 envelopes (low CD4 avidity, 2G12 sensitive). Entry was investigated for Env+ pseudovirions treated or untreated with 50 μg/ml 2G12 or 37G12 [isotope control; anti-p24 MAb of the same IgG1(κ) isotype]. Env+ pseudovirions were then inhibited at different times after addition to cells by the addition of excess amounts of T20 (50 μg/ml), and entry was evaluated by β-galactosidase-induced luminescence at 48 h. Escape from T20 represents virions that have either been internalized into endosomes before fusion (10) or undergone sufficient gp41 conformational changes to become insensitive to T20.
For all Env+ pseudovirions, entry increased with time, peaking by 24 h. Figure 3 shows that 2G12 did not affect the kinetics of B33 but substantially reduced infection by both LN40 and the B33-T283-HF-QP-RN386 mutant. 2G12 also affected the entry of the B33-D386N and B33-T283-SHFE-INQP-RN386 mutants. At early times, both the B33-N386 and B33-T283-SHFE-INQP-RN386 mutants are neutralized by 2G12. However, there is increasing escape with time. These observations show that 2G12 bound Env trimers on the B33-N386 and B33-T283-SHFE-INQP-RN386 mutants but failed to sustain neutralization. This effect is likely to be mediated by faster 2G12 “off rates” for these two Envs than for LN40 and the B33-T283-HF-QP-RN386 mutant. The escape phenotypes of the B33-N386 (high CD4 avidity) and B33-T283-SHFE-INQP-RN386 (low CD4 avidity) mutants confirm that an increased avidity for CD4 is not the primary determinant of the variation in 2G12 sensitivity. These observations strongly suggest that the glycans on these two envelopes are not optimally aligned for 2G12 binding, which results in reduced affinity and neutralization resistance.
FIG. 3.
Delayed entry kinetics indicate that 2G12 binds to envelopes that are substantially resistant to 2G12. Delayed entry kinetics were conferred by B33 envelope mutants that bind 2G12 as monomeric gp120 but are substantially resistant to neutralization. Entry kinetics were evaluated by estimating the time required to escape T20 added at different times after infection. Entry kinetics of B33, LN40, and the B33-N386, B33-T283-SHFE-INQP-RN386, and B33-T283-HF-QP-RN386 mutants were similar. In the presence of 2G12, B33 kinetics were unaffected, consistent with the lack of binding by this MAb, while LN40 and B33-T283-HF-QP-RN386 mutant infectivity was reduced. In contrast, entry kinetics for the B33-N386 and B33-T283-SHFE-INQP-RN386 mutants were delayed, with almost complete neutralization registered at early times, followed by escape. 37G12 is an anti-p24 MAb (Polymun, Inc.) of the same Ig isotype [IgG1(κ)] as 2G12. 37G12 had no impact on the entry kinetics of the envelopes tested. These observations show that 2G12 binds B33-N386 and B33-T283-SHFE-INQP-RN386 trimeric Envs but fails to maintain neutralization. Error bars represent standard deviations.
Our data indicate that Env determinants within and proximal to the CD4bs that affect the exposure of CD4 contact residues may act by shifting the orientation of proximal glycans and in turn impact 2G12 binding. Although we studied only two envelopes, our data are likely to be generally applicable to HIV-1. For example, the upstream flank of the CD4 binding loop (shown here to affect 2G12 glycans) is highly variable. Several groups have shown that this region acts to protect or expose CD4 contact residues (3, 4, 20, 21) and that this is applicable to many HIV-1 envelopes (21). The glycan at N386 has been implicated in the protection of the CD4bs from antibodies (5, 6, 16) and is a prime candidate for such orientation shifts (Fig. 4). We showed previously that this glycan is involved in the resistance of LN40 to the CD4bs MAb b12. Moreover, the introduction of the N386 glycan into the B33 envelope (3) or into different strains (5, 6, 16) increased resistance to b12. Yet, the glycan at N386 is also present on the crystal structure of gp120 from the HXBc2 strain (22), which is highly sensitive to b12. These observations are consistent with a wide variation in orientation of the N386 glycan, sometimes allowing CD4 and b12 to bind and sometimes restricting access.
FIG. 4.
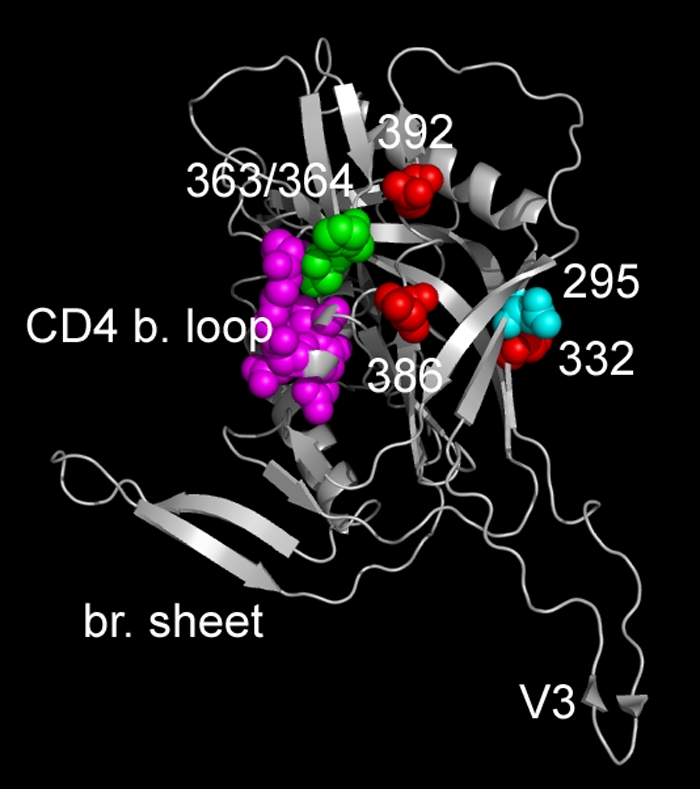
Positions of PNGSs and CD4 binding loop on a V3 loop containing CD4-bound HIV-1 gp120. The structure of a JR-FL core gp120 containing a V3 loop (CD4-bound form) is shown (8). Residues 295, 332, 386, and 392, which are sites for PNGSs important for LN40 neutralization (Fig. 1), are shown as either red (332, 386, and 392) or cyan (295) spheres. CD4 contact residues on the CD4 binding loop are shown as pink spheres, while adjacent residues (363 and 364) on the N-terminal flank of this loop are shown in green. For both B33 and LN40, substitutions at the latter residues altered sensitivity to 2G12. These residues are close to PNGSs at residues 386 and 392. br. sheet, bridging sheet.
In summary, we show that envelope residues not involved in glycosylation impact the sensitivity of 2G12 neutralization. Thus, determinants within and proximal to the CD4bs act to reorient glycans, resulting in altered 2G12 binding and neutralization. Our results highlight the constantly evolving configuration and structure of the glycan shield of gp120.
Acknowledgments
We thank Paul Peters for his insightful comments and input for this study.
Our work was supported by NIH grants R01 MH64408 and P01 AI082274.
We acknowledge the University of Massachusetts Center for AIDS Research (CFAR), the NIH AIDS Research and Reference Reagent Program, and the Centre for AIDS Reagents, NIBSC, United Kingdom, for services and reagents. We thank Sabine Hadulco at Roche Palo Alto for supplying T-20. We also thank Peter Simmonds (University of Edinburgh) and Jeanne Bell (MRC HIV Brain and Tissue Bank, University of Edinburgh) for providing the initial tissue that gave rise to the envelopes investigated here and for participation in the earlier studies of these envelopes.
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 2.Chen, H., X. Xu, A. Bishop, and I. M. Jones. 2005. Reintroduction of the 2G12 epitope in an HIV-1 clade C gp120. AIDS 19:833-835. [DOI] [PubMed] [Google Scholar]
- 3.Duenas-Decamp, M. J., P. Peters, D. Burton, and P. R. Clapham. 2008. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J. Virol. 82:5807-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duenas-Decamp, M. J., P. J. Peters, D. Burton, and P. R. Clapham. 2009. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J. Virol. 83:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunfee, R. L., E. R. Thomas, P. R. Gorry, J. Wang, J. Taylor, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. U. S. A. 103:15160-15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunfee, R. L., E. R. Thomas, J. Wang, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2007. Loss of the N-linked glycosylation site at position 386 in the HIV envelope V4 region enhances macrophage tropism and is associated with dementia. Virology 367:222-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray, E. S., P. L. Moore, R. A. Pantophlet, and L. Morris. 2007. N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. J. Virol. 81:10769-10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, W. R., W. J. Syu, B. Du, M. Matsuda, S. Tan, A. Wolf, M. Essex, and T. H. Lee. 1992. Nonrandom distribution of gp120 N-linked glycosylation sites important for infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 89:2213-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyauchi, K., Y. Kim, O. Latinovic, V. Morozov, and G. B. Melikyan. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137:433-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuochi, T., M. W. Spellman, M. Larkin, J. Solomon, L. J. Basa, and T. Feizi. 1988. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochem. J. 254:599-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters, P. J., J. Bhattacharya, S. Hibbitts, M. T. Dittmar, G. Simmons, J. Bell, P. Simmonds, and P. R. Clapham. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J. Virol. 78:6915-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters, P. J., M. J. Duenas-Decamp, W. M. Sullivan, R. Brown, C. Ankghuambom, K. Luzuriaga, J. Robinson, D. R. Burton, J. Bell, P. Simmonds, J. Ball, and P. Clapham. 2008. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters, P. J., W. M. Sullivan, M. J. Duenas-Decamp, J. Bhattacharya, C. Ankghuambom, R. Brown, K. Luzuriaga, J. Bell, P. Simmonds, J. Ball, and P. R. Clapham. 2006. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J. Virol. 80:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poon, A. F., F. I. Lewis, S. L. Pond, and S. D. Frost. 2007. Evolutionary interactions between N-linked glycosylation sites in the HIV-1 envelope. PLoS Comput. Biol. 3:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders, R. W., E. van Anken, A. A. Nabatov, I. M. Liscaljet, I. Bontjer, D. Eggink, M. Melchers, E. Busser, M. M. Dankers, F. Groot, I. Braakman, B. Berkhout, and W. A. Paxton. 2008. The carbohydrate at asparagine 386 on HIV-1 gp120 is not essential for protein folding and function but is involved in immune evasion. Retrovirology 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. O. Saphire, D. Calarese, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, D. R. Burton, and P. M. Rudd. 2003. The carbohydrate epitope of the neutralizing anti-HIV-1 antibody 2G12. Adv. Exp. Med. Biol. 535:205-218. [DOI] [PubMed] [Google Scholar]
- 20.Sterjovski, J., M. J. Churchill, A. Ellett, L. R. Gray, M. J. Roche, R. L. Dunfee, D. F. Purcell, N. Saksena, B. Wang, S. Sonza, S. L. Wesselingh, I. Karlsson, E. M. Fenyo, D. Gabuzda, A. L. Cunningham, and P. R. Gorry. 2007. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology 4:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, X., T. Zhou, S. O'Dell, R. T. Wyatt, P. D. Kwong, and J. R. Mascola. 2009. Mechanism of HIV-1 resistance to monoclonal antibody b12 that effectively targets the site of CD4 attachment. J. Virol. 83:10892-10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]