Abstract
The tegument of all herpesviruses contains a high-molecular-weight protein homologous to herpes simplex virus (HSV) UL36. This large (3,164 amino acids), essential, and multifunctional polypeptide is located on the capsid surface and present at 100 to 150 copies per virion. We have been testing the idea that UL36 is important for the structural organization of the tegument. UL36 is proposed to bind directly to the capsid with other tegument proteins bound indirectly by way of UL36. Here we report the results of studies carried out with HSV type 1-derived structures containing the capsid but lacking a membrane and depleted of all tegument proteins except UL36 and a second high-molecular-weight protein, UL37. Electron microscopic analysis demonstrated that, compared to capsids lacking a tegument, these capsids (called T36 capsids) had tufts of protein located at the vertices. Projecting from the tufts were thin, variably curved strands with lengths (15 to 70 nm) in some cases sufficient to extend across the entire thickness of the tegument (∼50 nm). Strands were sensitive to removal from the capsid by brief sonication, which also removed UL36 and UL37. The findings are interpreted to indicate that UL36 and UL37 are the components of the tufts and of the thin strands that extend from them. The strand lengths support the view that they could serve as organizing features for the tegument, as they have the potential to reach all parts of the tegument. The variably curved structure of the strands suggests they may be flexible, a property that could contribute to the deformable nature of the tegument.
All herpesviruses have a tegument, a layer of protein located between the virus capsid and membrane. The tegument accounts for a substantial proportion of the overall virus structure. Its thickness (30 to 50 nm), for example, may be comparable to the capsid radius, and tegument proteins can account for 40% or more of the total virion protein. Herpesvirus tegument proteins are thought to function promptly after initiation of infection, before expression of virus genes can take place (11, 13, 14, 21, 33, 37).
Electron microscopic analysis of virions has demonstrated that the tegument is not highly structured (9, 22). It does not have icosahedral symmetry like the capsid, and it may be uniformly or asymmetrically arranged around the capsid (26). Tegument structure is described as fibrous or granular, and its morphology is found to change as the virus matures. Studies with herpes simplex virus type 1 (HSV-1), for example, indicate that the tegument structure is altered in cell-associated compared to extracellular virus (26).
The tegument has been most thoroughly studied in HSV-1, where biochemical analyses indicate that it is composed of approximately 20 distinct, virus-encoded protein species. The predominant components are the products of the genes UL47, UL48, and UL49, with each protein present in 800 or more copies per virion (12, 40). Other tegument proteins can occur in 100 or fewer copies, and trace amounts of cell-encoded proteins are also present (17). Tegument proteins are classified as inner or outer components based on their association with the capsid after it enters the host cell cytoplasm. The inner tegument proteins (UL36, UL37, and US3) are those that remain bound to the capsid after entry, while the others (the outer tegument proteins) become detached (7, 18).
The HSV-1 UL36 protein has the potential to play a central role in organizing the overall structure of the tegument. With a length of 3,164 amino acids, UL36 could span the thickness of the tegument multiple times. One hundred to 150 UL36 molecules are present in the tegument (12), and they are bound to the capsid by way of an essential C-terminal domain (2, 16). UL36 is able to bind the major tegument components by way of documented direct (UL37 and UL48) and indirect (UL46, UL47, and UL49) contacts (6, 15, 24, 38).
Here we describe the results of studies designed to test the idea that UL36 serves to organize the tegument structure. Beginning with infectious virus, a novel method has been used to isolate capsids that contain UL36 and UL37 but lack the virus membrane and are depleted of all other tegument proteins. These capsids (T36 capsids) were examined by electron microscopy to clarify the structure of UL36 and UL37 molecules and their location on the capsid surface.
MATERIALS AND METHODS
Virus growth and titration.
The KOS strain of HSV-1 was grown on monolayer cultures of Vero cells as previously described, in Dulbecco's modified Eagle's medium with antibiotics (35). Virus titers were determined by growth of 1:10 virus dilutions on Vero cell monolayers produced in 24-well plates. One-milliliter virus samples were tested by both endpoint dilution and plaque counting methods. For calculation of the virus particle/PFU ratio, the particle number was estimated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis of an undiluted stock of purified virus with a known titer, followed by Coomassie staining to determine the amount of the major capsid protein (VP5, UL19 gene) present. It was assumed that each virion contains 955 VP5 molecules (molecular weight, 149,075).
Virus purification.
Virus for T36 capsid preparation was grown on Vero cells in 175-cm2 flasks or 850-cm2 roller bottles. Cells were infected at a multiplicity of infection of 10, incubated for 18 h at 37°C, harvested by scraping, and pelleted by low-speed centrifugation (3,000 × g for 5 min). Virus was isolated beginning with batches of 2 × 108 to 5 × 108 infected cells. These were washed once in phosphate-buffered saline (PBS; 30 ml, 4°C), pelleted, resuspended in 30 ml TNE (0.01 M Tris-HCl, 0.5 M NaCl, 1 mM EDTA, pH 7.5), and incubated for 15 min at 4°C to detach cell-associated virus. TNE extraction was repeated once, and the virus was harvested by centrifugation into a pellet through a layer (5 ml) of 20% sucrose in TNE (Beckman SW28 rotor operated at 23,000 rpm for 45 min at 4°C). The supernatant was removed, the virus-containing pellet was resuspended in 1 ml TNE, and the virus was purified by sucrose density gradient centrifugation (20 to 50% sucrose in TNE, centrifugation for 1 h at 23,000 rpm in an SW41 rotor). After centrifugation, the virus band (∼1 ml) was removed by aspiration and used for capsid isolation. The virus yield was 1 to 2 mg.
T36 capsid isolation.
T36 capsids were isolated from the purified virus by extraction with 0.5% Triton X-100 (TX-100) in TNE as described previously (26). Virus (1 ml) was added to 9 ml TNE containing 0.5% TX-100, incubated for 5 min at 4°C, and centrifuged into a pellet in an SW41 tube containing a 0.5-ml cushion of 35% sucrose in TNE (23,000 rpm for 45 min). The capsid-containing pellet was resuspended in 0.2 ml TNE and purified by centrifugation on a 0.6-ml gradient of 20 to 50% sucrose (no Triton) in an SW55 rotor (centrifugation at 23,000 rpm for 45 min). Capsids were removed from the gradient by aspiration and used for further analysis as described below. The yield was ∼200 μg capsids beginning with 1 ml of concentrated virus. Staining with Gel Red (27) confirmed that nearly all of the capsids contained DNA. Recently described methods were used for isolation of capsids from infected cell nuclei (35), SDS-polyacrylamide gel electrophoresis (30), and photographic recording of sucrose density gradients (26). Quantitative measurement of virus and capsids in sucrose density gradients was performed beginning with digital photographs, which were scanned with UN-SCAN-IT (version 5.2; Silk Scientific).
Sonication of T36 capsids.
Experiments were carried out beginning with HSV-1 harvested as described above by TNE extraction of cells infected for 18 h at 37°C and purified by sucrose density gradient centrifugation. Virus samples (100 μl, ∼0.25 mg/ml) were adjusted to 0.5% TX-100 at 4°C and subjected immediately to sonication for 3 s in the “sweet spot” of a bath sonicator. Following sonication, capsids were isolated by sucrose density gradient centrifugation on 0.6-ml gradients of 20 to 50% sucrose (in TNE, no TX-100) as described above. After the gradients were photographed, the capsids were removed and analyzed by SDS-polyacrylamide gel electrophoresis and electron microscopy as described below. Control experiments were done without the sonication step.
Electron microscopy.
Previously described procedures were used for preparing specimens for electron microscopy by thin sectioning (19) and by negative staining with 1% uranyl acetate (28, 29). All electron microscopy was performed with a Philips 400T instrument operated at 80,000 eV. Images were recorded on film, digitized in a flat-bed scanner, and measured with ImageJ and Photoshop CS3. Measurements were plotted with SigmaPlot 10.
RESULTS
Virus isolation.
The studies described here were carried out with capsids prepared by TX-100 extraction of infectious virus (22). TX-100 extraction removes the membrane, the membrane glycoproteins, and a variable amount of tegument, depending on the time the virus has been detached from the host cell. Previous studies demonstrated that less tegument is solubilized the longer the virus stays in the extracellular medium before extraction (26). The goal of our study was to examine capsids prepared promptly after virus release with the idea that while such capsids should be depleted of tegument proteins, they might retain components involved in establishing tegument structure.
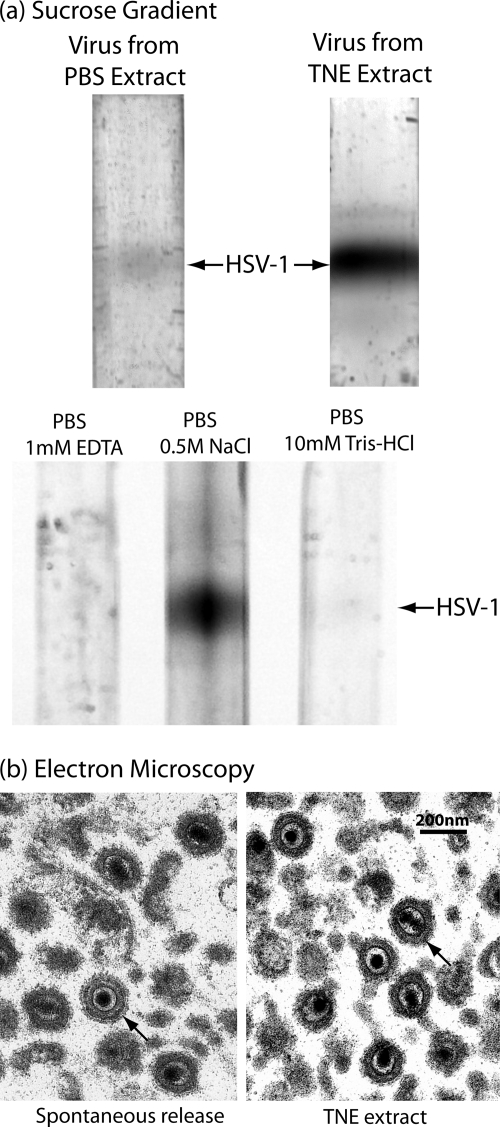
A novel method was used to harvest progeny virus from the host cell surface. Harvest began with monolayer cultures of Vero cells that were infected with the KOS strain of HSV-1 for 18 h, a time when little progeny virus is spontaneously released from the cell. A much greater quantity was found to be released, however, when cells were treated with TNE (for 15 min at 4°C). This is illustrated in Fig. 1a, in which sucrose density gradient analysis was used to compare the amount of virus released from 18-h cells with PBS (top gradients, left lane) and TNE (right lane). Quantitative determination of the virus yield by light scattering from the bands indicated that the yield differed by 22.7- and 15.6-fold in two determinations.
FIG. 1.
Characterization of HSV-1 harvested from the infected cell surface by treatment with TNE. (a) Sucrose density gradient ultracentrifugation and (b) electron microscopy are shown. The top two gradients in panel a compare the amounts of virus released by treatment of cells infected for 18 h with TNE (right gradient) and PBS (left). Note that substantially more virus was released with TNE. The lower three gradients in panel a compare the amounts of virus released with PBS containing the individual components of TNE, 1 mM EDTA, 0.5 M NaCl, and 10 mM Tris-HCl, pH 7.4. Note that the amount of virus released was greatest with PBS containing 0.5 M NaCl. Micrographs in panel b show HSV-1 spontaneously released from Vero cells infected for 18 h (left) and HSV-1 released from similar cells by treatment with TNE. Note that in both cases the tegument has the uniform arrangement characteristic of early-release virus (27). Arrows indicate representative virions in both cases.
Similar experiments were performed to identify the component of TNE responsible for detaching virus from the host cell surface. Sucrose density gradient analysis was used to measure the virus released from Vero cells infected for 18 h and treated with PBS containing 1 mM EDTA, 0.5 M NaCl, or 1 mM Tris-HCl, pH 7.4 (Fig. 1a, lower three gradients). Visual inspection of the gradients indicated that it was the NaCl component of TNE that was primarily responsible for detaching cell-associated virus. Quantitative determination indicated that the amounts released were proportional to 1:135:8 for the PBS/1 mM EDTA, PBS/0.5 M NaCl, and PBS/1 mM Tris-HCl treatments, respectively.
HSV-1 extracted from the cell surface with TNE was found to be qualitatively similar to the smaller amount released spontaneously. For instance, analysis by electron microscopy demonstrated that the tegument layer in TNE-extracted and spontaneously released HSV-1 was symmetrically arranged around the capsid, as it is in cell-associated virus (Fig. 1b). It was rare to see the asymmetric tegument distribution characteristic of virus harvested 30 h or more after infection (26).
The infectivity of TNE-released and spontaneously released virus was also found to be similar. When measured by particle/PFU ratio, the infectivity of TNE-released and spontaneously released virus was 105 and 61 particles/PFU, respectively (Table 1). In contrast, the infectivity of late-harvest (72 h) virus was 1,325 particles/PFU (average of two determinations; Table 1).
TABLE 1.
Infectivity of extracellular and TNE-released HSV-1
| Infection time (h) | Virus | Titer (PFU/ml) |
No. of particles/ml |
No. of particles/PFU |
|||
|---|---|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 1 | Expt 2 | Expt 1 | Expt 2 | ||
| 20 | Extracellulara | 1 × 108 | 5 × 108 | 6.1 × 109 | 3.4 × 109 | 61 | 6.8 |
| 20 | TNE extract | 8 × 108 | NDb | 8.4 × 1010 | ND | 105 | ND |
| 72 | Extracellular | 8 × 107 | 1 × 108 | 13.4 × 1011 | 9.7 × 1010 | 1,680 | 970 |
Virus harvested from the culture supernatant.
ND, not determined.
T36 capsids.
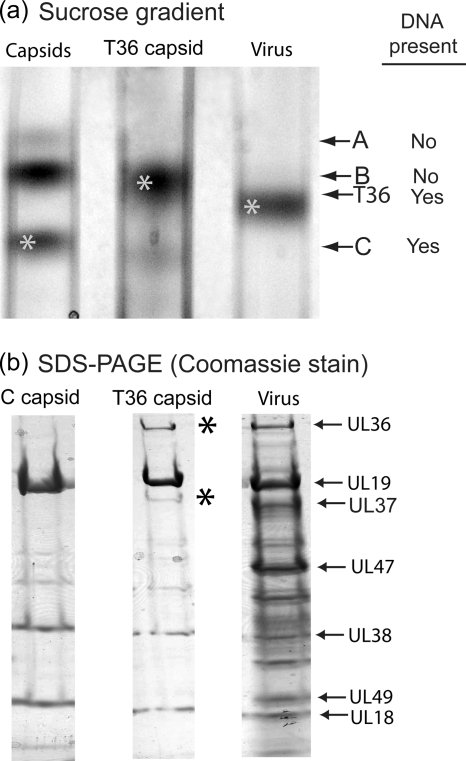
Virus released from the infected cell with TNE was treated promptly (within <15 min) with 0.5% TX-100, and the resulting capsids were examined by sucrose density gradient centrifugation, SDS-polyacrylamide gel electrophoresis, and electron microscopy. Sucrose density gradient centrifugation revealed the presence of a single band of capsids migrating between the bands of nuclear B capsids and virus (Fig. 2a, middle lane). All further studies were carried out with capsids recovered from this band. The capsids are identified as T36 capsids because they were derived by TX-100 extraction of virions and because of their UL36 content (see below).
FIG. 2.
Characterization of T36 capsids by sucrose density gradient ultracentrifugation (a) and SDS-polyacrylamide gel electrophoresis (b). T36 capsids were prepared as described in Materials and Methods, beginning with HSV-1 released by TNE treatment of Vero cells infected for 18 h. Note that during sucrose density gradient centrifugation, T36 capsids migrated more rapidly than nuclear B capsids (left gradient) but more slowly than HSV-1 (right). SDS-polyacrylamide gel electrophoresis was carried out with material harvested from the starred bands in panel a. Note that the protein composition of T36 capsids (middle gel) was similar to that of nuclear C capsids (left gel), except that T36 capsids contained two additional protein species, UL36 and UL37 (asterisks).
SDS-polyacrylamide gel analysis revealed that T36 capsids had a protein composition similar to that of nuclear C capsids (Fig. 2b, compare middle and left lanes). Prominent components of both were the major capsid protein (UL19, VP5) and the two triplex proteins Tri1 and Tri2 (UL38 and UL18). T36 capsids, however, had two additional proteins, UL36 and UL37, not present in C capsids. Visual inspection of Coomassie-stained gels indicated that the amounts of UL36 and UL37 were comparable to the amounts present in virions (Fig. 2b, compare the middle and right lanes). Apart from UL36 and UL37, no other major differences were observed in the protein contents of C and T36 capsids. In particular, there was no evidence of US3, an inner tegument protein (molecular weight, 52,835).
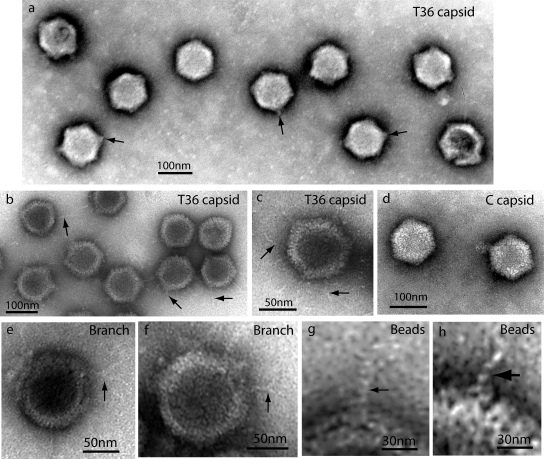
Electron microscopy of T36 capsids was carried out with negatively stained specimens. Images showed that T36 capsids resembled nuclear C capsids in that there was little evidence of tegument in either case (compare the capsids in Fig. 3 a with those in panel d). T36 and C capsids differed, however, because a short tuft or stem of density was observed at one or more vertices in most of the T36 capsid images (Fig. 3a). No comparable feature was observed in C capsids (Fig. 3d). For instance, of 131 T36 capsid images examined, tufts were seen in 115 (88%) and not in 16 (12%). One tuft projected radially outward from one vertex; it was very rare to see tufts originating from nonvertex sites. Multiple tufts were observed in most capsids. For example, the average number of tufts/capsid observed in negatively stained specimens was 2.76 in 131 T36 capsid images examined. Tufts were not seen in T capsids made from late-harvest HSV (26).
FIG. 3.
Electron microscopy of T36 capsids (a to c and e to h) and nuclear C capsids (d). All specimens were prepared by negative staining. Heavily stained images are shown in panel a, while other specimens are more lightly stained. Note that in all cases, T36 capsids had projections not found in C capsids. Projections appeared as tufts after heavy staining (a) and strands when staining was lighter (b, c, and e to h). Some strands were found to branch, as shown in panels e and f. Substructure was observed in strands, suggesting a coiled or beaded composition (g and h).
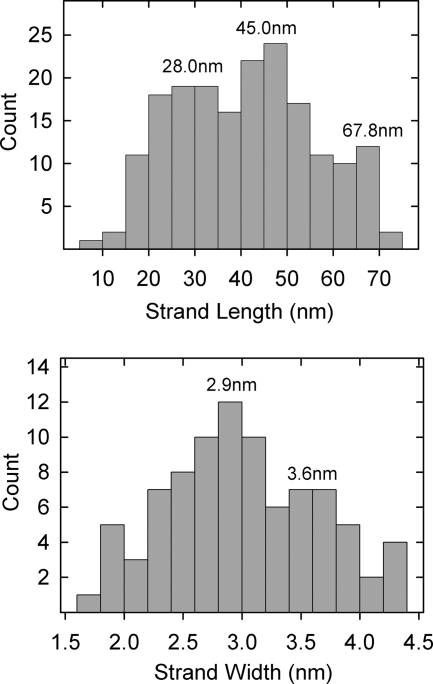
In lightly stained specimens, thin strands or fibers could be seen to project outward from T36 capsids (Fig. 3b and c). Most of the strands extended from capsid vertices, although some appeared to arise from edges. An example of an edge strand is shown at ∼6 o'clock in Fig. 3c. Most strands were straight or gently curved. They differed greatly in length, with the longest (∼70 nm) comparable to the capsid radius and the shortest ∼15 nm. Length measurements of 184 strands showed concentrations centered at lengths of 28.0 nm, 45.0 nm, and 67.8 nm (Fig. 4, top). Branched strands were observed, but this was a rare finding. No strand was seen to have more than a single branch (Fig. 3e and f).
FIG. 4.
Histograms showing strand lengths (top) and widths (bottom) measured in electron micrographs of T36 capsids similar to those shown in Fig. 3b and c.
Electron microscopic images showed that the strands were thin compared to the diameter of capsomers or the tufts found to project from vertices. Measured diameters of 84 strands showed a range of 2.0 to 4.0 nm, with concentrations at diameters of 2.9 and 3.6 nm (Fig. 4, bottom). In some cases, strands could be seen to be composed of distinct subunits consistent with a beaded structure or one composed of multiple intertwined threads (Fig. 3g and h).
Sonication of T36 capsids.
The length of the fibers and their small diameter suggested that the shear forces produced by brief sonication might be sufficient to detach them from the capsid surface. This expectation was tested beginning with T36 capsids generated by TX-100 treatment of virus but not isolated from other virus components. Virus-TX-100 mixtures were subjected to brief (3-s) sonication in a bath sonicator. Capsids were then isolated by sucrose density gradient centrifugation and analyzed by SDS-polyacrylamide gel electrophoresis and electron microscopy.
Sucrose density gradient analysis showed that most of the sonicated capsids migrated more rapidly than control, unsonicated specimens (Fig. 5a, band 1). Further analysis was performed with this predominant capsid species. Minor, more slowly migrating species were found to correspond to capsids induced by sonication to lose some or all of their DNA (bands 2 to 5).
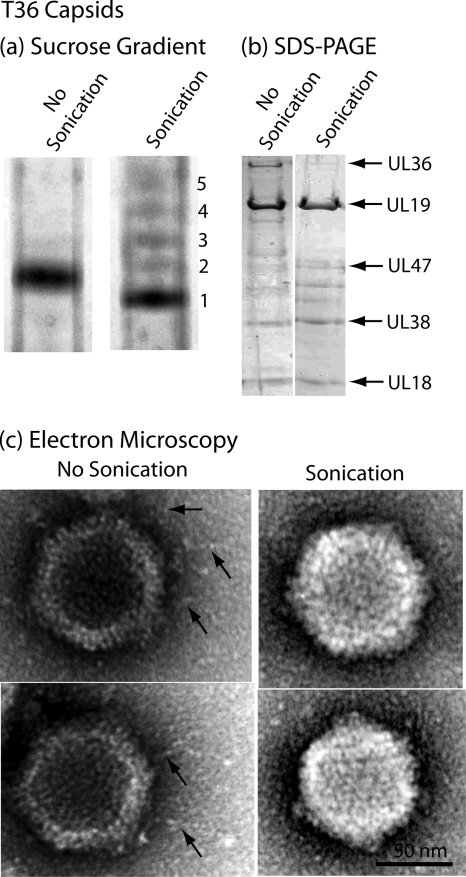
FIG. 5.
Characterization of sonicated T36 capsids by sucrose density gradient centrifugation (a), SDS-polyacrylamide gel electrophoresis (b), and electron microscopy (c). For the procedures used to produce T36 capsids and to expose them to brief sonication, see Materials and Methods. Note that sucrose density gradient analysis demonstrated that sonication resulted in the formation of a predominant band of capsids (band 1) that migrated slightly more rapidly than control, unsonicated T36 capsids. Minor bands of more slowly migrating capsids are suggested to have lost DNA as a result of sonication. SDS-polyacrylamide gel and electron microscopic analyses demonstrated that sonication caused loss of UL36 and UL37 and also of the fibers projecting from the T36 capsid surface. The findings are interpreted to support the view that the fibers are composed of UL36 and/or UL37.
Protein analysis by SDS-polyacrylamide gel electrophoresis revealed that sonication removed both UL36 and UL37, with removal complete or nearly so in both cases (Fig. 5b). In contrast, there was little change in other prominent T36 capsid proteins, including the major capsid protein (UL19) and the two triplex proteins (UL38 and UL18). Minor protein species migrating between UL47 and UL38 were found to be affected to various extents, with no pattern observed in multiple experiments.
Electron microscopic analysis demonstrated that fibers were observed associated with control T36 capsids as described above. It was rare, however, to observe them in sonicated preparations. Representative images are shown in Fig. 5c. Counts made from electron microscope negatives showed that fibers were seen in 32 of 32 control T36 capsid images but in only 1 of 64 sonicated capsids. Micrographs therefore support the correlation of fibers with the presence of the UL36 and UL37 proteins.
DISCUSSION
The tegument is central to the assembly of HSV-1 and other herpesviruses. Most HSV-1 tegument proteins become associated with the capsid during secondary envelopment in the cytoplasm (23). A mature, DNA-containing capsid buds into a cytoplasmic vesicle beginning in a region coated with a thick layer of tegument. As it buds, the capsid becomes coated with tegument and an envelope membrane. Vesicles involved in budding are located near the nucleus in a structure called the assembly compartment (1, 34), maturation compartment (3), or assembly site (36). Vesicle membranes are derived from the trans-Golgi network, but they may have input from other vesicle types as well (4, 5, 10).
Most of the tegument becomes detached from the capsid promptly after the infection of a new host cell (8, 20). Fusion of virus and host cell membranes, as required for virus entry, results in deposition of the tegument and capsid into the peripheral cytoplasm. The capsid then detaches from the tegument and traffics to the cell nucleus, with the inner tegument proteins UL36, UL37, and US3 remaining bound (7, 18). In contrast, the bulk of the tegument remains structurally intact and adjacent to the cell membrane for a short time after entry. Promptly, however, the tegument becomes disassembled and is no longer recognizable as a unit (8, 20).
It is relevant to emphasize here that the tegument behaves as a more-or-less intact unit throughout its functioning in virion assembly, egress, and infection (up until the time it is disassembled). The capsid binds to a tegument-sized region of membrane during secondary envelopment, the tegument is recognizable as a unit in the intact virion, and it retains its identity for a short time in the peripheral cytoplasm of a newly infected cell (20). As noted above, the goal of the present study was to test the idea that UL36 is involved in maintaining the structural integrity of the tegument until the overall structure becomes dismantled.
Isolation and biochemical analysis of T36 capsids.
Studies were carried out with virus-derived capsids that contained UL36 and UL37 but were depleted of all other tegument proteins. It was observed that such capsids (T36 capsids) could be isolated by a two-step procedure involving (i) release of mature HSV-1 from the infected cell surface with TNE and (ii) treatment of purified virions with TNE containing 0.5% TX-100 to remove the virion membrane and most of the tegument proteins (Fig. 2). Virion release from the cell surface with TNE yielded the greatest quantity of virus when extraction was performed 18 to 20 h postinfection. After longer periods, most of the virus was spontaneously released into the culture supernatant. The large amount of virus extracted with TNE from the surface of cells infected for 18 h (Fig. 1a) suggests that there is a delay in the release of mature HSV-1 virions after they are first exposed on the host cell surface.
Further analysis of virion release from the cell surface with TNE indicated that it is the 0.5 M NaCl component of TNE that is responsible for virion detachment (Fig. 1a). This observation suggests that cell-associated virus is attached by a noncovalent interaction that is disrupted by 0.5 M NaCl.
HSV-1 detached from the cell surface with TNE was found to be infectious and to have the uniform distribution of tegument characteristic of early-harvest compared to late-harvest virus (26). Measurement of virus infectivity by the particle/PFU ratio indicated that TNE-extracted virus more closely resembled the high infectivity of early-release virus compared to the lower infectivity found at late harvest times (Table 1). This similarity is reasonable, as cell-associated virus is the immediate precursor of virus detached from the cell with TNE.
The uniform tegument distribution observed in TNE-extracted virus (Fig. 1b) was expected, as a uniform distribution is found in cell-associated virus (26). The uniform distribution contrasts with the asymmetric tegument arrangement characteristic of late-harvest HSV-1 (8, 9, 25, 31). The correlation of asymmetric tegument and reduced infectivity in late-harvest virus suggests that the two phenomena may be related. An asymmetric tegument may not be compatible with the highest virion infectivity.
It was expected that most tegument proteins would be removed by treatment of TNE-released virus with 0.5% TX-100. Nearly complete removal was observed with cell-associated, but not late-harvest, virus (22, 26). The selective resistance of UL36 and UL37 to extraction was novel and suggests that, compared to other tegument proteins, UL36 and UL37 are more strongly anchored to the capsid. Selective resistance to the removal of UL36 and UL37 is also observed in late-harvest HSV-1 after extraction with TX-100 containing 1 M KCl (39). The strong association of UL36 and UL37 with the capsid, as suggested in both studies, may be related to the need of inner tegument proteins to remain bound to the capsid after entry of the tegument-capsid complex into the host cell cytoplasm. Evidence has been presented indicating that UL36 is attached to the capsid by way of an interaction involving capsid protein UL25 and the C-terminal 63 amino acids of UL36 (2, 16).
It was consistently observed that despite their greater mass, T36 capsids migrated more slowly than C capsids during sucrose density gradient centrifugation. We suggest that this effect may be due to hydrodynamic drag produced by projecting UL36 and UL37 molecules. It is consistent with the above interpretation that removal of UL36 and UL37 by sonication or by trypsin treatment produced capsids that migrate coincidentally with C capsids (Fig. 5a) (26).
Electron microscopy.
Electron microscopic examination of T36 capsids showed that they contain short tufts or stems and extended threads not observed in nuclear C capsids (Fig. 3). Both stems and threads were found to correlate with the presence of UL36 and UL37. We therefore suggest that UL36 and UL37 (1,123 amino acids) are the subunits of the stems and threads. The difference between the tuft and thread morphologies is suggested to be due to the conditions of T36 capsid staining and drying for electron microscopy. For example, a higher stain concentration may lead to strand condensation to form stems. While electron micrographs showed that most threads originate at capsid vertices, some were observed at nonvertex sites. The appearance of nonvertex threads indicates that some may arise from capsid edges or faces or, alternatively, from an obscured vertex. We favor the view that nonvertex threads arise from obscured vertices, as such threads are only rarely seen in electron micrographs.
Assignment of UL36 and UL37 to the tufts and threads is supported by the results of studies in which T36 capsids were subjected to brief sonication. Sonic treatment caused removal of the threads (Fig. 5c), and removal was found to correlate with loss of UL36 and UL37, as judged by SDS-polyacrylamide gel analysis of sonicated capsids (Fig. 5b). UL36 and UL37 were also found to be removed by brief treatment of capsids with 0.1 μg/ml trypsin (26). Sensitivity to trypsin treatment is consistent with the idea that UL36 and UL37 are exposed on the T36 capsid surface. Also consistent with this assignment are the results of protein secondary structure prediction that suggest thin, extended structures for both UL36 and UL37 (see, for example, http://www.sbg.bio.ic.ac.uk/phyre/html/index.html and http://bioinf4.cs.ucl.ac.uk:3000/psipred/). Both UL36 and UL37 have the potential to form extended structures with the length observed for the T36 capsid threads (Fig. 4, top panel).
Tegument structure.
The threads or strands were found to vary considerably in length, with a range of 15 to 70 nm (Fig. 4, top panel). The longest of the strands have the potential to span the thickness of the tegument (30 to 50 nm). They therefore suggest themselves as structures that could function in organizing the tegument as a whole since they have the capacity to reach all parts of the overall tegument mass. We suggest that the tegument may be composed of flexible UL36 and UL37 molecules that extend to all accessible regions near the capsid surface. The remainder of the tegument (most of its mass) is suggested to be created by proteins that bind directly or indirectly to UL36 or UL37.
The observed variability in strand length may be an authentic part of their nature. Alternatively, it could be due to strand breakage as T36 capsids are prepared for electron microscopy, or it could result from a variable amount of UL36/UL37 protein condensed in tufts. We favor the idea that UL36 and UL37 may be condensed in tufts to a variable extent because SDS-polyacrylamide gels show no evidence of truncated forms of UL36 or UL37 (Fig. 2b).
Features of strand morphology suggest that at least some are molecular oligomers. For instance, there are two distinct populations of strands that differ in thickness (Fig. 4, bottom panel). These are interpreted to correspond to molecular monomers (2.9 nm in diameter) and dimers (3.6 nm), respectively. Occasional strand images show evidence of branching (Fig. 3e and f), indicating that the parent strand is likely to be a dimer or higher oligomer. Strands composed of more than one component strand could be homooligomers of either UL36 or UL37, or they could be heterooligomers containing UL36 and UL37 component strands. The documented interaction between UL36 and UL37 suggests that at least some are heterooligomers (24, 32, 38).
Although electron micrographs demonstrated that most of the strands were straight, projecting radially outward from the capsid, others were found to be gently curved indicating a degree of flexibility in the structure (Fig. 3). Such flexibility may underlie the way the tegument is able to change its morphology in progressing from a uniform distribution found in cell-associated virus to the asymmetric form found in late-harvest virions (26).
In the future, the ready availability of capsids (T36 capsids) with a small number of tegument proteins may make it possible to examine events of tegument formation with in vitro experiments. For instance, addition of tegument proteins to T36 capsids may result in assembly events that would be revealing about tegument formation as it occurs in infected cells.
Acknowledgments
We gratefully acknowledge Dean Kedes and Rebecca Mingo for helpful comments on the manuscript.
This work was supported by NIAID award AI041644.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Buchkovich, N. J., T. G. Maguire, A. W. Paton, J. C. Paton, and J. C. Alwine. 2009. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. J. Virol. 83:11421-11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coller, K. E., J. I. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farnsworth, A., and D. C. Johnson. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J. Virol. 80:3167-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 7.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granzow, H., F. Weiland, A. Jons, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grünewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 10.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, B., M. Gross, and B. Roizman. 1997. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishiyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 80:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, K., T. Daikoku, F. Goshima, H. Kume, K. Yamaki, and Y. Nishiyama. 2000. Synthesis, subcellular localization and VP16 interaction of the herpes simplex virus type 2 UL46 gene product. Arch. Virol. 145:2149-2162. [DOI] [PubMed] [Google Scholar]
- 15.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loret, S., G. Guay, and R. Lippe. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605-8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matusick-Kumar, L., W. W. Newcomb, J. C. Brown, P. J. McCann, W. Hurlburt, S. P. Weinheimer, and M. Gao. 1995. The C-terminal 25 amino acids of the protease and its substrate ICP35 of herpes simplex virus type 1 are involved in formation of sealed capsids. J. Virol. 69:4347-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurer, U. E., B. Sodeik, and K. Grünewald. 2008. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc. Natl. Acad. Sci. U. S. A. 105:10559-10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKnight, J. L., T. M. Kristie, and B. Roizman. 1987. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc. Natl. Acad. Sci. U. S. A. 84:7061-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73:269-276. [DOI] [PubMed] [Google Scholar]
- 23.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mijatov, B., A. L. Cunningham, and R. J. Diefenbach. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26-31. [DOI] [PubMed] [Google Scholar]
- 25.Morgan, C., H. M. Rose, and B. Mednis. 1968. Electron microscopy of herpes simplex virus. I. Entry. J. Virol. 2:507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newcomb, W. W., and J. C. Brown. 2009. Time-dependent transformation of the herpesvirus tegument. J. Virol. 83:8082-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newcomb, W. W., S. K. Cockrell, F. L. Homa, and J. C. Brown. 2009. Polarized DNA ejection from the herpesvirus capsid. J. Mol. Biol. 392:885-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2005. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J. Virol. 79:10540-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 31.Rixon, F. J. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4:135-144. [Google Scholar]
- 32.Roberts, A. P., F. Abaitua, P. O'hare, D. McNab, F. J. Rixon, and D. Pasdeloup. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo, J. Y., and W. J. Britt. 2008. Multimerization of tegument protein pp28 within the assembly compartment is required for cytoplasmic envelopment of human cytomegalovirus. J. Virol. 82:6272-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stylianou, J., K. Maringer, R. Cook, E. Bernard, and G. Elliott. 2009. Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J. Virol. 83:5204-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taddeo, B., W. Zhang, and B. Roizman. 2006. The U(L)41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. U. S. A. 103:2827-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfstein, A., C. H. Nagel, K. Radtke, K. Dohner, V. J. Allan, and B. Sodeik. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227-237. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, Y., and J. L. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 67:1482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]