Abstract
Nanoviruses are multipartite single-stranded DNA (ssDNA) plant viruses that cause important diseases of leguminous crops and banana. Little has been known about the variability and molecular evolution of these viruses. Here we report on the variability of faba bean necrotic stunt virus (FBNSV), a nanovirus from Ethiopia. We found mutation frequencies of 7.52 × 10−4 substitutions per nucleotide in a field population of the virus and 5.07 × 10−4 substitutions per nucleotide in a laboratory-maintained population derived thereof. Based on virus propagation for a period of more than 2 years, we determined a nucleotide substitution rate of 1.78 × 10−3 substitutions per nucleotide per year. This high molecular evolution rate places FBNSV, as a representative of the family Nanoviridae, among the fastest-evolving ssDNA viruses infecting plants or vertebrates.
Viruses face continuous challenges by their hosts and in their vectors, which require them to cope with numerous different antiviral reactions activated upon infection. As a result, virus genomes vary in sequence to an extent that in some cases makes it difficult to define a single unique prototype, e.g., for bacteriophage Qβ or human immunodeficiency virus (HIV) (13, 74). Evidence has accumulated that rapid virus evolution due to genome sequence variation is general (18) and not restricted to RNA viruses and retroviruses, where this was amply documented (15, 43, 62, 74). The main generators of sequence variation in these viruses are their RNA-dependent RNA polymerases or reverse transcriptases due to their elevated error rates and lack of proofreading activity (4, 6, 14, 15). Viruses with a double-stranded DNA (dsDNA) genome are generally less variable in their sequences, since they encode less-error-prone polymerases or rely on proofreading host-encoded DNA polymerases and repair systems (18, 33, 48, 55). Viruses with a single-stranded DNA (ssDNA) genome depend for replication exclusively on host DNA polymerases yet exhibit a degree of sequence variability similar to that of RNA viruses (26, 52, 65, 66). ssDNA plant virus variability and rapid sequence evolution were documented for geminiviruses with monopartite and bipartite genomes (7, 16, 17, 34, 40, 42, 68, 71). The polymerases that replicate ssDNA plant virus genomes are not known; hence, their respective contributions to the sequence diversity of this virus group remain elusive.
A group of ssDNA viruses in which the genomic sequence information is distributed over multiple genome components is the Nanoviridae, a family of aphid-transmitted plant viruses that individually encapsidate six or eight ssDNAs of about 1 kb, which do not replicate in the aphid vectors (73). This family comprises two genera with three and four species, respectively. The genomes of banana bunchy top virus (BBTV) (8) and abacá bunchy top virus (ABTV) (67) in the genus Babuvirus and of faba bean necrotic yellows virus (FBNYV) (45), faba bean necrotic stunt virus (FBNSV) (37), milk vetch dwarf virus (MDV) (63), and subterranean clover stunt virus (SCSV) (10) in the genus Nanovirus have been sequenced. Only five of the six to eight nanovirus-encoded proteins have been functionally characterized (38, 73). Nanovirus molecular biology is still in its infancy, owing mainly to the fact that the viruses are not transmissible. Purified virions or virion-derived DNA becomes infectious with a low level of efficiency upon particle bombardment (27). Infectivity of cloned DNAs of FBNYV was demonstrated in 2006, yet the virus produced upon infection was not transmissible by the natural aphid vectors (70).
The recent establishment of cloned FBNSV DNAs that are fully infectious and also lead to the production of virions that can be readily and sustainably transmitted by the pea aphid (Acyrthosiphon pisum) and the cowpea aphid (Aphis craccivora), two natural insect vectors, represents a considerable accomplishment for nanovirus research (37). It was based on the sequencing of more than 200 individual genomic DNA molecules and the choice of suitable molecules from the eight heterogeneous ssDNA swarms that constitute the FBNSV genome.
Here we present a detailed analysis of the sequence heterogeneity in two FBNSV populations that were sampled about 2.4 years apart. The first sample is a field isolate from Holetta, Ethiopia, collected in 1997. The second sample represents a virus population derived from the 1997 field population after more than 30 successive acquisition and inoculation cycles using large numbers (>100) of pea aphids (37). In addition, we determined the sequence variation in a virus population resulting from the most severe possible bottleneck, a single-molecule infection using cloned DNAs.
MATERIALS AND METHODS
Characterization of FBNSV by rolling-circle amplification (RCA) and PCR-based cloning and sequencing.
FBNSV-infected faba bean (Vicia faba) plants were collected in October 1997 near Holetta, Ethiopia, and the virus was maintained by serial vector transmissions using elevated numbers (>100 per plant) of the pea aphid (Acyrthosiphon pisum Harris) at the Julius Kühn-Institute (JKI), Braunschweig, Germany, until 2004 (37). During 1998 to 1999, DNA of samples collected over almost 1 year was amplified by several immunocapture-PCRs (IC-PCRs) using primer pairs indicated in Table S1 in the supplemental material, cloned, and sequenced as described previously (1). These sequences (GenBank accession numbers AJ749894 to AJ749901) are referred to here as isolate FBNSV-[ET:Hol;JKI-1998/99].
DNA of the original field isolate FBNSV-[ET:Hol;1997] from Holetta, Ethiopia, collected in October 1997, and from the isolate FBNSV-[ET:Hol;JKI-2000], derived from the field population and maintained in faba bean for about 2.4 years by >30 sequential transmissions by pea aphid at JKI, was isolated, amplified by RCA, cloned, and sequenced as described previously, taking care to analyze plasmids from as many independent transformation events as possible (37). In addition, the population of genomic DNAs of virus reconstituted by agroinoculation of faba bean with a mixture of eight individually cloned single genomic components, followed at about 3 weeks postinoculation (p.i.) by one transmission (T1) using cowpea aphids, was analyzed the same way. This sample was termed isolate FBNSV-[ET:Hol;ISV-T1]. Standard Sanger dideoxy BigDye terminator cycle sequencing on an ABI 3730xl instrument using M13 direct and reverse primers was carried out at GATC Biotech GmbH, Konstanz, Germany. Read lengths usually covered both DNA strands of an entire cloned FBNSV genome component; in some cases, component-specific primers were used to close gaps, thus ensuring ∼2× base coverage of all cloned viral DNAs used for analysis. In cases where cloning of the restricted RCA product did not immediately yield a sufficient number of individual clones (>20) for each of the eight genome components, we used component-specific primers (see Table S1 in the supplemental material) for PCR amplification by the thermostable and high-fidelity proofreading Pfu DNA polymerase (Fermentas). Subsequently, we cloned the PCR products representing the respective individual complete genomic DNAs and determined their sequences. The FBNSV DNAs thus cloned represent 8.6%, 9.5%, and 32.5%, respectively, of the FBNSV-[ET:Hol;1997], FBNSV-[ET:Hol;JKI-2000], and FBNSV-[ET:Hol;ISV-T1] populations.
In order to assess the contribution of potential errors introduced during RCA by the φ29 polymerase and in the process of passage through Escherichia coli and cycle sequencing of the amplified products, we reamplified, recloned, and resequenced the same recombinant plasmids of all eight FBNSV-[ET:Hol;JKI-2000] DNAs that had been used to reconstitute the FBNSV genome (37). For that purpose, we separately cultured E. coli harboring the recombinant Litmus 28 or 38 plasmid, each carrying one of the eight FBNSV DNAs, for >18 h in LB medium. We injected ∼200 μl of each culture into noninfected faba bean leaves, which were then immediately frozen in liquid nitrogen. Eight total DNA preparations thereof were subjected to individual RCAs exactly as described for FBNSV-infected tissue (37). We restricted the amplified DNA with ScaI endonuclease, which cuts in the β-lactamase gene of Litmus 28 or 38 plasmid; electrophoresed the product on 1% agarose gels; excised the resulting fragments (Litmus plus the respective FBNSV DNA insert) from the gel; circularized them by ligation; and introduced them into chemically competent E. coli strain Top10 (Invitrogen). The inserts of at least 10 independent recombinant plasmids (colonies) derived from each of the eight individual RCA, restriction, ligation and transformation experiments, along with a control sample of the respective bacterial cultures used to inject the faba bean leaves, were sequenced as described above.
Sequence analysis.
Sequences were analyzed with the Genetics Computer Group (GCG) (Madison, WI) software package, version 10.0 (11), and DNASTAR of Lasergene, version 8.0.2 (DNASTAR, Inc. Madison, WI). DNA sequences were aligned by ClustalW using the IUB DNA weight matrix. Phylogenetic analyses were conducted in PhyML (39) (maximum likelihood), and the resulting trees were plotted using MEGA version 4 (69). All positions containing gaps were eliminated from the data set. Mutation frequencies and Shannon entropy values were calculated as described by Domingo et al. (12). Genetic distances were estimated using the Tamura-Nei model implemented in MEGA, version 4 (69), with standard deviations calculated by bootstrapping with 1,000 replicates. Synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dN) were calculated according to single-likelihood ancestor counting (SLAC) analysis (47), as implemented in Datamonkey (http://www.datamonkey.org/) (46). Two-by-two chi-square (χ2) statistics (71) were applied to validate whether the observed substitutions differed from the expected frequency for a given substitution type calculated under the assumption that all substitution types were equally likely.
Nucleotide sequence accession numbers.
The FBNSV sequences reported here were deposited in GenBank under the accession numbers AJ749894 to AJ749901 for FBNSV-[ET:Hol;JKI-1998/99] and GU983866 to GU983873 for FBNSV-[ET:Hol;1997]. All sequences used for analysis are provided in the supplemental material.
RESULTS
Intragenome relationship of FBNSV DNAs.
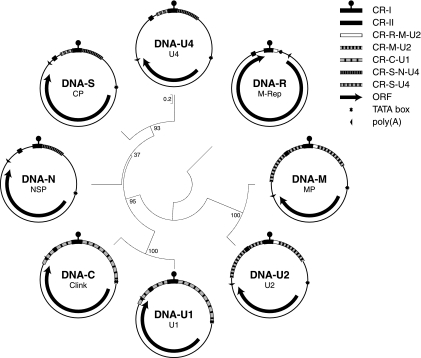
Grigoras et al. molecularly characterized the eight FBNSV genome components (37). Like those of all nanoviruses, the FBNSV DNAs have a 71- or 73-base common region (CR-I) flanking inverted repeat sequences, which potentially form a stem-loop and contain the viral (+) strand origin of replication (Fig. 1). CR-I is 100% identical between DNA-R, -S, -M, -N, -U2, and -U4 and between DNA-C and -U1, but there is only 83.6% identity between the two groups of CR-I sequences. A second region conserved in all eight genomic DNAs of FBNSV, referred to as the second common region (CR-II) (45), is 20 nucleotides (nt) long and located 5′ of CR-I. The sequence identity between components in this region varies from 75 to 100%. FBNSV DNAs form two groups irrespective of whether CR-I or CR-II is used for comparison: DNA-R, -S, -M, -N, -U2, and -U4 group together, whereas DNA-C and -U1 group separately. Figure 1 illustrates the relationship of the eight genomic FBNSV DNAs, arranged according their positions on a maximum-likelihood phylogenetic tree. If only the noncoding sequences are used, their relationship and the topology of the tree are the same. There are at least two instances where conserved sequences overlap with coding sequences: (i) CR-II within the 3′ part of the m-rep gene and (ii) the region common between DNA-C and DNA-U1 (CR-C-U1) with the 3′end of open reading frame (ORF) U1. However, in the case of DNA-U1 and DNA-C of the Moroccan isolate FBNSV-[MA;Mor5] (1) there is no overlap between CR-C-U1 and the respective protein-coding sequences. Also, the carboxy termini of the predicted U1 proteins of FBNSV-[MA;Mor5], FBNSV-[ET:Hol;1997], FBNYV, and MDV, in which CR-C-U1 also does not overlap the 3′ end of the ORF, vary considerably. Therefore, we consider conservation of the CR-C-U1 sequence as being dominant over its feature of also encoding part of a protein. Similar arguments may be raised for CR-II and the protein sequence of M-Rep. Stretches of sequence similarity occur in the entire noncoding regions, with sequence identities ranging from 44% to 95%. Several almost perfectly conserved regions (∼99% identity) are shared by all or by a subset of the eight DNAs and are highlighted in Fig. 1.
FIG. 1.
Genome organization of FBNSV. The eight viral DNAs are grouped according to their phylogenetic relationships. Branch lengths are drawn to scale with base substitutions per nucleotide as units, and branch support is shown by bootstrap values (500 replicates). Inside each circle the name of the genomic DNA is given, indicating the respective encoded proteins. M-Rep, master replication initiator protein; CP, capsid protein; Clink, cell cycle link protein; MP, movement protein; NSP, nuclear shuttle protein; U1, U2, and U4, proteins of as-yet-unknown function; CR-I, common region I (inverted repeat sequences flanking the replication origin are symbolized by a knob); CR-II, second common region. Other highly similar sequences (common regions) in DNAs are denoted as CR- followed by the name of the respective genomic DNA.
Variability of FBNSV is illustrated by intraisolate sequence heterogeneity.
Sequencing of between 20 and 40 individual clones of each of the eight genomic DNAs of FBNSV from Holetta, Ethiopia (isolate FBNSV-[ET:Hol;JKI-2000]) had revealed considerable intraisolate variability of the viral genome (37). To ensure that the observed variability was not artificially inflated by potential φ29 polymerase errors during RCA and the process of subsequent cloning in E. coli and cycle sequencing, a control experiment of reamplification, recloning, and resequencing 10 individual recombinant plasmids of each of the eight DNAs of FBNSV-[ET:Hol;JKI-2000] was carried out as described in Materials and Methods. For a total of 79,070 nucleotides sequenced, we found two independent mutations in DNA-N, one nonsynonymous transversion and one synonymous transition, corresponding to a mutation frequency of 2.53 × 10−5. Compared to the reported in vitro error frequency of the φ29 polymerase, ranging from 4 × 10−6 to 2.2 × 10−5 (24), and that of combined φ29 multiple displacement amplifications with subsequent PCR-based sequencing without cloning, determined as 9.5 × 10−6 per nucleotide sequenced (58), the experimental error frequency of 2.53× 10−5 in our amplification system, including cloning in E. coli, was only slightly higher. The mutation frequencies of the three FBNSV populations analyzed here were corrected for the experimental error frequency of 2.53 × 10−5, which marginally lowered the uncorrected values (Table 1). Mutation frequencies of individual genomic DNAs were not corrected.
TABLE 1.
Variability of field-isolated and laboratory-propagated FBNSV strains
| Genome component | FBNSV-[ET:Hol;1997] |
FBNSV-[ET:Hol;JKI-2000] |
FBNSV-[ET:Hol;ISV-T1] |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (nt) | No. of: |
Mutation frequency (×10−4) | Length (nt) | No. of: |
Mutation frequency (×10−4) | Length (nt) | No. of: |
Mutation frequency (×10−4) | ||||
| Number sequenced clones | Variable sitesa | Number sequenced clones | Variable sites | Number sequenced clones | Variable sites | |||||||
| DNA-R | 1,003 | 20 | 8 | 3.99 | 1,003 | 22 | 8 | 3.63 | 1,003 | 20 | 4 | 2.00 |
| DNA-S | 992 | 26 | 12 | 4.65 | 992 | 29 | 13 | 4.52 | 992 | 20 | 5 | 2.52 |
| DNA-C | 994 | 20 | 20 | 10.06 | 994 | 20 | 5 | 2.52 | 994 | 20 | 3 | 1.51 |
| DNA-M | 979 | 20 | 18 | 9.41 | 980 | 27 | 19 | 7.18 | 980 | 20 | 4 | 2.04 |
| DNA-N | 980 | 20 | 32 | 16.33 | 981 | 40 | 18 | 4.59 | 981 | 42 | 4 | 0.97 |
| DNA-U1 | 985 | 39 | 16 | 4.18 | 986 | 29 | 24 | 8.40 | 986 | 21 | 1 | 0.48 |
| DNA-U2 | 983 | 22 | 18 | 8.35 | 984 | 26 | 16 | 6.26 | 984 | 20 | 7 | 3.56 |
| DNA-U4 | 987 | 20 | 19 | 9.63 | 987 | 39 | 19 | 4.94 | 987 | 43 | 11 | 2.59 |
| Entire genome (correctedb) | 7,903 | 187 | 143 | 7.77 (7.52) | 7,907 | 232 | 122 | 5.33 (5.07) | 7,907 | 206 | 39 | 1.92 (1.66) |
| Total no. of nt sequenced | 184,000 | 229,049 | 203,392 | |||||||||
Variable sites include bases changes, indels, and recombination events.
The experimental error frequency of our amplification-cloning system of 2.53 × 10−05 was subtracted from the observed mutation frequency to yield the corrected mutation frequency.
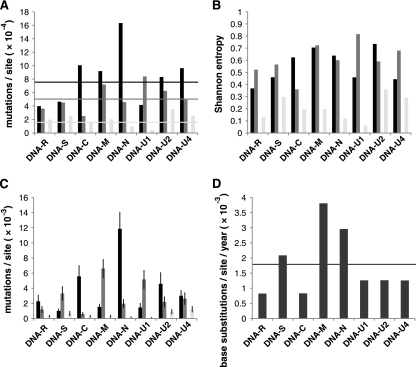
As the FBNSV-[ET:Hol;JKI-2000] isolate was sampled after about 30 successive aphid transmissions of the virus in the laboratory, we were interested in determining the extent of heterogeneity in the original field isolate from Holetta, Ethiopia (FBNSV-[ET:Hol;1997]). We analyzed a total of 187 individually cloned genomic DNAs of the 1997 field sample (184,000 nt sequenced), which exhibited an overall variability of 7.52 × 10−4 alterations per site (base changes, deletions, insertions, or recombination events combined) (Table 1 and Fig. 2 A). A total of 143 deviations from the respective consensus sequences and 77 distinct types of molecules were identified (see Table S2 in the supplemental material). The overall sequence heterogeneity of the 1997 FBNSV field sample was about 1.5-fold higher than that of the laboratory-maintained virus population, for which an overall (entire genome) mutation frequency of 5.07 × 10−4 changes per nucleotide sequenced was determined, (Table 1). When considering each of the eight different genomic DNAs of the original field sample individually, up to a 4-fold variation in sequence diversity between them was apparent. DNA-N was the most polymorphic genome component, with 16.33 × 10−4 changes per nucleotide, and DNA-R the least variable one, with 3.99 × 10−4 changes per nucleotide (Table 1). In contrast, in the FBNSV-[ET:Hol;JKI-2000] sample of the laboratory-maintained population the most conserved genome component was DNA-C. Only five single-base deviations from the consensus were observed within 19,880 nt (20 clones of individual DNA-C molecules, each 994 bp in length), equivalent to a frequency of 2.52 × 10−4 mutations per nucleotide sequenced (Table 1), whereas DNA-U1 had accumulated 24 mutations per 28,585 nt (29 clones of individual DNA-U1 molecules, 20 of 986 bp and 9 of 985 bp in length) (Table 1), resulting in an about 3-fold-higher mutation frequency of 8.40 × 10−4 changes per nucleotide. Considering the entire genome, about a 1.5-fold reduction of sequence heterogeneity, reflected by a drop in mutation frequency in the FBNSV-[ET:Hol;JKI-2000] population, had occurred upon the successive passaging by aphid transmission over a period of about 2.4 years (Fig. 2A).
FIG. 2.
Variability and evolution rates of a field isolate and laboratory-propagated isolates of FBNSV. Mutation frequencies (A), genetic diversities (B), genetic distances (C), and substitution rates (D) of the eight individual genomic DNAs within each of the three FBNSV populations are shown as bar diagrams. Black, FBNSV-[ET:Hol;1997] (original field isolate); dark gray, FBNSV-[ET:Hol;JKI-2000] (laboratory-maintained population); light gray, FBNSV-[ET:Hol;ISV-T1] (bottleneck-derived population). Horizontal lines in panel A indicate the mutation frequencies of the entire FBNSV genome (average of those for the eight individual DNAs) in the respective three populations: 7.52 × 10−4 mutations/site for FBNSV-[ET:Hol;1997], 5.07 × 10−4 mutations/site for FBNSV-[ET:Hol;JKI-2000], and 1.66 × 10−4 mutations/site for the single-transfer FBNSV-[ET:Hol;ISV-T1] population. Error bars in panel C refer to standard deviations of the genetic distances. The horizontal line in panel D indicates the overall nucleotide substitution rate of FBNSV (1.78 × 10−3 substitutions/site/year). The data and numerical values used for calculations and graphic display are provided in Tables S2 and S3 in the supplemental material.
This reduction in variability is also clearly seen when following the maintenance over time of distinct sequence subgroups. By looking at sequence covariations, represented as clustered deviations from the respective consensus sequences in individual cloned molecules from the 1997 field sample, distinct subgroups of sequence variants within the DNAs-C, -N, and -U2 populations are evident (see Fig. S1 in the supplemental material). Despite using large numbers of aphids for vector transmission of the virus in the laboratory, only some of these subgroups were maintained through passaging in the laboratory, as well illustrated by the substantial drop in the genetic distance among DNA-C, -N, and -U2 population members (Fig. 2C; see Table S2 in the supplemental material).
Whereas the sequence heterogeneity of seven of the FBNSV genomic DNAs diminished upon repeated passaging in faba bean, that of DNA-U1 increased about 2-fold between 1997 and 2000 (Table 1 and Fig. 2, A). When the genetic complexity of the virus population in the 1997 field sample was compared with that of the laboratory-maintained population, reflected by the Shannon entropy of the eight individual genomic DNAs, again DNA-U1 showed an about 2-fold increase (Fig. 2B; see Table S2 in the supplemental material). A similar increase in Shannon entropy was calculated for DNA-U4, despite a reduction in its mutation frequency (Fig. 2A and B). For the moment, it appears to be difficult to explain such an increase in entropy, in particular as the functions of both the U1 and U4 proteins are not known.
The FBNSV genome evolves rapidly.
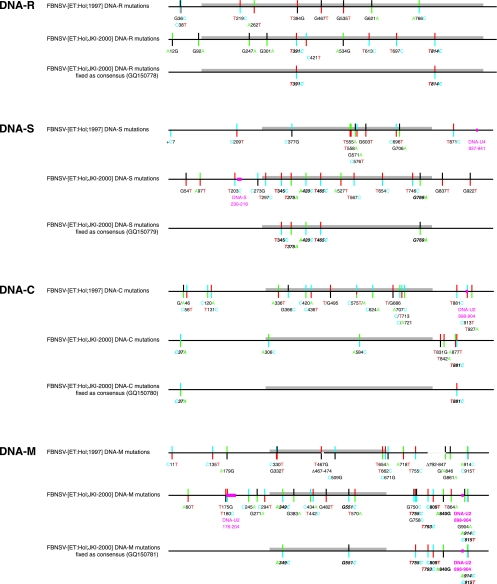
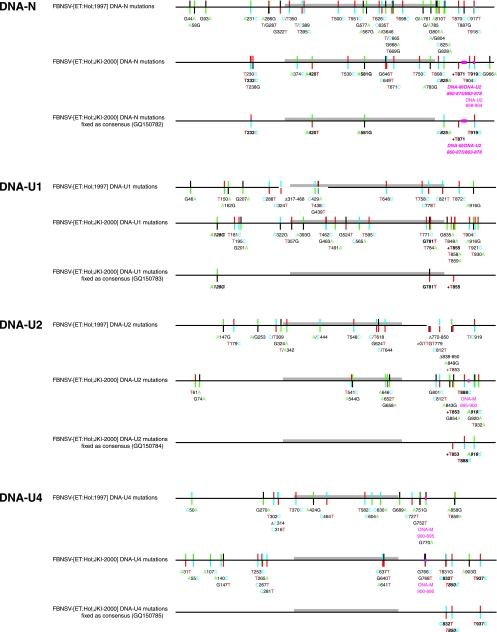
Figure 3 provides a comprehensive graphic summary of all sequence variations shown as color-coded deviations from the respective consensus sequences of the eight genomic FBNSV DNAs. The consensus of the 1997 field sample population serves as a reference. Mutations that were found completely fixed in the laboratory-maintained FBNSV-[ET:Hol;JKI-2000] population (constituting 100% of the consensus) are shown in bold italics; mutations found in the majority of cases (>50% and <100%) are shown in bold. This distinction is important, as the former changes do not contribute to the mutation frequency of the FBNSV-[ET:Hol;JKI-2000] sample whereas the latter do (Table 1).
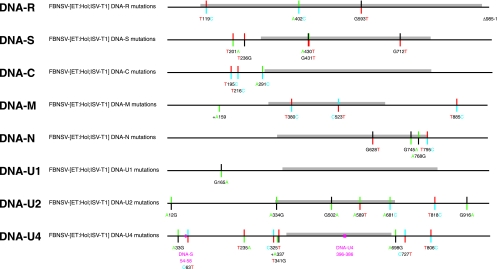
FIG. 3.
Summary of sequence alterations in the field and laboratory-propagated FBNSV populations. Nanovirus DNAs are shown in a linear fashion as scaled line drawings, with ORF sequences represented as gray bars. Positions of variable sites are indicated by bicolored vertical lines: the upper parts of the vertical lines denote the nucleotide present in the FBNSV-[ET:Hol;1997] consensus sequence and the bottom part the allele. Mutations that were found completely fixed in the FBNSV-[ET:Hol;JKI-2000] population are shown in bold italics, and mutations found in the majority of cases are shown in bold. Nucleotides are color coded as in Applied Biosystems BigDye terminator sequencing: A, green; T, red; C, blue; and G, black. Deletions (Δ) or recombinant sequences are represented by blank or magenta sections of the scaled lines, respectively. Insertions are marked by a +.
Based on a total of 34 mutations that became fixed after 2.4 years as the consensus sequence of the FBNSV-[ET:Hol;JKI-2000] population compared with the original Holetta field isolate FBNSV-[ET:Hol;1997], FBNSV evolved at an overall rate of 1.78 × 10−3 substitutions per nucleotide per year. The substitution rate for the combined noncoding regions was about twice that for the combined coding regions: 2.25 × 10−3 versus 1.29 × 10−3 (see Table S3 in the supplemental material). When considering the eight genomic DNAs individually, about a 4.6-fold variation in substitution rates between them became apparent, revealing DNA-M as the fastest-changing genome component (Fig. 2D; see Table S3 in the supplemental material).
Consensus sequences of FBNSV were also determined in 1998 to 1999. As these sequences originated from several samples taken over a period of more than 6 months and were based on a lower coverage of the genome, we refrained from using them for rate calculations. Their respective degrees of similarity with DNAs of either the FBNSV-[ET:Hol;1997] field population or the FBNSV-[ET:Hol;JKI-2000] laboratory population can be assessed from Fig. S1 in the supplemental material. A 55-nt deletion in the noncoding region of DNA-M found in 8 out of 20 cloned molecules from the FBNSV-[ET:Hol;1997] sample was also present in the two available 1998/1999 consensus sequences but absent in the FBNSV-[ET:Hol;JKI-2000] population. Whether this mutant type of DNA-M is biologically active in an infection along with the other genome components is not known.
Distribution of mutations.
Although the extent of heterogeneity varied between different genome components (e.g., DNA-C and DNA-N), no particular hot or cold spots for mutations were observed within a given genomic DNA. Mutations were distributed almost equally between noncoding and coding sequences, i.e., 52% in the noncoding regions of FBNSV-[ET:Hol;1997] field population and 61% in those of the FBNSV-[ET:Hol;JKI-2000] laboratory population, revealing only a slight mutation bias toward noncoding sequences. The mutations in the coding sequences of the FBNSV-[ET:Hol;1997] field population comprised a total of 36 synonymous and 28 nonsynonymous ones (Table 2). Synonymous mutations per synonymous site (dS) and nonsynonymous mutations per synonymous site (dN) were calculated by SLAC analysis using Datamonkey (46), with dN/dS ratios ranging from 0.055 (DNA-N) to 0.566 (DNA-C). All ratios are <1, indicating various degrees of conservation bias for the respective proteins. No particular individual sites subject to either positive or negative selection (significance level of 0.1) were identified.
TABLE 2.
Distribution of mutations and diversity for synonymous (S) and nonsynonymous (N) sites of field-isolated and laboratory-propagated FBNSV strains
| Genome component | FBNSV-[ET:Hol;1997] |
FBNSV-[ET:Hol;JKI-2000] |
FBNSV-[ET:Hol;ISV-T1] |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of mutations |
dN/dS | No. of mutations |
dN/dS | No. of mutations |
dN/dS | |||||||||||||
| Total | Noncoding region | Coding region |
Total | Noncoding region | Coding region |
Total | Noncoding region | Coding region |
||||||||||
| S | N | ORF length changing | S | N | ORF length changing | S | N | ORF length changing | ||||||||||
| DNA-R | 8 | 2 | 3 | 2 | 1 | 0.173 | 8 | 2 | 5 | 1 | 0 | 0.053 | 4 | 1 | 0 | 3 | 0 | NDa |
| DNA-S | 12 | 4 | 4 | 4 | 0 | 0.270 | 13 | 7 | 4 | 2 | 0 | 0.139 | 5 | 2 | 0 | 3 | 0 | ND |
| DNA-C | 20 | 8 | 3 | 8 | 1 | 0.566 | 5 | 3 | 0 | 2 | 0 | ND | 3 | 3 | 0 | 0 | 0 | ND |
| DNA-M | 18 | 11 | 4 | 2 | 1 | 0.133 | 19 | 14 | 3 | 2 | 0 | 0.181 | 4 | 2 | 1 | 1 | 0 | 0.272 |
| DNA-N | 32 | 16 | 13 | 3 | 0 | 0.055 | 18 | 9 | 4 | 5 | 0 | 0.423 | 4 | 0 | 0 | 3 | 1 | ND |
| DNA-U1 | 16 | 8 | 3 | 3 | 2 | 0.151 | 24 | 13 | 5 | 5 | 1 | 0.191 | 1 | 1 | 0 | 0 | 0 | ND |
| DNA-U2 | 18 | 12 | 3 | 3 | 0 | 0.228 | 16 | 11 | 0 | 5 | 0 | ND | 7 | 3 | 0 | 4 | 0 | ND |
| DNA-U4 | 19 | 13 | 3 | 3 | 0 | 0.274 | 19 | 16 | 0 | 3 | 0 | ND | 11 | 10 | 0 | 0 | 1 | ND |
| Entire genome | 143 | 74 | 36 | 28 | 5 | ND | 122 | 75 | 21 | 25 | 1 | ND | 39 | 22 | 1 | 14 | 2 | ND |
ND, not determined.
In the FBNSV-[ET:Hol;JKI-2000] population, there were fewer synonymous than nonsynonymous mutations (21 versus 25). Where possible, the dN/dS ratios were calculated; they ranged from 0.053 (DNA-R) to 0.423 (DNA-N). The increase of the dN/dS ratios for DNA-N between the 1997 and the 2000 samples might indicate that the encoded protein is not (yet) well adapted to faba bean as a continuous host. DNA-R, -S, -M, and -U1 had the lowest dN/dS ratios throughout, consistent with the fact that the encoded proteins are essential for infectivity (70).
Variability generated after a severe bottleneck.
Having compared the variability of a field population of FBNSV with that of one derived thereof and maintained for 2.4 years in the laboratory, we were interested in the degree of sequence variation occurring after infection of faba bean by a single molecule of each of the eight virus DNAs. For this purpose we analyzed the virus population resulting from agroinoculation of faba bean with eight cloned FBNSV DNAs representing the FBNSV-[ET:Hol;JKI-2000] population consensus, followed by a single aphid transfer of the virus (37). From this sample, FBNSV-[ET:Hol;ISV-T1], we cloned and sequenced 206 individual genomic DNA molecules representing a total of 203,392 nt. Mutation frequencies, genetic diversity, and genetic distance were determined as described above and are summarized in Table 1, Fig. 2, Table S2C in the supplemental material, and Fig. 4. Remarkably, we observed a frequency of 1.66 × 10−4 mutations per nucleotide sequenced, corresponding to about 22% of the values found in the 1997 field population. This indicated that within the limited number of replication cycles during about 6 weeks, already a considerable number of sequence variants were generated from single unique molecules. As noted above for the virus population from the field and the population maintained for 2.4 years in the laboratory, variation was about the same for noncoding and coding sequences, i.e., 56% in the noncoding parts of the genome (Table 2). Compared to the synonymous mutations, the number of nonsynonymous mutations was rather elevated (1 versus 14). However, none of the mutations had yet become fixed as consensus (see Fig. S1 in the supplemental material), suggesting either that the time for the mutations to become fixed was not long enough or that the FBNSV-[ET:Hol;JKI-2000] clones used for agroinoculation were sufficiently fit to be maintained for the multiplication period of about 6 weeks.
FIG. 4.
Mutations after one transmission of single-molecule-derived virus. Mutations detected in the FBNSV-[ET:Hol;ISV-T1] virus sample taken 6 weeks after agroinoculation using eight individually cloned single genomic DNAs followed by one aphid transmission are shown. Variable sites are color coded as in Fig. 3.
Types of sequence alterations.
In general, in FBNSV-[ET:Hol;JKI-2000], transitions were more common than transversions (50.8% versus 41.8%), but depending on the component, the transition/transversion ratio (Ts/Tv) varied from 0/5 in DNA-C to 8/0 in DNA-R (Table 3; see Table S4 in the supplemental material). The most frequent base substitution was T → C (23.8%), followed by G → A (11.5%), whereas the transversions C → A (2.5%), C → G (1.6%), and G → C (3.3%) were rare. Table 3 summarizes the type of mutations and their respective probabilities, assuming no transition/transversion bias imposed by DNA structure, the biochemistry of the way in which the mutations were generated, or whether they occurred in coding or noncoding sequences. The Ts/Tv for the noncoding sequences in the FBNSV-[ET:Hol;1997] field isolate was 3.00, compared to 1.31 in the respective coding sequences of the same isolate. The Ts/Tv was more uniform in the FBNSV-[ET:Hol;JKI-2000] laboratory-maintained population, with ratios of 1.47 for the noncoding sequences and 1.06 for the coding sequences.
TABLE 3.
Mutant spectra of field-isolated and laboratory-propagated FBNSV strains
| Sequence alteration | FBNSV-[ET:Hol;1997] |
FBNSV-[ET:Hol;JKI-2000] |
FBNSV-[ET:Hol;ISV-T1] |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. observeda | No. expected | χ2 | P value | No. observed | No. expected | χ2 | P value | No. observed | No. expected | χ2 | P value | |
| Transitions | ||||||||||||
| Total | 86 | 43.67 | 61.5611 | 4.29 × 10−15 | 62 | 38 | 23.5820 | 1.20 × 10−6 | 21 | 11.33 | 12.3689 | 0.0004 |
| A → G | 11 (+7) | 12.72 | 0.2576-2.4273 | 0.6118-0.1192 | 10 | 10.93 | 0.0873 | 0.7677 | 5 | 3.29 | 0.9851 | 0.3209 |
| G → A | 16 (+7) | 9.58 | 4.6409-20.2801 | 0.0312-6.69 × 10−6 | 14 | 8.28 | 4.2718 | 0.0387 | 4 | 2.49 | 0.9879 | 0.3203 |
| C → T | 22 (+8) | 7.34 | 31.0509-74.1723 | 2.51 × 10−8-7.16 × 10−18 | 9 | 6.40 | 1.1234 | 0.2892 | 4 | 1.92 | 2.3765 | 0.1232 |
| T → C | 22 (+8) | 14.03 | 5.0704-20.3583 | 0.0243-6.42 × 10−6 | 29 | 12.06 | 26.6105 | 2.49 × 10−7 | 8 | 3.63 | 5.8885 | 0.0152 |
| Transversions | ||||||||||||
| Total | 45 | 87.33 | 61.5611 | 4.29 × 10−15 | 51 | 75 | 23.5820 | 1.20 × 10−6 | 13 | 22.66 | 12.3689 | 0.0004 |
| A → C | 4 (+2) | 12.72 | 6.6208-3.9321 | 0.0101-0.0474 | 7 | 10.93 | 1.5633 | 0.2112 | 3 | 3.29 | 0.0282 | 0.8667 |
| C → A | 9 (+2) | 7.34 | 0.3998-1.9386 | 0.5272-0.1638 | 3 | 6.40 | 1.9119 | 0.1668 | 0 | 1.92 | 2.0390 | 0.1533 |
| A → T | 4 (+1) | 12.72 | 6.6208-5.1894 | 0.0101-0.0227 | 8 | 10.93 | 0.8687 | 0.3513 | 2 | 3.29 | 0.5595 | 0.4545 |
| T → A | 6 (+1) | 14.03 | 5.1473-3.9451 | 0.0233-0.0470 | 12 | 12.06 | 0.0004 | 0.9840 | 2 | 3.63 | 0.8198 | 0.3652 |
| C → G | 3 | 7.34 | 2.7149 | 0.0994 | 2 | 6.40 | 3.2034 | 0.0735 | 0 | 1.92 | 2.0390 | 0.1533 |
| G → C | 2 | 9.58 | 6.4712 | 0.0110 | 4 | 8.28 | 2.3843 | 0.1226 | 0 | 2.49 | 2.6872 | 0.1012 |
| G → T | 7 (+3) | 9.58 | 0.7499-0.0198 | 0.3865-0.8880 | 9 | 8.28 | 0.0683 | 0.7938 | 4 | 2.49 | 0.9879 | 0.3203 |
| T → G | 4 (+3) | 14.03 | 8.0306-3.9451 | 0.0046-0.0470 | 6 | 12.06 | 3.4144 | 0.0646 | 2 | 3.63 | 0.8198 | 0.3652 |
| Ts/Tvb | 1.91 | 0.5 | 1.22 | 0.5 | 1.62 | 0.5 | ||||||
| Deletions | 6 | 0 | 1 | |||||||||
| Insertions | 3 | 3 | 2 | |||||||||
| Recombination events | 3 | 6 | 2 | |||||||||
Numbers in parentheses refer to potential additional mutations resulting from 50% ± 10% ambiguities of the consensus (reference) base and the respective change. Note that these numbers have to be identical for reciprocal changes (e.g., A → G and G → A, etc.), and they are counted only once for the total number of transitions or transversions.
Ts/Tv, transition/transversion ratio.
Recombination between FBNSV genome components.
Recombination has been shown to be frequent in a diverse range of ssDNA viruses (49), yet with only few examples of the Nanoviridae (30). Two of the 27 DNA-M sequences of the FBNSV-[ET:Hol;JKI-2000] isolate were found to be recombinant. The recombination occurred between DNA-M and DNA-U2 at a site localized after the end of CR-M-U2 (positions 176 to 205 in DNA-M) where stretches of identical sequences in the two DNAs begin to alternate with stretches of very divergent sequences (Fig. 3; see Fig. S1 in the supplemental material).
A second clear recombination event affected the CR-II of DNA-N in the FBNSV-[ET:Hol;JKI-2000] population, where a stretch of at least 16 nt was replaced by the corresponding sequence from DNA-M or DNA-U2 (Fig. 3; see Fig. S2 in the supplemental material).
A third and quite interesting type of sequence alteration due sequence conversion by recombination and recombinational sequence inversion was found between DNA-M or DNA-U2 and DNA-U4. There was a group of three informative nucleotide changes in the common region (CR-M-U2-U4) of FBNSV-[ET:Hol;JKI-2000] DNA-M, represented as either the sequence 895AG-TCTT900 or 895TGCTCTC901, and DNA-U2, where the identical alterations occurred (sequences 898 to 904/905, respectively). In DNA-U4 both alterations also occurred, but as the reverse complement sequence 766GAGAGCA772 or 766AAGA-CT771, respectively, because these changes are part of a 60- or 61-nt inversion comprising nt 883 to 942 of DNA-M, nt 886 to 946 of DNA-U2, and nt 724 to 784 of DNA-U4. In the FBNSV-[ET:Hol;1997] field population, DNA-M sequences were 100% of the 895AG-TCTT900 type. At equivalent positions the DNA-U2 sequences of the FBNSV-[ET:Hol;1997] field isolate were also 100% homogeneous, but as the other nucleotide combination of the alterations, 898TGCTCTC904. In contrast, both sequence alternatives were represented in DNA-U4 of the FBNSV-[ET:Hol;1997] field population as part of the 61-nt inversion. For further details, see Fig. S2 and Table S5 in the supplemental material.
Finally, there were several cases in which particular base changes might have been created by replication and/or repair errors or introduced by intracomponent recombination (e.g., see A916G in DNA-U1 in Fig. S1 and Table S5 in the supplemental material).
A detailed compilation of all mutations is provided in Table S5 in the supplemental material.
DISCUSSION
We analyzed the variability of the nanovirus FBNSV as a natural field isolate and as an isolate derived thereof that had been maintained in the laboratory over a period of about 2.4 years. Moreover, we also determined the mutation frequency apparent after infection by eight distinct individually cloned single genomic DNAs, representing a severe bottleneck, followed by only one aphid transfer. The observed variability ranged from 7.52 × 10−4 mutations per nucleotide sequenced for the FBNSV-[ET:Hol;1997] field population to 5.07 × 10−4 substitutions per nucleotide for the laboratory-maintained FBNSV-[ET:Hol;JKI-2000] population. These data allowed determination of an evolution rate of 1.78 × 10−3 substitutions/site/year for FBNSV, suggesting that the molecular evolution rate of this virus is at the upper end of the rates determined for a variety of RNA and DNA viruses (18, 43).
Numerous studies of the variability and molecular evolution of RNA plant viruses are available (for reviews, see references 25, 29, 32, and 60). In contrast, nucleotide substitution rates for ssDNA plant viruses have only recently been determined, either by evaluating geminivirus sequences available in databases (16, 17) or by directly sequencing parts of or the entire genomes of individual members of virus populations (34, 40, 71). These studies indicated an evolution rate of 2.9 × 10−4 nucleotide substitutions/site/year for the tomato yellow leaf curl viruses (TYLCVs) over a period of almost 20 years (16), while those for DNA-A and DNA-B of East African cassava mosaic viruses (EACMVs) over about 5 years were 1.6 × 10−3 and 1.3 × 10−4 substitutions/site/year, respectively (17). Analysis of a “short-term” evolution over a 60-day multiplication period of tomato yellow leaf curl China virus in muntju (Nicotiana benthamiana) revealed about 3.5 × 10−4 substitutions/site, resulting in an extrapolated evolution rate of about 2.1 × 10−3 substitutions/site/year (34). Maize streak virus (MSV) evolution over an experimental period of 5 or 6 years was similar, with a rate of 7.4 to 7.9 × 10−4 substitutions/site/year (71) or 2.0 × 10−4 substitutions/site/year (40), in line with earlier findings that determined about 2.6 × 10−4 substitutions/site/year for an MSV isolate maintained for about 4 years in the perennial host Coix lacryma-jobi (42). A 32-year heterochronous sampling experiment employing sugarcane streak Réunion virus (SSRV) revealed a comparable mutation rate of about 3.5 × 10−4 substitutions/site/year (40). Moreover, those authors clearly demonstrated that there is no codivergence of mastreviruses with their hosts, contrary to an earlier claim by Wu and coworkers (76). Another striking example of a very high rate of 1.2 × 10−3 substitutions/site/year and a noncodivergence between a circovirus and a vertebrate host is provided by the recent assessment of the evolutionary history of porcine circovirus 2 (26).
Concerning Nanoviridae, only one phylogenetic study of the local evolution of BBTV in Hawaii determined a mutation of 1.4 × 10−4 substitutions/site/year (2). From work under experimental conditions over a period of 1 year, the authors inferred an evolution rate of 3.9 × 10−4 substitutions/site/year. It should be noted, however, that this rate was based on a single base change.
The observed elevated mutation rate of FBNSV is further supported by the frequency of 1.66 × 10−4 alterations per nucleotide sequenced determined after a single passage by cowpea aphids of the virus reconstituted following agroinoculation with eight individually cloned genomic DNAs, notwithstanding the fact that none of the sequence changes identified had yet become fixed as consensus after about 6 weeks. Using a single cloned molecule of each genomic DNA represents the narrowest possible bottleneck, yet no obvious decrease in virus fitness in faba bean as the host has been observed during a further period of more than 18 months of successive aphid transfers. Clear bottleneck effects were also apparent when comparing the FBNSV-[ET:Hol;1997] field population with that of the laboratory-maintained FBNSV-[ET:Hol;JKI-2000]: three distinct subpopulations of DNA-N molecules were present in the field population, yet only one of them had been passed through the multiple successive aphid acquisitions and transmissions that led to the FBNSV-[ET:Hol;JKI-2000] population (see Fig. S1 in the supplemental material). Similar distinct subpopulations could be discerned for the FBNSV-[ET:Hol;1997] DNA-C and DNA-U2 molecules (see Fig. S1 in the supplemental material).
When considering the eight FBNSV genome DNAs individually, clear differences in variability were evident, with DNA-R and DNA-S being the least variable and DNA-N the most variable genome components (Fig. 2 and Tables 1 and 2; see Table S3 in the supplemental material). Comparable profound differences in variability were found between DNA-A and DNA-B of the EACMVs (17) and between the two genomic RNAs of the crinivirus tomato chlorosis virus (53).
In contrast to the variability in the geminiviruses, where an elevated mutation frequency was observed in noncoding sequences (16, 34), we did not find such differences when comparing the overall mutation frequency as well as the types of mutation between noncoding and coding regions of the FBNSV DNAs (Table 2; see Table S3 in the supplemental material). This might suggest that the eight DNA molecules constituting the FBNSV genome are subject to variation and selection as individual entities, with no particularly relaxed sequence constraints for their noncoding regions. Possibly each ssDNA circle requires adoption of a particular secondary structure comprising coding and noncoding sequences alike. For at least one genome component, DNA-R, extended secondary structures are likely to be of importance (36).
Regarding the types of mutations, the transitions G → A and C → T were significantly more frequent in the FBNSV-[ET:Hol;1997] field population than in the laboratory-maintained FBNSV-[ET:Hol;JKI-2000] population (Table 3), probably reflecting the underlying influence on the mutation spectra of guanine and cytosine deamination of the ssDNA (9, 28). Duffy and Holmes (16, 17) detected a similar overrepresentation of the same transitions in their analyses of TYLCV and EACMV sequences from databases. The disappearance of the G → A and C → T transition bias in the laboratory-maintained FBNSV-[ET:Hol;JKI-2000] population may be due to the prolonged passage in a single host species. Faba bean is a cool season crop in Ethiopia or countries of North Africa, where it is grown for only a limited time period each year. FBNSV probably survives the hot and dry season in perennial leguminous (or nonleguminous) plants that act as reservoir hosts of FBNSV but have not been identified yet. Therefore, the shift in mutation spectra in FBNSV after the 2.4-year experimental passage in faba bean may reflect a host effect. A comparable effect of a host different from the “natural” one occurred upon prolonged multiplication of MSV in sugarcane as opposed to maize, where an elevated G → T transversion frequency was observed (71). Conceivably, the efficiency of protection against DNA damage by nucleotide excision repair systems (44) and/or translesion synthesis by specialized repair polymerases (55, 75) may have differed between the natural and laboratory host species employed. However, nothing is known about the nature of host polymerases replicating ssDNA plant viruses and the associated DNA damage repair processes. Another clear host effect is the enhancement of the overall mutation frequency that was observed, for instance, when using pepper (Capsicum annuum) as opposed to tomato (Solanum lycopersicum) or muntju (N. benthamiana) for propagation of the two alpha-like plant viruses tobacco mosaic virus and cucumber mosaic virus (64) or in turnip mosaic virus populations adapted from the susceptible host turnip (Brassica rapa) to radish (Raphanus sativus) as a new and almost insusceptible host (56).
In contrast to what has been described for MSV and the begomoviruses, we observed a mutation bias toward T → C transitions in both field and laboratory-maintained FBNSV populations (Table 3). This overrepresentation of T → C transitions is due to significantly more T → C transitions in DNA-S, DNA-N, and DNA-U1 (see Table S3 in the supplemental material), for which we have no obvious explanation yet.
In addition to elevated rates of nucleotide changes, recombination has been identified as another major driving force for the evolution of ssDNA viruses (31, 49, 50, 57, 59, 72), including the nanovirus BBTV (30). In the three FBNSV populations analyzed here, we detected 12 clear intragenomic recombination events between different individual DNA components, two of which became established as consensus (Fig. 3). One affects the CR-II of DNA-N, where a stretch of at least 16 nt was replaced by a corresponding sequence from DNA-M or DNA-U2 (see Fig. S2 in the supplemental material). This recombination might have occurred between 1997 and 2000, as the recombinant sequence was present in all DNA-N molecules of the FBNSV-[ET:Hol;JKI-2000] population. Another quite peculiar recombination event comprises a 60- or 61-nt sequence of DNA-M (nt 883 to 942), DNA-U2 (nt 887 to 945), and DNA-U4 (nt 726 to 784) harboring a triplet of informative mutations (see Fig. S2 in the supplemental material). In addition, in DNA-U4 the exchanged 61-nt sequence is inverted. A similar recombinant sequence of 34 nt can also be detected at equivalent positions in DNA-M, DNA-U2, and DNA-U4 of a recently described FBNSV isolate from Morocco (1). In this FBNSV isolate also, the recombinant 34-nt sequence is inverted in DNA-U4, suggesting that this sequence synteny is ancient in FBNSV evolution.
A potential genetic exchange between nanovirus genome components was proposed for common noncoding sequences of SCSV, implying a concerted evolution due to sequence conversion (41). The same type of conversion leads to the grouping of the FBNSV DNAs as shown in Fig. 1. With the increasing number of nanovirus sequences expected to become available in future, a search for interspecies recombinants will be possible and may yield further insights into the evolution of these viruses.
The fact that the FBNSV genetic information is distributed over eight individual DNA molecules of ∼1 kb each, in conjunction with the elevated mutation frequency, allows the virus to take additional advantage of the combinatorial power of reassortment. Nanovirus genomes can be considered swarms of six to eight distinct molecular quasispecies that combine several modes (mutation, recombination, and reassortment) for very efficiently exploiting the sequence space for their evolution (5). Models developed to describe the behavior and evolution of heterogeneous populations of macromolecules (19, 21) were found to be applicable to the variability and evolution of viral genome sequences (3, 20), and their predictions were experimentally tested and confirmed (see reference 12 and references therein).
Given the average mutation frequency of 0.75 sequence changes per 1 kb of DNA, or per genome component, the spread of genetic information over eight individual molecules rather than a single one allows for easy complementation and maintenance of suboptimal sequence variants within the heterogeneous genomic DNA swarms, facilitating avoidance of extinction by an error catastrophe (22, 23, 54).
With further application of the RCA technique, including environmental samples, more and more ssDNA viruses are being discovered (51, 61), and “time—the emerging dimension of (plant) virus studies” (35) will definitely provide us with further insights into the biology and evolution of the little-known nanoviruses also.
Supplementary Material
Acknowledgments
We thank K. M. Makkouk for providing us with the FBNSV sample from Holetta, Ethiopia, and A. Sieg-Müller and P. Lüddecke for excellent technical assistance. We are grateful to S. Mondy for assistance with statistics.
This work was supported through ERA-NET Plant Genomics 040B “RCA GENOMICS” and the Trilateral Cooperation GABI-GENOPLANTE-MEC, with funding from Agence Nationale de la Recherche (ANR), Ministerio de Educación y Ciencia and the European Regional Development Fund (ERDF), the CNRS, and BFU2007-65080BMC (Plan Nacional de I+D+I, Ministerio de Ciencia e Innovación, Micinn, Spain). A.G.-P. was also supported by a Ramón y Cajal contract from Micinn, the European Social Fund, and the Junta de Andalucía stay grant program.
Footnotes
Published ahead of print on 30 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abraham, A. D., B. Bencharki, V. Torok, L. Katul, M. Varrelmann, and H. J. Vetten. 2010. Two distinct nanovirus species infecting faba bean in Morocco. Arch. Virol. 155:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida, R. P., G. M. Bennett, M. D. Anhalt, C. W. Tsai, and P. O'Grady. 2009. Spread of an introduced vector-borne banana virus in Hawaii. Mol. Ecol. 18:136-146. [DOI] [PubMed] [Google Scholar]
- 3.Batschelet, E., E. Domingo, and C. Weissmann. 1976. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene 1:27-32. [DOI] [PubMed] [Google Scholar]
- 4.Bebenek, K., J. C. Boyer, and T. A. Kunkel. 1999. The base substitution fidelity of HIV-1 reverse transcriptase on DNA and RNA templates probed with 8-oxo-deoxyguanosine triphosphate. Mutat. Res. 429:149-158. [DOI] [PubMed] [Google Scholar]
- 5.Biebricher, C. K., and M. Eigen. 2006. What is a quasispecies? Curr. Top. Microbiol. Immunol. 299:1-31. [DOI] [PubMed] [Google Scholar]
- 6.Boyer, J. C., K. Bebenek, and T. A. Kunkel. 1992. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc. Natl. Acad. Sci. U. S. A. 89:6919-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bull, S. E., R. W. Briddon, W. S. Sserubombwe, K. Ngugi, P. G. Markham, and J. Stanley. 2006. Genetic diversity and phylogeography of cassava mosaic viruses in Kenya. J. Gen. Virol. 87:3053-3065. [DOI] [PubMed] [Google Scholar]
- 8.Burns, T. M., R. M. Harding, and J. L. Dale. 1995. The genome organization of banana bunchy top virus: analysis of six ssDNA components. J. Gen. Virol. 76:1471-1482. [DOI] [PubMed] [Google Scholar]
- 9.Caulfield, J. L., J. S. Wishnok, and S. R. Tannenbaum. 1998. Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J. Biol. Chem. 273:12689-12695. [DOI] [PubMed] [Google Scholar]
- 10.Chu, P. W., P. Keese, B. S. Qiu, P. M. Waterhouse, and W. L. Gerlach. 1993. Putative full-length clones of the genomic DNA segments of subterranean clover stunt virus and identification of the segment coding for the viral coat protein. Virus Res. 27:161-171. [DOI] [PubMed] [Google Scholar]
- 11.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domingo, E., V. Martín, C. Perales, A. Grande-Pérez, J. García-Arriaza, and A. Arias. 2006. Viruses as quasispecies: biological implications. Curr. Top. Microbiol. Immunol. 299:51-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingo, E., D. Sabo, T. Taniguchi, and C. Weissmann. 1978. Nucleotide sequence heterogeneity of an RNA phage population. Cell 13:735-744. [DOI] [PubMed] [Google Scholar]
- 14.Drake, J. W. 1993. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 90:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duffy, S., and E. C. Holmes. 2008. Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus tomato yellow leaf curl virus. J. Virol. 82:957-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duffy, S., and E. C. Holmes. 2009. Validation of high rates of nucleotide substitution in geminiviruses: phylogenetic evidence from East African cassava mosaic viruses. J. Gen. Virol. 90:1539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy, S., L. A. Shackelton, and E. C. Holmes. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9:267-276. [DOI] [PubMed] [Google Scholar]
- 19.Eigen, M. 1971. Self organization of matter and the evolution of biological macromolecules. Naturwissenschaften 58:465-523. [DOI] [PubMed] [Google Scholar]
- 20.Eigen, M., C. K. Biebricher, M. Gebinoga, and W. C. Gardiner. 1991. The hypercycle. Coupling of RNA and protein biosynthesis in the infection cycle of an RNA bacteriophage. Biochemistry 30:11005-11018. [DOI] [PubMed] [Google Scholar]
- 21.Eigen, M., and P. Schuster. 1977. The hypercycle. A principle of natural self-organization. A. Emergence of the hypercycle. Naturwissenschaften 64:541-565. [DOI] [PubMed] [Google Scholar]
- 22.Escarmís, C., G. Gómez-Mariano, M. Dávila, E. Lázaro, and E. Domingo. 2002. Resistance to extinction of low fitness virus subjected to plaque-to-plaque transfers: diversification by mutation clustering. J. Mol. Biol. 315:647-661. [DOI] [PubMed] [Google Scholar]
- 23.Escarmís, C., E. Lázaro, and S. C. Manrubia. 2006. Population bottlenecks in quasispecies dynamics. Curr. Top. Microbiol. Immunol. 299:141-170. [DOI] [PubMed] [Google Scholar]
- 24.Esteban, J. A., M. Salas, and L. Blanco. 1993. Fidelity of phi29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J. Biol. Chem. 268:2719-2726. [PubMed] [Google Scholar]
- 25.Fargette, D., A. Pinel-Galzi, D. Sereme, S. Lacombe, E. Hebrard, O. Traore, and G. Konate. 2008. Diversification of rice yellow mottle virus and related viruses spans the history of agriculture from the neolithic to the present. PLoS Pathog. 4:e1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firth, C., M. A. Charleston, S. Duffy, B. Shapiro, and E. C. Holmes. 2009. Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. J. Virol. 83:12813-12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franz, A. W., F. van der Wilk, M. Verbeek, A. M. Dullemans, and J. F. van den Heuvel. 1999. Faba bean necrotic yellows virus (genus Nanovirus) requires a helper factor for its aphid transmission. Virology 262:210-219. [DOI] [PubMed] [Google Scholar]
- 28.Frederico, L. A., T. A. Kunkel, and B. R. Shaw. 1990. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry 29:2532-2537. [DOI] [PubMed] [Google Scholar]
- 29.French, R., and D. C. Stenger. 2003. Evolution of Wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu. Rev. Phytopathol. 41:199-214. [DOI] [PubMed] [Google Scholar]
- 30.Fu, H. C., J.-M. Hu, T.-H. Hung, H.-J. Su, and H.-H. Yeh. 2009. Unusual events involved in Banana bunchy top virus strain evolution. Phytopathology 99:812-822. [DOI] [PubMed] [Google Scholar]
- 31.García-Andres, S., D. M. Tomás, S. Sánchez-Campos, J. Navas-Castillo, and E. Moriones. 2007. Frequent occurrence of recombinants in mixed infections of tomato yellow leaf curl disease-associated begomoviruses. Virology 365:210-219. [DOI] [PubMed] [Google Scholar]
- 32.García-Arenal, F., A. Fraile, and J. M. Malpica. 2001. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 39:157-186. [DOI] [PubMed] [Google Scholar]
- 33.García-Díaz, M., and K. Bebenek. 2007. Multiple functions of DNA polymerases. Crit. Rev. Plant Sci. 26:105-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge, L., J. Zhang, X. Zhou, and H. Li. 2007. Genetic structure and population variability of tomato yellow leaf curl China virus. J. Virol. 81:5902-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbs, A. J., D. Fargette, F. Garcia-Arenal, and M. J. Gibbs. 2010. Time—the emerging dimension of plant virus studies. J. Gen. Virol. 91:13-22. [DOI] [PubMed] [Google Scholar]
- 36.Grigoras, I., T. Timchenko, and B. Gronenborn. 2008. Transcripts encoding the nanovirus master replication initiator proteins are terminally redundant. J. Gen. Virol. 89:583-593. [DOI] [PubMed] [Google Scholar]
- 37.Grigoras, I., T. Timchenko, L. Katul, A. Grande-Pérez, H. J. Vetten, and B. Gronenborn. 2009. Reconstitution of authentic nanovirus from multiple cloned DNAs. J. Virol. 83:10778-10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gronenborn, B. 2004. Nanoviruses: genome organisation and protein function. Vet. Microbiol. 98:103-109. [DOI] [PubMed] [Google Scholar]
- 39.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 40.Harkins, G. W., W. Delport, S. Duffy, N. Wood, A. L. Monjane, B. E. Owor, L. Donaldson, S. Saumtally, G. Triton, R. W. Briddon, D. N. Shepherd, E. P. Rybicki, D. P. Martin, and A. Varsani. 2009. Experimental evidence indicating that mastreviruses probably did not co-diverge with their hosts. Virol. J. 6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes, A. L. 2004. Birth-and-death evolution of protein-coding regions and concerted evolution of non-coding regions in the multi-component genomes of nanoviruses. Mol. Phylogenet. Evol. 30:287-294. [DOI] [PubMed] [Google Scholar]
- 42.Isnard, M., M. Granier, R. Frutos, B. Reynaud, and M. Peterschmitt. 1998. Quasispecies nature of three maize streak virus isolates obtained through different modes of selection from a population used to assess response to infection of maize cultivars. J. Gen. Virol. 79:3091-3099. [DOI] [PubMed] [Google Scholar]
- 43.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:156-165. [DOI] [PubMed] [Google Scholar]
- 44.Jiricny, J. 2006. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 7:335-346. [DOI] [PubMed] [Google Scholar]
- 45.Katul, L., T. Timchenko, B. Gronenborn, and H. J. Vetten. 1998. Ten distinct circular ssDNA components, four of which encode putative replication-associated proteins, are associated with the faba bean necrotic yellows virus genome. J. Gen. Virol. 79:3101-3109. [DOI] [PubMed] [Google Scholar]
- 46.Kosakovsky Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 47.Kosakovsky Pond, S. L., and S. D. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 48.Kunkel, T. A., and D. A. Erie. 2005. DNA mismatch repair. Annu. Rev. Biochem. 74:681-710. [DOI] [PubMed] [Google Scholar]
- 49.Lefeuvre, P., J. M. Lett, A. Varsani, and D. P. Martin. 2009. Widely conserved recombination patterns among single-stranded DNA viruses. J. Virol. 83:2697-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefeuvre, P., D. P. Martin, M. Hoareau, F. Naze, H. Delatte, M. Thierry, A. Varsani, N. Becker, B. Reynaud, and J. M. Lett. 2007. Begomovirus ‘melting pot’ in the south-west Indian Ocean islands: molecular diversity and evolution through recombination. J. Gen. Virol. 88:3458-3468. [DOI] [PubMed] [Google Scholar]
- 51.López-Bueno, A., J. Tamames, D. Velázquez, A. Moya, A. Quesada, and A. Alcamí. 2009. High diversity of the viral community from an Antarctic lake. Science 326:858-861. [DOI] [PubMed] [Google Scholar]
- 52.López-Bueno, A., L. P. Villarreal, and J. M. Almendral. 2006. Parvovirus variation for disease: a difference with RNA viruses? Curr. Top. Microbiol. Immunol. 299:349-370. [DOI] [PubMed] [Google Scholar]
- 53.Lozano, G., A. Grande-Pérez, and J. Navas-Castillo. 2009. Populations of genomic RNAs devoted to the replication or spread of a bipartite plant virus differ in genetic structure. J. Virol. 83:12973-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manrubia, S. C., C. Escarmís, E. Domingo, and E. Lázaro. 2005. High mutation rates, bottlenecks, and robustness of RNA viral quasispecies. Gene 347:273-282. [DOI] [PubMed] [Google Scholar]
- 55.McCulloch, S. D., and T. A. Kunkel. 2008. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18:148-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohshima, K., S. Akaishi, H. Kajiyama, R. Koga, and A. J. Gibbs. 2010. Evolutionary trajectory of turnip mosaic virus populations adapting to a new host. J. Gen. Virol. 91:788-801. [DOI] [PubMed] [Google Scholar]
- 57.Padidam, M., S. Sawyer, and C. M. Fauquet. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218-225. [DOI] [PubMed] [Google Scholar]
- 58.Paez, J. G., M. Lin, R. Beroukhim, J. C. Lee, X. Zhao, D. J. Richter, S. Gabriel, P. Herman, H. Sasaki, D. Altshuler, C. Li, M. Meyerson, and W. R. Sellers. 2004. Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 32:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pita, J. S., V. N. Fondong, A. Sangaré, G. W. Otim-Nape, S. Ogwal, and C. M. Fauquet. 2001. Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 82:655-665. [DOI] [PubMed] [Google Scholar]
- 60.Roossinck, M. J., and W. L. Schneider. 2006. Mutant clouds and occupation of sequence space in plant RNA viruses. Curr. Top. Microbiol. Immunol. 299:337-348. [DOI] [PubMed] [Google Scholar]
- 61.Rosario, K., S. Duffy, and M. Breitbart. 2009. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. Virol. 90:2418-2424. [DOI] [PubMed] [Google Scholar]
- 62.Sala, M., and S. Wain-Hobson. 2000. Are RNA viruses adapting or merely changing? J. Mol. Evol. 51:12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sano, Y., M. Wada, Y. Hashimoto, T. Matsumoto, and M. Kojima. 1998. Sequences of ten circular ssDNA components associated with the milk vetch dwarf virus genome. J. Gen. Virol. 79:3111-3118. [DOI] [PubMed] [Google Scholar]
- 64.Schneider, W. L., and M. J. Roossinck. 2001. Genetic diversity in RNA virus quasispecies is controlled by host-virus interactions. J. Virol. 75:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shackelton, L. A., and E. C. Holmes. 2006. Phylogenetic evidence for the rapid evolution of human B19 erythrovirus. J. Virol. 80:3666-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shackelton, L. A., C. R. Parrish, U. Truyen, and E. C. Holmes. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. U. S. A. 102:379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharman, M., J. E. Thomas, S. Skabo, and T. A. Holton. 2008. Abacá bunchy top virus, a new member of the genus Babuvirus (family Nanoviridae). Arch. Virol. 153:135-147. [DOI] [PubMed] [Google Scholar]
- 68.Sserubombwe, W. S., R. W. Briddon, Y. K. Baguma, G. N. Ssemakula, S. E. Bull, A. Bua, T. Alicai, C. Omongo, G. W. Otim-Nape, and J. Stanley. 2008. Diversity of begomoviruses associated with mosaic disease of cultivated cassava (Manihot esculenta Crantz) and its wild relative (Manihot glaziovii Mull. Arg.) in Uganda. J. Gen. Virol. 89:1759-1769. [DOI] [PubMed] [Google Scholar]
- 69.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 70.Timchenko, T., L. Katul, M. Aronson, J. C. Vega-Arreguín, B. C. Ramirez, H. J. Vetten, and B. Gronenborn. 2006. Infectivity of nanovirus DNAs: induction of disease by cloned genome components of Faba bean necrotic yellows virus. J. Gen. Virol. 87:1735-1743. [DOI] [PubMed] [Google Scholar]
- 71.van der Walt, E., D. P. Martin, A. Varsani, J. E. Polston, and E. P. Rybicki. 2008. Experimental observations of rapid maize streak virus evolution reveal a strand-specific nucleotide substitution bias. Virol. J. 5:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van der Walt, E., E. P. Rybicki, A. Varsani, J. E. Polston, R. Billharz, L. Donaldson, A. L. Monjane, and D. P. Martin. 2009. Rapid host adaptation by extensive recombination. J. Gen. Virol. 90:734-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vetten, H. J., P. W. G. Chu, J. L. Dale, R. Harding, J. Hu, L. Katul, M. Kojima, J. W. Randles, Y. Sano, and J. E. Thomas. 2005. Nanoviridae, p. 343-352. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 74.Wain-Hobson, S. 1996. Running the gamut of retroviral variation. Trends Microbiol. 4:135-141. [DOI] [PubMed] [Google Scholar]
- 75.Waters, L. S., B. K. Minesinger, M. E. Wiltrout, S. D'Souza, R. V. Woodruff, and G. C. Walker. 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73:134-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu, B., U. Melcher, X. Guo, X. Wang, L. Fan, and G. Zhou. 2008. Assessment of codivergence of mastreviruses with their plant hosts. BMC Evol. Biol. 8:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.