Abstract
Hepatitis C virus (HCV) core protein forms the nucleocapsid of the HCV particle. Although many functions of core protein have been reported, how the HCV particle is assembled is not well understood. Here we show that the nucleocapsid-like particle of HCV is composed of a disulfide-bonded core protein complex (dbc-complex). We also found that the disulfide-bonded dimer of the core protein (dbd-core) is formed at the endoplasmic reticulum (ER), where the core protein is initially produced and processed. Mutational analysis revealed that the cysteine residue at amino acid position 128 (Cys128) of the core protein, a highly conserved residue among almost all reported isolates, is responsible for dbd-core formation and virus-like particle production but has no effect on the replication of the HCV RNA genome or the several known functions of the core protein, including RNA binding ability and localization to the lipid droplet. The Cys128 mutant core protein showed a dominant negative effect in terms of HCV-like particle production. These results suggest that this disulfide bond is critical for the HCV virion. We also obtained the results that the dbc-complex in the nucleocapsid-like structure was sensitive to proteinase K but not trypsin digestion, suggesting that the capsid is built up of a tightly packed structure of the core protein, with its amino (N)-terminal arginine-rich region being concealed inside.
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma, affecting approximately 200 million people worldwide (13, 29, 44). Current treatment strategies, including interferon coupled with ribavirin, are not effective for all patients infected with HCV. An error-prone replication strategy allows HCV to undergo rapid mutational evolution in response to immune pressure and thus evade adaptive immune responses (10). New approaches to HCV therapy include the development of specifically targeted antiviral therapies for hepatitis C (STAT-Cs) which target such HCV proteins as the nonstructural 3/4A (NS3/4A), serine protease, and RNA-dependent RNA polymerase NS5B proteins (3). Despite the potent antiviral activities of some of these approaches, many resistant HCV strains have been reported after treatment with existing STAT-Cs (23, 48, 51). Therefore, identification of new targets that are common to all HCV strains and that are associated with low mutation rates is an area of active research.
HCV has a 9.6-kb, plus-strand RNA genome composed of a 5′ untranslated region (UTR), an open reading frame that encodes a single polyprotein of about 3,000 amino acids, and a 3′ UTR. The polyprotein is processed by host and viral proteases to produce three structural proteins (the core, envelope 1 [E1], and E2 proteins) and seven nonstructural proteins (the p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B proteins) (14, 16, 17, 22, 49). The HCV core protein is produced cotranslationally via carboxyl (C)-terminal cleavage to generate an immature core protein, 191 amino acids in length, on the endoplasmic reticulum (ER) (16). This protein consists of three predicted domains: the N-terminal hydrophilic domain (D1), the C-terminal hydrophobic domain (D2), and the tail domain (33), which serves as a signal peptide for the E1 protein. D1 includes a number of positively charged amino acids responsible for viral RNA binding (amino acids 1 to 75) (43) and the region involved in multimerization of the core protein via homotypic interactions (amino acids 36 to 91 and 82 to 102) (32, 40) (see Fig. S1 in the supplemental material). Hydrophobic D2 includes the region responsible for core protein association with lipid droplets (LDs; amino acids 125 to 144) (7, 18, 37), which accumulate in response to core protein production (1, 6).
Many functions of the core protein have been reported (13, 38, 50), yet because infectious HCV particles cannot be appropriately produced in currently available experimental systems, HCV particle assembly has not been elucidated to date. A cell culture system that reproduces the complete life cycle of HCV in vitro was developed by Wakita et al. using a cloned HCV genome (JFH1) (53). Using this system, the assembly of infectious HCV particles was found to occur near LDs and ER-derived LD-associated membranes (36, 47). Neither the structures nor the functions of the virus proteins involved in virus particle assembly are known, however. To elucidate this point, we have analyzed the biochemical characteristics of the proteins within the fraction containing the HCV particle and found a disulfide-bonded core protein complex (dbc-complex). We revealed that the disulfide-bonded dimer of core protein (dbd-core) was formed by a single cysteine residue at amino acid position 128 on the ER. The roles of the disulfide bond of the core protein in virus-like particle formation are discussed in this paper.
MATERIALS AND METHODS
Cell culture.
Cells of the HuH-7 and HuH-7.5 human hepatoma cell lines were grown in Dulbecco's modified Eagle's medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum, 100 U/ml nonessential amino acids (Invitrogen, Carlsbad, CA), and 100 μg/ml each penicillin and streptomycin sulfate (Invitrogen).
Antibodies.
The antibodies used for immunoblotting and indirect immunofluorescence analysis were specific for core protein (antibody 32-1), FLAG M2 (Sigma-Aldrich, St. Louis, MO), c-myc (Sigma-Aldrich), NS5A protein (CL1), adipocyte differentiation-related protein (ADRP; StressGen, Victoria, British, Columbia, Canada), calnexin (Calnexin-NT; StressGen), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Chemicon, Temecula, CA). Antibodies specific for core protein (antibody 32-1) were a gift from M. Kohara (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). Rabbit polyclonal anti-NS5A protein CL1 antibodies have been described previously (36).
Plasmid construction.
All plasmids were generated by inserting PCR-amplified fragments into expression plasmids. The plasmids, primer sequences, templates for the PCRs, and restriction enzyme sites used to construct the plasmids are listed in Table S1 in the supplemental material. Plasmids pJFH1E2FL (encoding the full-length HCV genome with the FLAG epitope in the E2 hypervariable region), pJFH1AAA99 (encoding a NS5A mutant of JFH1E2FL, resulting in noninfectious HCV particles), pJFH1PP/AA (encoding a core protein mutant of JFH1E2FL, which allows replication in cells but prevents HCV particle production), and pcDNA3-coreWT (an expression plasmid encoding the full-length core protein of JFH1) have been described previously (36). Plasmid pJ6/JFH1, which contains the full-length HCV genome encoding structural proteins from the J6 strain and nonstructural proteins from the JFH1 strain, was kindly provided by Charles M. Rice (The Rockefeller University, New York, NY).
In vitro transcription.
RNA for transfection was synthesized as described previously (36). In brief, plasmids carrying the HCV RNA sequence were linearized with XbaI and used as templates for in vitro transcription with MEGAscript T7 (Ambion, Austin, TX).
Transfection.
Ten micrograms of JFH1E2FL, JFH1C128A, JFH1C184A, JFH1C128/184A, JFH1PP/AA, or JFHAAA99 and J6/JFH1 or J6/JFH1AAA99 RNAs were transfected into HuH-7 and HuH-7.5 cells (1.0 × 107 cells) by electroporation (260 V, 0.95 μF) using a Gene Pulser II system (Bio-Rad, Hercules, CA). Core protein expression plasmids were transfected into HuH-7 cells using Lipofectamine LTX (Invitrogen), according to the manufacturer's protocol.
HCV particle precipitation.
Culture medium from HCV RNA-transfected cells were concentrated using Amicon Ultra-15 centrifugal filters with Ultracell-100 membranes (Millipore, Billerica, MA) and mixed with sucrose solution in phosphate-buffered saline (PBS) to a final sucrose concentration of 2%. This mixture was ultracentrifuged (100,000 × g, 4°C for 2 h), and the HCV particles were obtained as a pellet. The pellet was then suspended in culture medium for infection experiments or PBS for immunoblot analysis.
Indirect immunofluorescence analysis.
Indirect immunofluorescence analyses of HCV infection and the cellular localization of the HCV proteins were performed as described previously (36).
Protease protection assay.
Concentrated culture medium from JFH1E2FL RNA-transfected HuH-7 cells was fractionated using 20 to 50% sucrose density gradients, and the HCV RNA titer was measured in quantitative reverse transcription-PCRs (RT-PCRs) as described below. Fractions with high HCV RNA titers were collected, and JFH1E2FL particles were obtained as a pellet after ultracentrifugation (100,000 × g, 4°C for 2 h). The pellet was suspended in PBS and treated with 10 μg/ml trypsin or 5 μg/ml proteinase K in the presence or absence of 1% Nonidet P-40 (NP-40) at 37°C for 15 min, unless otherwise indicated. The reaction was quenched by the addition of protease inhibitor cocktail (Nacalai Tesque), followed by SDS-PAGE under nonreducing conditions and immunoblotting specific for core protein.
Immunoblot analysis.
Samples were subjected to SDS-PAGE in sample buffer (62.5 mM Tris-HCl [pH 7.8], 1% SDS, 10% glycerol) with or without 5% β-mercaptoethanol (β-ME) or 50 mM dithiothreitol (DTT) for reducing and nonreducing conditions, respectively. N-Ethylmaleimide (NEM; Nacalai Tesque) was added to the sample buffer to a final concentration of 5 mM for the indicated samples. Proteins were transferred to a polyvinylidene difluoride membrane and blocked in blocking buffer for 1 h at room temperature with gentle agitation. After incubation with primary antibodies overnight at 4°C, the membrane was washed three times for 5 min in washing buffer at room temperature with gentle agitation. The membrane was then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. After three washes in washing buffer, proteins were detected using Western Lightning reagent (PerkinElmer, Waltham, MA) or ECL Advance (GE Healthcare, Buckinghamshire, England) and Kodak MXJB Plus medical X-ray film (Kodak, Rochester, NY) or an LAS-4000 system (Fujifilm, Tokyo, Japan).
Preparation of LDs.
LDs were prepared as described previously (36).
Preparation of MMFs.
Microsomal membrane fractions (MMFs) were collected as described previously (15) with some modifications. In brief, cells were collected in homogenization buffer (20 mM Tris-HCl [pH 7.8], 250 mM sucrose, and 0.1% ethanol supplemented with protease inhibitor cocktail) and homogenized on ice using 40 strokes of a Dounce homogenizer. The samples were then centrifuged at 1,000 × g for 10 min at 4°C. The supernatant was collected in a new tube and centrifuged again at 16,000 × g for 20 min at 4°C. The supernatant was further centrifuged at 100,000 × g for 1 h at 4°C. The MMF precipitate was homogenized in lysis buffer (1% NP-40, 0.1% SDS, 20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, and 10% glycerol supplemented with protease inhibitor cocktail) using a Dounce homogenizer.
qRT-PCR analysis.
Quantitative RT-PCR (qRT-PCR) analysis for determination of the HCV RNA titer was performed as described previously (36).
ELISA specific for core protein.
The core protein in culture medium was quantified using an enzyme-linked immunosorbent assay (ELISA; HCV antigen ELISA; Ortho-Clinical Diagnostics, Raritan, NJ), according to the manufacturer's protocol.
RNA-protein binding precipitation assay.
CoreWT or coreC128A was translated in vitro from pcDNA3-coreWT and pcDNA3-coreC128A, respectively, using a TNT-coupled rabbit reticulocyte lysate system (Promega, Madison, WI), according to the manufacturer's protocol. These proteins were incubated with poly(U) agarose (Sigma) in binding buffer (50 mM HEPES (pH 7.4)-100 mM NaCl-0.1% NP-40-20 U RNase inhibitor) at 4°C for 2 h with or without RNase A. After five washes, the resin-bound core proteins were immunoblotted.
RESULTS
The HCV particle contains core protein complex formed by a disulfide bond.
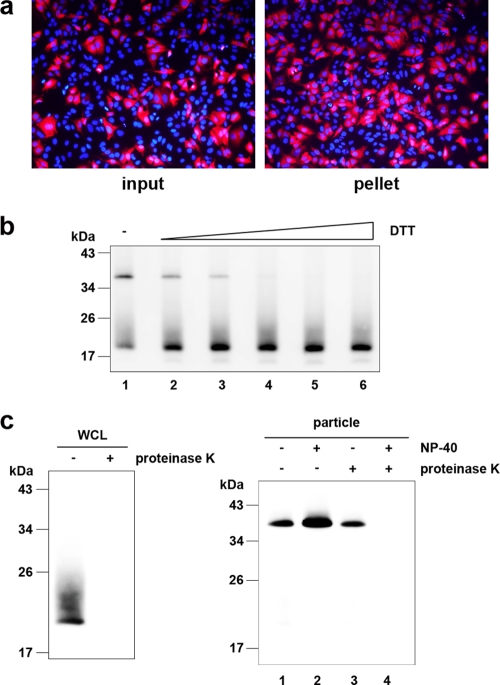
To analyze the core protein of the HCV particle, we first subjected the concentrated culture medium of HuH-7 cells transfected with in vitro-transcribed JFH1E2FL RNA to ultracentrifugation. After the resulting pellet was resuspended in culture medium, we confirmed the presence of infectious HCV particles on the basis of the infectivity of HuH-7.5 cells (Fig. 1 a). The infectious JFH1E2FL particle-containing pellet was separated by SDS-PAGE under nonreducing conditions, and immunoblot analysis showed the presence of a core antibody-reactive protein that was approximately twice the size of the core protein (38 kDa), in addition to the expected 19-kDa core protein (Fig. 1b, lane 1). Because treatment with DTT eliminated the larger core protein antibody-reactive band while the levels of the core protein monomer increased (Fig. 1b, lanes 2 to 6), the larger protein likely represented a dbc-complex. This complex was also found in J6/JFH1-derived particles (see Fig. S2 in the supplemental material), indicating that the complex was not specific for JFH1E2FL.
FIG. 1.
The HCV-like particle consists of a core complex formed by a disulfide bond. (a) The infectivity of the pellet fraction collected from concentrated culture medium from JFH1E2FL RNA-transfected HuH-7 cells was analyzed as described in Materials and Methods. Input indicates the same volume of concentrated culture medium used to pellet the virus-like particles. (b) Immunoblot analysis of the core protein in pellets containing JFH1E2FL virus particles treated with various levels of DTT (lanes 1, 2, 3, 4, 5, and 6, 0, 1.56, 3.13, 6.25, 12.5, and 25 mM DTT, respectively). (c) Immunoblot analysis of the core protein in JFH1E2FL particles collected from sucrose density gradient fractions with high HCV RNA titers (particle) (Fig. 2a, fractions 8 to 13) and treated with 5 μg/ml proteinase K at 37°C for 15 min in the presence or absence of 1% NP-40 (right panel). As a positive control, WCL prepared from JFH1E2FL RNA-transfected HuH-7 cells in lysis buffer was treated with 5 μg/ml proteinase K at 37°C for 15 min (left panel). Data are representative of three independent experiments.
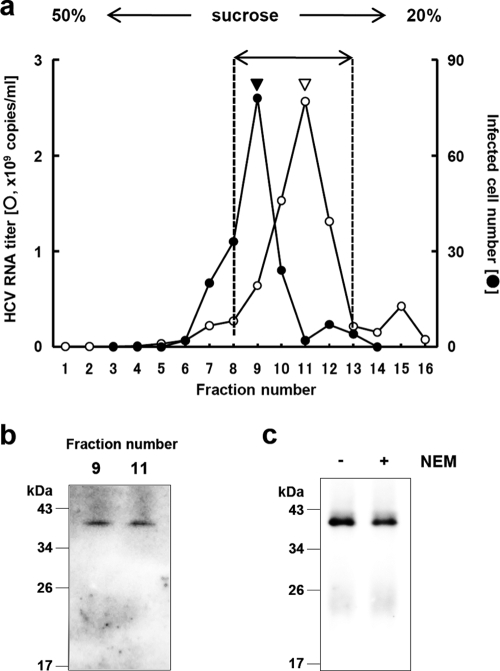
To determine whether the dbc-complex is a component of the HCV particle, a protease protection assay was performed using RNase-resistant HCV particles fractionated on the basis of their buoyant densities. Concentrated culture medium from HuH-7 cells transfected with in vitro-transcribed JFH1E2FL RNA was fractionated using a 20 to 50% sucrose density gradient; and JFH1E2FL particles, which were presumed to contain both infectious and noninfectious particles, were collected from fractions with high HCV RNA titers using ultracentrifugation (Fig. 2 a, fractions 8 to 13). The core protein from the collected fractions was analyzed by immunoblotting after SDS-PAGE under nonreducing conditions and showed only the dbc-complex (Fig. 1c, right panel).
FIG. 2.
HCV nucleocapsid-like particle consists of core complex. (a) HCV RNA titer in culture medium separated on a 20 to 50% sucrose density gradient. Concentrated culture medium from JFH1E2FL RNA-transfected HuH-7 cells were treated with RNase and separated on a 20 to 50% sucrose density gradient. Fractions 1 to 16 were obtained from the bottom to the top of the tube, respectively. The HCV RNA titer and infectivity of each fraction were analyzed by real-time qRT-PCR (for fractions 1 to 16) and counting the number of cells infected with HCV-like particle detected by immunofluorescence (for fractions 3 to 14), respectively, as described in Materials and Methods. In brief, each fraction was diluted with 1× PBS and HCV-like particles were collected by ultracentrifugation, and then the pellets were suspended in culture medium and used for infection. (b) HCV-like particle collected from the infectious HCV peak (from panel a, filled arrowhead) and the HCV RNA peak (from panel a, open arrowhead) were collected by ultracentrifugation, subjected to nonreducing SDS-PAGE, and detected by immunoblotting against the core protein. (c) HCV-like particles collected from fractions 8 to 13 (a) were subjected to nonreducing SDS-PAGE in the presence (lane +) or absence (lane −) of 5 mM NEM and analyzed by immunoblotting against the core protein. Data are representative of two (a, infectivity of fractions) or three independent experiments.
To examine whether the complex contributes to the infectivity of the particles, we analyzed the dbc-complex in the fractions containing infectious and noninfectious HCV particles (fractions 9 and 11 of Fig. 2a, filled and open arrowheads, respectively). Both the infectious and noninfectious HCV particle-containing fractions contained the dbc-complex (Fig. 2b). To confirm this further, a pellet containing particles of mutant JFH1AAA99—a mutant of JFH1E2FL that primarily produces noninfectious particles (36)—was analyzed in a similar manner. These dbc-complexes were found in pelleted particles of both JFH1AAA99 and J6/JFH1AAA99, which was a mutant J6/JFH1 with a similar substitution to JFH1AAA99 (see Fig. S2 in the supplemental material). These results indicated that the dbc-complex was present in both the infectious and noninfectious HCV-like particles.
The core protein monomer observed in the pellet samples (Fig. 1b) may be from the secreted core protein or the debris of apoptotic cells, because the core protein is known to be secreted from cells expressing this protein under particular conditions (42) and strain JFH1 is known to cause apoptosis (45). The dbc-complex-specific signals in the HCV particles seem to be increased in the NP-40-treated samples for some unknown reason (Fig. 1c, lanes 1 and 2). Although the intermolecular disulfide bond is known to be artificially formed in denaturing SDS-PAGE in the absence of reducing agents, the dbc-complex was still observed even in the presence of NEM, which is the alkylating agent for free sulfhydryls, during sample preparation (Fig. 2c), indicating that the dbc-complex was naturally present in the virus-like particles.
The HCV nucleocapsid is covered with lipid membranes and E1 and E2 proteins, making it resistant to proteases. As expected, in the absence of NP-40, the dbc-complex was resistant to proteinase K (Fig. 1c, lane 3), whereas proteinase K was able to digest core protein in whole-cell lysates (WCLs) collected from JFH1E2FL-transfected HuH-7 cells (Fig. 1c, left panel). Disrupting the envelope structure with NP-40 made the dbc-complex susceptible to proteinase K treatment (Fig. 1c, lane 4), indicating that the dbc-complex was indeed a component of the HCV particle.
The dbc-complex forms on the ER.
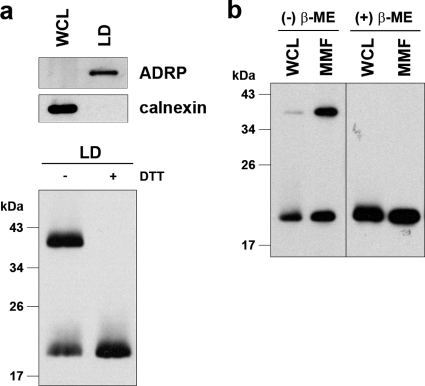
To investigate the subcellular site at which the dbc-complex forms, LDs and MMFs from JFH1E2FL replicating HuH-7 cells were analyzed by immunoblotting. We first analyzed the dbc-complex in LDs, because LDs are involved in infectious HCV particle formation (36, 47). The purity of the LD fraction was determined using immunoblot analysis of calnexin and ADRP, ER and LD marker proteins, respectively (Fig. 3 a, upper panel). The core protein was then analyzed in the LD fraction. As shown in Fig. 3a (lower panel), the dbc-complex was observed in the LD fraction from JFH1E2FL RNA-transfected HuH-7 cells. We next analyzed the core protein in the ER-containing MMF, because the core protein is first translated and processed on the ER (16). As shown in Fig. 3b, the dbc-complex was observed in the MMF from JFH1E2FL RNA-transfected HuH-7 cells. These results suggest that the dbc-complex is first formed at the ER. To assess the possibility that dbc-complex-containing HCV particles were also assembled on the ER, the sensitivity of the dbc-complex to protease treatment was analyzed. The dbc-complex in the MMF was susceptible to protease treatment in the absence of NP-40, indicating that the dbc-complex on the ER was not yet part of a HCV particle (data not shown).
FIG. 3.
The core complex is formed at the LD and ER. (a) The LD fraction and WCL were collected from JFH1E2FL RNA-transfected HuH-7 cells on day 5 posttransfection. (upper panel) Immunoblot analysis of the LD marker ADRP and the ER marker calnexin in the LD fraction; (lower panel) immunoblot analysis of the core protein in the LD fraction treated or not treated with 50 mM DTT. (b) Immunoblot analysis of the core protein in the MMF and WCL collected from JFH1E2FL-producing HuH-7 cells on day 5 posttransfection in the presence or absence of 5% β-ME. Data are representative of those from three independent experiments.
dbc-complex is most likely a disulfide-bonded dimer form of the core.
In order to examine whether the core protein itself has the potential to form a dbc-complex, we analyzed the dbc-complex formation of the full-length wild-type core protein (coreWT) expressed from pcDNA3-coreWT (36), the expression plasmid encoding the 191-amino-acid full-length core protein of JFH1 strain. We used this expression plasmid because the core protein from this plasmid was likely to be processed correctly enough to produce infectious HCV particles when it was cotransfected with the RNA of JFH1dc3, which is a core protein deletion mutant of JFH1 (36). As a result, the dbc-complex formation was observed from the MMF of coreWT-expressing cells both in the absence and in the presence of NEM (Fig. 4 b; lane 2 and data not shown, respectively). We next investigated the effect of the amino acid region of E1 on the production of the dbc-complex, because it has been reported that the efficient processing of core protein is dependent on the presence of some E1 sequence to ensure the insertion of the signal sequence for E1 in the translocon/membrane machinery (34). The dbc-complex was also observed when the core protein was expressed from pcDNA3-C-E1/25, which encodes the full-length core protein followed by the N-terminal 25-amino-acid sequence of E1 to ensure that the core protein is processed properly (see Fig. S3a in the supplemental material). These data showed that the dbc-complex was formed by expression of the core protein only in the cells.
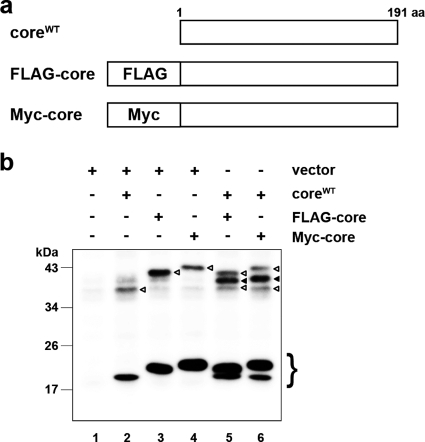
FIG. 4.
The core complex consists of a core dimer. (a) Schematic of wild-type, FLAG-tagged (FLAG-core), and Myc-tagged (Myc-core) core proteins. (b) Immunoblot analysis of the core protein in the MMF collected from HuH-7 cells transfected with combinations of pcDNA3 (vector) and/or core expression plasmids (e.g., encoding coreWT, FLAG-core, and Myc-core), as indicated. The experiment was performed under nonreducing conditions. The lower bands represent core monomer (marked on the right with a brace). White arrowheads, bands corresponding to dbd-core; black arrowheads, positions of the intermediately sized core complex formed by coreWT and the tagged core. Data are representative of those from three independent experiments.
Next, we examined the structural components of the dbc-complex. Because the dbc-complex was twice the size of the core protein monomer, it was likely dbd-core. So, we investigated whether the core protein molecules with different tags were able to form the dbd-core. We first generated expression plasmids encoding core protein with the N-terminal FLAG and Myc tags (pcDNA3-FLAG-core and pcDNA3-Myc-core, respectively; Fig. 4a). The tagged core proteins were expressed or coexpressed with coreWT in HuH-7 cells, and the MMF was analyzed by immunoblotting. The FLAG or Myc tag shifted the positions of the monomer and the complex bands (Fig. 4b, lanes 3 and 4) compared with the position of coreWT (Fig. 4b, lane 2). When coreWT was coexpressed with FLAG-core or Myc-core, the core protein complex of an intermediate size was observed, in addition to the bands obtained when the constructs were independently expressed (Fig. 4b, lanes 5 and 6, filled arrowheads); the intermediate-sized band disappeared after treatment with β-ME (see Fig. S3b, lanes 11 and 12, filled arrows, in the supplemental material), indicating that coreWT and tagged core protein formed a heteromeric disulfide-bonded dimer. These results demonstrated that the dbc-complex on the ER is a dbd-core. Although we tried to detect the hetero- or homodimer consisting of the tagged core protein by using anti-FLAG or anti-Myc antibodies, these dimers were not detected, possibly because of the lower levels of sensitivity and specificity of the antibodies compared to those of the anti-core protein antibody that we used, especially against epitopes in the dbd-core. The results presented above, coupled with the similarities of the molecular sizes and sensitivities to β-ME and DTT, suggested that the dbc-complex in the HCV particle is most likely a dbd-core.
Core protein Cys128 mediates dbd-core formation.
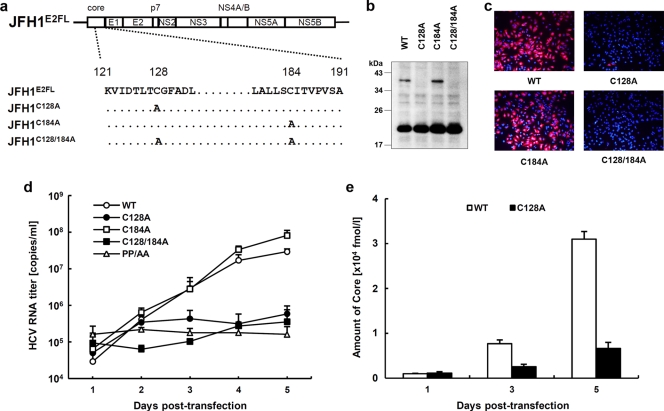
Our results showed that core protein from JFH1E2FL forms a disulfide-bonded dimer on the ER. A search for cysteine residues in the JFH1E2FL core protein identified amino acid positions 128 (Cys128) and 184 (Cys184) (see Fig. S1 in the supplemental material). These residues are highly conserved in core proteins from the approximately 2,000 reported HCV strains (HCVdb, http://www.hcvdb.org/, Hepatitis C Virus Database; http://s2as02.genes.nig.ac.jp/). To determine which cysteine residue mediated disulfide bond formation, we generated point mutations in JFH1E2FL that replaced Cys128 and/or Cys184 with alanine (Ala) (C128A, C184A, and C128/184A in JFH1C128A, JFH1C184A, and JFH1C128/184A, respectively; Fig. 5 a). As shown in Fig. 5b, the core proteins from JFH1C128A and JFH1C128/184A failed to form a dbd-core under nonreducing condition, whereas the core protein from JFH1C184A formed the dimer, indicating that Cys128 was the residue responsible. Similar results were observed when Cys was replaced by serine (Ser) instead of Ala (see Fig. S5c in the supplemental material). Recently, Majeau et al. reported that the core protein of the J6/JFH1 strain with Cys128 replacements by Ala or Ser were unstable in both Pichia pastoris and human hepatoma cell line HuH-7.5 (31), although we did not detect any noticeable degradation of the mutant core proteins of strain JFH1 (Fig. 5b; see also Fig. S5c in the supplemental material). This difference may have resulted from the difference in sample preparation methods, as we used the full-length genome of JFH1E2FL strain bearing the strain JFH1 core protein and HuH-7 cells instead of a core protein-expressing plasmid for the J6 strain and HuH-7.5 cells.
FIG. 5.
The core dimer is formed via a bond between cysteine residues at amino acid position 128. (a) Site-directed mutagenesis of JFH1E2FL. (b) Immunoblot analysis of the core protein in MMFs collected from HuH-7 cells under nonreducing conditions 3 days post-transfection with JFH1E2FL (WT), JFH1C128A (C128A), JFH1C184A (C184A), or JFH1C128/184A (C128/184A) RNA. (c) Infectivity of culture medium collected and concentrated on day 5 posttransfection from HuH-7 cells transfected with WT, C128A, C184A, or C128/184A RNA. (d) Real-time qRT-PCR analysis of HCV RNA titers in culture medium collected at the indicated time points from HuH-7 cells transfected with WT, C128A, C184A, C128/184A, or PP/AA (JFH1PP/AA) RNA. (e) ELISAs of core protein levels in culture medium collected at the indicated time points from HuH-7 cells transfected with WT or C128A RNA. Data are representative of those from three independent experiments (b and c) or are the means ± standard deviations from three independent experiments (d and e).
To exclude the possibility that mutation of Cys128 inhibited dbd-core formation by creating a conformational change, T127A and G129A core protein mutants (JFH1T127A and JHF1G129A, respectively) were created and examined for their effects on dbd-core formation and infectious virus particle production. These mutants formed dbd-core, and infectious HCV particles were detected in the culture medium (see Fig. S4a to c in the supplemental material), supporting an essential role for Cys128 in dbd-core and particle formation.
dbd-core contributes to HCV particle production.
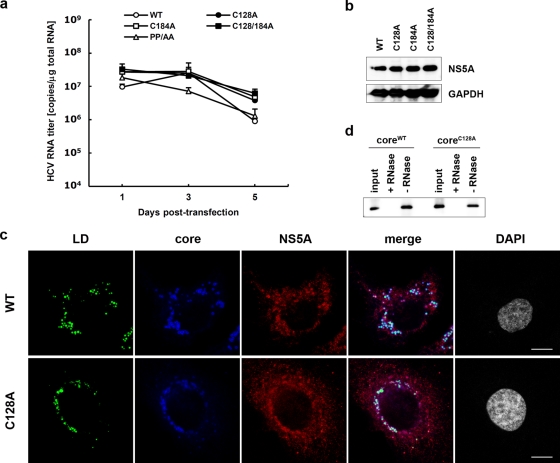
To examine the functional roles of dbd-core, infectious HCV particle production, HCV replication efficiency, colocalization of the core protein and LDs, and RNA binding of mutant and wild-type (JFH1E2FL) core protein were evaluated. Culture medium from HuH-7 cells transfected with JFH1C128A or JFH1C128/184A RNA contained significantly fewer infectious HCV particles compared with the numbers obtained with JFH1E2FL or JFH1C184A RNA (Fig. 5c). We also found significant decreases in the levels of HCV RNA and core protein in the culture medium of HuH-7 cells transfected with JFH1C128A or JFH1C128/184A RNA (Fig. 5d and e). Similar results were observed with J6/JFH1 C128A or the C128/184A mutant strain (data not shown). To investigate whether these results were due to suppressed HCV replication, the HCV RNA and protein levels in cells transfected with mutant RNA were analyzed using qRT-PCR and immunoblot analyses, respectively. Compared with the results obtained with JFH1E2FL, no significant changes in the cellular HCV RNA titer at days 1, 3, and 5 posttransfection or in the expression of HCV nonstructural protein NS5A were observed (Fig. 6 a and b). This indicated that substitution of Cys128 did not significantly affect HCV RNA genome replication or viral protein production, demonstrating that the dbd-core functions during HCV particle production rather than HCV genome replication. Similar results were observed using RNA of JFH1 mutant strain JFH1C128S, in which the cysteine at position 128 was replaced by Ser instead of Ala (see Fig. S5a, b, and d in the supplemental material).
FIG. 6.
Site-directed mutagenesis has no effect on HCV replication. (a) Real-time qRT-PCR analysis of the HCV RNA titer using total cellular RNA collected at the indicated time points from cells transfected with JFH1E2FL (WT), JFH1C128A (C128A), JFH1C184A (C184A), JFH1C128/184A (C128/184A), or JFH1PP/AA (PP/AA) RNA. (b) Immunoblot analysis of NS5A protein and GAPDH in whole-cell lysate collected from cells transfected with WT, C128A, C184A, or C128/184A RNA at day 3 posttransfection. (c) Confocal microscopy of the subcellular localization of the LD (green), core (blue), NS5A protein (red), and nucleus (4′,6-diamidino-2-phenylindole [DAPI]) (gray) in WT and C128A replicating cells on day 3 posttransfection. Bars, 10 μm. (d) An RNA-protein binding precipitation assay was performed with in vitro-translated coreWT and coreC128A using poly(U) agarose as the resin. +RNase and −RNase, samples with and without RNase treatment, respectively, as described in Materials and Methods. Input indicates that 1/40 of the amount of translated product was used in this assay. Data represent the means ± standard deviations from three independent experiments (a) or are representative of those from three independent experiments (b to d).
The subcellular localizations of the core protein and NS5A protein in HuH-7 cells transfected with HCV RNA were investigated using indirect immunofluorescence and confocal microscopy, because recruiting HCV proteins to LDs is an important step in infectious HCV particle production (36, 47) and core trafficking to LDs is dependent on signal peptide peptidase (SPP)-mediated cleavage of the tail region (34, 41). JFH1C128A mutant core protein and NS5A protein were efficiently trafficked to LDs, as was observed with wild-type JHF1E2FL (Fig. 6c), suggesting that SPP cleavage and core protein maturation were not affected by the C128A mutation. Similar results were obtained with the core proteins derived from JFH1C184A and JFH1C128/184A (see Fig. S6 in the supplemental material) and also Ser mutant JFH1C128S (see Fig. 5e in the supplemental material).
Because HCV core protein can bind to RNA, including the HCV genome, during viral particle assembly (43), we analyzed RNA binding by the core protein using in vitro-translated coreC128A, coreWT, and poly(U) agarose resin. CoreC128A and coreWT bound similarly to the poly(U) resin (Fig. 6d).
dbd-core is important for HCV particle assembly.
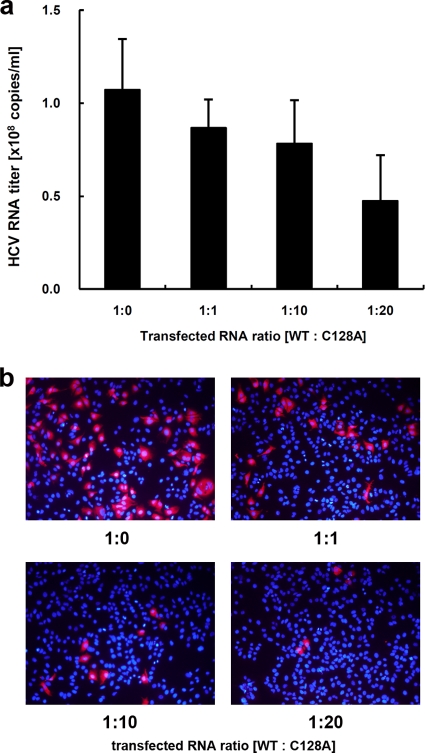
The mutational analysis of the core protein indicated that coreC128A and coreWT similarly localize to LDs, recruit NS proteins to the LD, and bind to RNA. Moreover, this mutation did not markedly affect HCV genome replication. How does coreC128A affect the production of HCV particles? An important function of the core protein is multimerization, which is followed by capsid construction and packaging of the RNA genome in the viral particles. We therefore determined whether coreC128A had a dominant negative effect on virus-like particle production. Wild-type JFH1E2FL RNA and different amounts of JFH1C128A RNA were cotransfected into HuH-7 cells, and the HCV RNA titer and infectivity of the virus-like particles in the culture medium were analyzed. As expected, the HCV RNA titer in the cells increased with higher levels of transfected RNA (see Fig. S7a in the supplemental material). In contrast, the HCV RNA titer and infectivity in the culture medium decreased in a JFH1C128A RNA dose-dependent manner when this mutant RNA was cotransfected with wild-type RNA (Fig. 7 a, b). This suppressive effect was not observed when either wild-type RNA or core deletion mutant JFH1dc3 RNA was used instead of mutant RNA in a similar experiment (see Fig. S7b to e in the supplemental material), indicating that higher levels of HCV RNA alone did not inhibit HCV particle production. Thus, coreC128A had a dominant negative effect on HCV particle production. Together, these results suggest that dbd-core is involved in the assembly of HCV particles.
FIG. 7.
JFH1C128A core inhibits JFH1E2FL particle assembly. A competitive inhibitory assay was performed with JFH1E2FL (WT) and JFH1C128A (C128A). (a) Real-time qRT-PCR analysis of the HCV RNA titer in HuH-7 cell culture medium 3 days after the cells were transfected with the indicated ratio of WT and C128A RNA. (b) The infectivity of culture medium collected from HuH-7 cells that had been transfected with the indicated ratio of WT and C128A RNA was analyzed as described in Materials and Methods. Data represent the means ± standard deviations from three independent experiments (a) or are representative of those from three independent experiments (b).
The nucleocapsid-like particle of HCV was resistant to trypsin treatment.
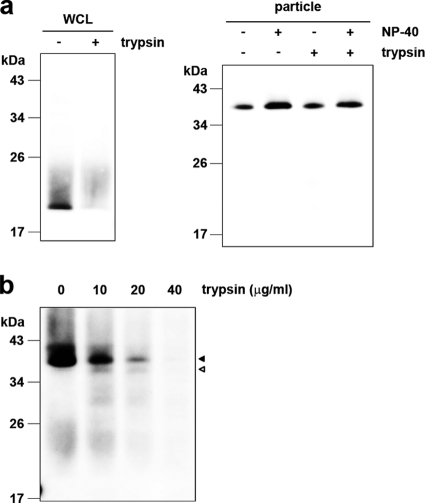
To further investigate the structure of the HCV nucleocapsid-like particle most likely formed by dbd-core, we examined the sensitivity of the particle to trypsin, which cleaves polypeptides at the C-terminal end of basic residues. Whereas trypsin digested the core protein in the whole-cell lysates (Fig. 8 a, left panel), dbd-core from buoyant density-fractionated JFH1E2FL particles was resistant to digestion, despite NP-40 treatment (Fig. 8a, right panel), although it was sensitive to proteinase K, which has a broad specificity (Fig. 1c). The N-terminal hydrophilic domain of the core protein (from residues 6 to 121) contains a number of trypsin cleavage sites (25 sites in strain JHF1) (see Fig. S1 in the supplemental material), suggesting that the N-terminal domain faces inward and/or that the conformation prevents protease access. To address this idea, buoyant density-fractionated JFH1E2FL particles were treated with trypsin under stricter conditions in the presence of NP-40. Cleavage of dbd-core by various levels of trypsin correlated with the appearance of a shorter molecule (Fig. 8b, white arrowhead). The shorter molecule was presumed to be partially digested dbd-core with an intact N-terminal region because it was recognized by anti-core protein antibodies, which bind to an epitope located from amino acids 20 to 40 of the core protein (M. Kohara, The Tokyo Metropolitan Institute of Medical Science, personal communication). These results suggest that dbd-core is assembled into the nucleocapsid-like particle such that most of the N-terminal domain is inside.
FIG. 8.
The nucleocapsid-like particle of JFH1E2FL is assembled with the C-terminal region of the core protein on the outer surface. (a) Immunoblot analysis of the core protein in JFH1E2FL particles collected from sucrose density gradient fractions with high HCV RNA titers (particle) (Fig. 2a, fractions 8 to 13). The fractions were treated with 10 μg/ml trypsin at 37°C for 15 min in the presence or absence of 1% NP-40 (right panel). As a positive control, WCL prepared from JFH1E2FL RNA-transfected HuH-7 cells in lysis buffer was treated with 10 μg/ml trypsin at 37°C for 15 min (left panel). (b) Immunoblot analysis of the core protein in JFH1E2FL particles collected from sucrose density gradient fractions with high HCV RNA titers (Fig. 2a, fractions 8 to 13). The fractions were treated with the indicated concentrations of trypsin at 37°C for 10 min in the presence of 1% NP-40. Open and filled arrows indicate the positions of dbd-core and the trypsin-digested fragment, respectively. Data are representative of those from three independent experiments.
DISCUSSION
In the present study, we have shown that the nucleocapsid-like particle of HCV most likely contains a dimer of core protein that is stabilized by a disulfide bond. Mutational analysis revealed that Cys128 forms a disulfide bond between core monomers. Several reports have shown that disulfide bonds in the capsid proteins of some viruses are involved in virus particle assembly and stabilization of the viral capsid structure (4, 21, 27, 28, 57); these viruses are characterized by icosahedral nucleocapsids. Because, like these viruses, the HCV virion is spherical (2, 20), it has been suggested that HCV may contain a nucleocapsid with a similar structure (20). We found the dbc-complex, which is most likely to be the dbd-core in JFH1E2FL virus-like particles (Fig. 1c and 8a). The dbd-core in the capsid structure was digested by proteinase K but not trypsin in the presence of NP-40 (Fig. 1c and 8a, lane 4). The resistance to trypsin suggested a tight conformation for dbd-core in the capsid and no exposed trypsin cleavage sites. The truncated form of dbd-core that was observed under certain trypsin treatment conditions likely resulted from cleavage in the C-terminal portion of the protein (e.g., arginine residues at positions 149 and 156) (see Fig. S1 in the supplemental material), although it is possible that the truncation of dbd-core was due to nonspecific cleavage by trypsin. These results imply that dbd-core is configured such that the N- and C-terminal ends are located at the inner and outer surfaces of the capsid, respectively. Because the N-terminal region of the core protein includes the RNA binding domain (43), the HCV RNA genome likely interacts with the core protein as it is packed in the nucleocapsid. On the other hand, the C-terminal hydrophobic domain probably faces the lipid membranes to form the envelope structure. Only part of the N-terminal hydrophilic region of the core protein has been structurally examined using X-ray crystal structural analysis (35) and structural bioinformatics and nuclear magnetic resonance analysis (11). Although the C-terminal half of the core protein has been structurally investigated by the use of bioinformatics (8), the three-dimensional structure containing the Cys128 residue is unknown. Thus, determination of the structure of the core protein in the nucleocapsid containing the Cys128 residue should be required for understanding the whole structure of this protein in the virus particles.
Because cotransfection of JFH1C128A RNA with wild-type JFH1E2FL RNA inhibited particle production in a mutant RNA dose-dependent manner (Fig. 7a and b), the C128A core variant clearly inhibited HCV particle formation by wild-type core protein. Cys128 was also previously reported to be a residue included in the region important for the production of infectious HCV (39). This residue is located near the N-terminal end of the hydrophobic region of the core (amino acids 122 to 177) and belongs to the hydrophilic side of an amphipathic helix expected to interact in the plane of the membrane interface (7). Therefore, it is possible to think that dbd-core formation via Cys128 can stabilize the interaction between the core protein and the membranes. The N-terminal half of the core protein (amino acids 1 to 124) reportedly assembles into nucleocapsid-like particles in the presence of the 5′ UTR from HCV RNA (24), suggesting that some nucleocapsid-like particles may assemble via only homotypic interactions from the core protein. In addition to weak homotypic interactions, the HCV core protein forms a disulfide bond to stabilize the capsid structure, thus making dbd-core indispensable in the stable virus-like particle. We observed that culture medium from JFH1C128A- or JFH1C128S-transfected cells included slight infectivity (Fig. 5c; see also Fig. S5d in the supplemental material). This made us speculate that this mutant may produce some infective particle-like structure formed by a homotypic interaction of the core. Such a slight infectivity may have reflected the optimized in vitro culture conditions compared with the in vivo conditions, allowing relatively unstable virus particles to survive.
A nucleocapsid must be resistant to environmental degradation yet still be able to disassemble after infection. Disulfide bonds could help with this process by switching between a stable and unstable virus capsid on the basis of different intracellular and extracellular oxidation conditions (12, 30). During the virus life cycle, the disulfide bond strengthens the viral capsid structure and protects the viral genome from oxidative conditions and cellular nucleases when virus particles are formed. Upon infection, the disulfide bond may be cleaved under cytoplasmic reducing conditions, thereby releasing the viral genome into the cell for replication. HCV may utilize the core protein disulfide bond in this way as HCV enters the host cell via clathrin-mediated endocytosis (5) into a low-pH, endosomal compartment (25, 52); this is presumably followed by endosomal membrane fusion and release of the viral capsid into the cytoplasm (38).
Treatment of HCV infection with pegylated interferon in combination with ribavirin is not effective for all patients. Recently, drugs targeting viral proteins NS3/4A and NS5B have been examined in clinical trials. Although these drugs are relatively specific, resulting in fewer side effects and potent antiviral activity, monotherapy can be complicated by rapidly emerging resistant variants carrying mutations that reduce drug efficacy, perhaps due to conformational changes in the target (23, 48, 51). Therefore, viral proteins that are highly conserved among strains and those characterized by low mutation rates may be better targets for drug development. Because the core protein is the most conserved HCV protein and Cys128 is conserved among almost all HCV strains examined, drugs that interact with Cys128 and/or the region around or near this residue will likely show broad-spectrum efficacy to block stable infectious particle formation. Structural analysis of dbd-core should aid with the development of new STAT-Cs that target Cys128 by direct interaction with the sulfide group and/or region around this residue. Until now and still, the mechanism of disulfide bond formation of the core protein on the ER is unknown. Dimerization of the capsid protein by disulfide bond has been reported in some enveloped viruses (9, 19, 54, 56), although some were shown not to be important for virus particle formation (26, 55). Unlike vaccinia virus (46), no redox system of its own has been reported for these viruses. Therefore, further investigations addressing the mechanisms underlying dbd-core formation on the ER may reveal a new mechanism for disulfide bond formation of viral proteins in infected cells.
Supplementary Material
Acknowledgments
This work was supported by grants-in-aid from the Ministry of Health, Labor, and Welfare of Japan and by grants-in-aid from the Japan Health Sciences Foundation.
Footnotes
Published ahead of print on 30 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abid, K., V. Pazienza, A. de Gottardi, L. Rubbia-Brandt, B. Conne, P. Pugnale, C. Rossi, A. Mangia, and F. Negro. 2005. An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J. Hepatol. 42:744-751. [DOI] [PubMed] [Google Scholar]
- 2.Aly, H. H., Y. Qi, K. Atsuzawa, N. Usuda, Y. Takada, M. Mizokami, K. Shimotohno, and M. Hijikata. 2009. Strain-dependent viral dynamics and virus-cell interactions in a novel in vitro system supporting the life cycle of blood-borne hepatitis C virus. Hepatology 50:689-696. [DOI] [PubMed] [Google Scholar]
- 3.Asselah, T., Y. Benhamou, and P. Marcellin. 2009. Protease and polymerase inhibitors for the treatment of hepatitis C. Liver Int. 29(Suppl. 1):57-67. [DOI] [PubMed] [Google Scholar]
- 4.Baron, M. D., and K. Forsell. 1991. Oligomerization of the structural proteins of rubella virus. Virology 185:811-819. [DOI] [PubMed] [Google Scholar]
- 5.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulant, S., M. W. Douglas, L. Moody, A. Budkowska, P. Targett-Adams, and J. McLauchlan. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 9:1268-1282. [DOI] [PubMed] [Google Scholar]
- 7.Boulant, S., R. Montserret, R. G. Hope, M. Ratinier, P. Targett-Adams, J. P. Lavergne, F. Penin, and J. McLauchlan. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281:22236-22247. [DOI] [PubMed] [Google Scholar]
- 8.Boulant, S., C. Vanbelle, C. Ebel, F. Penin, and J. P. Lavergne. 2005. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 79:11353-11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornillez-Ty, C. T., and D. W. Lazinski. 2003. Determination of the multimerization state of the hepatitis delta virus antigens in vivo. J. Virol. 77:10314-10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dustin, L. B., and C. M. Rice. 2007. Flying under the radar: the immunobiology of hepatitis C. Annu. Rev. Immunol. 25:71-99. [DOI] [PubMed] [Google Scholar]
- 11.Duvignaud, J. B., C. Savard, R. Fromentin, N. Majeau, D. Leclerc, and S. M. Gagne. 2009. Structure and dynamics of the N-terminal half of hepatitis C virus core protein: an intrinsically unstructured protein. Biochem. Biophys. Res. Commun. 378:27-31. [DOI] [PubMed] [Google Scholar]
- 12.Freedman, R. B., B. E. Brockway, and N. Lambert. 1984. Protein disulphide-isomerase and the formation of native disulphide bonds. Biochem. Soc. Trans. 12:929-932. [DOI] [PubMed] [Google Scholar]
- 13.Giannini, C., and C. Brechot. 2003. Hepatitis C virus biology. Cell Death Differ. 10(Suppl. 1):S27-S38. [DOI] [PubMed] [Google Scholar]
- 14.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higashi, Y., H. Itabe, H. Fukase, M. Mori, Y. Fujimoto, R. Sato, T. Imanaka, and T. Takano. 2002. Distribution of microsomal triglyceride transfer protein within sub-endoplasmic reticulum regions in human hepatoma cells. Biochim. Biophys. Acta 1581:127-136. [DOI] [PubMed] [Google Scholar]
- 16.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. U. S. A. 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kimura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hope, R. G., and J. McLauchlan. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913-1925. [DOI] [PubMed] [Google Scholar]
- 19.Hu, H. M., K. N. Shih, and S. J. Lo. 1996. Disulfide bond formation of the in vitro-translated large antigen of hepatitis D virus. J. Virol. Methods 60:39-46. [DOI] [PubMed] [Google Scholar]
- 20.Ishida, S., M. Kaito, M. Kohara, K. Tsukiyama-Kohora, N. Fujita, J. Ikoma, Y. Adachi, and S. Watanabe. 2001. Hepatitis C virus core particle detected by immunoelectron microscopy and optical rotation technique. Hepatol. Res. 20:335-347. [DOI] [PubMed] [Google Scholar]
- 21.Jeng, K. S., C. P. Hu, and C. M. Chang. 1991. Differential formation of disulfide linkages in the core antigen of extracellular and intracellular hepatitis B virus core particles. J. Virol. 65:3924-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. U. S. A. 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieffer, T. L., A. D. Kwong, and G. R. Picchio. 2010. Viral resistance to specifically targeted antiviral therapies for hepatitis C (STAT-Cs). J. Antimicrob. Chemother. 65:202-212. [DOI] [PubMed] [Google Scholar]
- 24.Kim, M., Y. Ha, and H. J. Park. 2006. Structural requirements for assembly and homotypic interactions of the hepatitis C virus core protein. Virus Res. 122:137-143. [DOI] [PubMed] [Google Scholar]
- 25.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. Y., D. Hwang, and S. Gillam. 1996. Dimerization of rubella virus capsid protein is not required for virus particle formation. Virology 216:223-227. [DOI] [PubMed] [Google Scholar]
- 27.Li, M., P. Beard, P. A. Estes, M. K. Lyon, and R. L. Garcea. 1998. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J. Virol. 72:2160-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, P. P., A. Nakanishi, S. W. Clark, and H. Kasamatsu. 2002. Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 Vp1 in the cytoplasm. Proc. Natl. Acad. Sci. U. S. A. 99:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, T. J., L. J. Jeffers, K. R. Reddy, M. De Medina, I. T. Parker, H. Cheinquer, V. Idrovo, A. Rabassa, and E. R. Schiff. 1993. Viral pathogenesis of hepatocellular carcinoma in the United States. Hepatology 18:1326-1333. [PubMed] [Google Scholar]
- 30.Liljas, L. 1999. Virus assembly. Curr. Opin. Struct. Biol. 9:129-134. [DOI] [PubMed] [Google Scholar]
- 31.Majeau, N., R. Fromentin, C. Savard, M. Duval, M. J. Tremblay, and D. Leclerc. 2009. Palmitoylation of hepatitis C virus core protein is important for virion production. J. Biol. Chem. 284:33915-33925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumoto, M., S. B. Hwang, K. S. Jeng, N. Zhu, and M. M. Lai. 1996. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology 218:43-51. [DOI] [PubMed] [Google Scholar]
- 33.McLauchlan, J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral Hepat. 7:2-14. [DOI] [PubMed] [Google Scholar]
- 34.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menez, R., M. Bossus, B. H. Muller, G. Sibai, P. Dalbon, F. Ducancel, C. Jolivet-Reynaud, and E. A. Stura. 2003. Crystal structure of a hydrophobic immunodominant antigenic site on hepatitis C virus core protein complexed to monoclonal antibody 19D9D6. J. Immunol. 170:1917-1924. [DOI] [PubMed] [Google Scholar]
- 36.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 37.Moradpour, D., C. Englert, T. Wakita, and J. R. Wands. 1996. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology 222:51-63. [DOI] [PubMed] [Google Scholar]
- 38.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453-463. [DOI] [PubMed] [Google Scholar]
- 39.Murray, C. L., C. T. Jones, J. Tassello, and C. M. Rice. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nolandt, O., V. Kern, H. Muller, E. Pfaff, L. Theilmann, R. Welker, and H. G. Krausslich. 1997. Analysis of hepatitis C virus core protein interaction domains. J. Gen. Virol. 78(Pt 6):1331-1340. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto, K., Y. Mori, Y. Komoda, T. Okamoto, M. Okochi, M. Takeda, T. Suzuki, K. Moriishi, and Y. Matsuura. 2008. Intramembrane processing by signal peptide peptidase regulates the membrane localization of hepatitis C virus core protein and viral propagation. J. Virol. 82:8349-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabile, A., G. Perlemuter, F. Bono, K. Kohara, F. Demaugre, M. Kohara, Y. Matsuura, T. Miyamura, C. Brechot, and G. Barba. 1999. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology 30:1064-1076. [DOI] [PubMed] [Google Scholar]
- 43.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seeff, L. B., and J. H. Hoofnagle. 2003. Appendix: The National Institutes of Health Consensus Development Conference Management of Hepatitis C 2002. Clin. Liver Dis. 7:261-287. [DOI] [PubMed] [Google Scholar]
- 45.Sekine-Osajima, Y., N. Sakamoto, K. Mishima, M. Nakagawa, Y. Itsui, M. Tasaka, Y. Nishimura-Sakurai, C. H. Chen, T. Kanai, K. Tsuchiya, T. Wakita, N. Enomoto, and M. Watanabe. 2008. Development of plaque assays for hepatitis C virus-JFH1 strain and isolation of mutants with enhanced cytopathogenicity and replication capacity. Virology 371:71-85. [DOI] [PubMed] [Google Scholar]
- 46.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2000. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl. Acad. Sci. U. S. A. 97:12068-12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shavinskaya, A., S. Boulant, F. Penin, J. McLauchlan, and R. Bartenschlager. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158-37169. [DOI] [PubMed] [Google Scholar]
- 48.Shimakami, T., R. E. Lanford, and S. M. Lemon. 2009. Hepatitis C: recent successes and continuing challenges in the development of improved treatment modalities. Curr. Opin. Pharmacol. 9:537-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tellinghuisen, T. L., M. J. Evans, T. von Hahn, S. You, and C. M. Rice. 2007. Studying hepatitis C virus: making the best of a bad virus. J. Virol. 81:8853-8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tellinghuisen, T. L., and C. M. Rice. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419-427. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, A. J., and J. G. McHutchison. 2009. Antiviral resistance and specifically targeted therapy for HCV (STAT-C). J. Viral Hepat. 16:377-387. [DOI] [PubMed] [Google Scholar]
- 52.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wootton, S. K., and D. Yoo. 2003. Homo-oligomerization of the porcine reproductive and respiratory syndrome virus nucleocapsid protein and the role of disulfide linkages. J. Virol. 77:4546-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, S., and D. N. Standring. 1992. Cys residues of the hepatitis B virus capsid protein are not essential for the assembly of viral core particles but can influence their stability. J. Virol. 66:5393-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, S., and D. N. Standring. 1992. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc. Natl. Acad. Sci. U. S. A. 89:10046-10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zweig, M., C. J. Heilman, Jr., and B. Hampar. 1979. Identification of disulfide-linked protein complexes in the nucleocapsids of herpes simplex virus type 2. Virology 94:442-450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.