Abstract
High-risk cohorts in East Africa and the United States show rates of dual HIV-1 infection—the concomitant or sequential infection by two HIV-1 strains—of 50% to 100% of those of primary infection, and our normal-risk HIV-positive cohort in Cameroon exhibits a rate of dual infection of 11% per year, signifying that these infections are not exceptional. Little is known regarding the effect of dual infections on host immunity, despite the fact that they provide unique opportunities to investigate how the immune response is affected when challenged with diverse HIV-1 antigens. Using heterologous primary isolates, we have shown here that dual HIV-1 infection by genetically distant strains correlates with significantly increased potency and breadth of the anti-HIV-1 neutralizing antibody response. When the neutralization capacities of sequential plasma obtained before and after the dual infection of 4 subjects were compared to those of matched plasma obtained from 23 singly infected control subjects, a significant increase in the neutralization capacity of the sequential sample was found for 16/28 dually infected plasma/virus pairs, while only 4/159 such combinations for the control subjects exhibited a significant increase (P < 0.0001). Similarly, there was a significant increase in the plasma dilution capable of neutralizing 50% of virus (IC50) for 18/24 dually infected plasma/virus pairs, while 0/36 controls exhibited such an increase (P < 0.0001). These results demonstrate that dual HIV-1 infection broadens and strengthens the anti-HIV-1 immune response, suggesting that vaccination schemes that include polyvalent, genetically divergent immunogens may generate highly protective immunity against any HIV-1 challenge strain.
The HIV pandemic is unique in the extreme genetic diversity that comprises its viral strains. Never before has the scientific community been faced with so great a challenge as designing an efficacious, prophylactic vaccine against such a wide array of viruses. To date, 12 subtypes and subsubtypes have been classified within the Main group of HIV-1, which are as much as 30% distant from one another in amino acid sequence. Complicating the HIV landscape significantly are the recombinants of these (sub)subtypes, which are believed to result from the dual HIV infection of the host, be it concomitant or sequential, and ultimately of single cells. During reverse transcription, recombination between copackaged heterogeneous genomes of these genetically divergent viruses results in unique recombinant forms (URFs) of the parental strains. Countless URFs of virtually every subtype combination have been identified globally (13). URFs that are competitive in the host and capable of spreading in a population are known as circulating recombinant forms (CRFs), 43 of which have been identified over the past decade. Clearly, dual infection is a potential source of rapid viral evolution, since it facilitates recombination, increasing the genetic complexity of the pandemic.
Recently we and others identified various dually infected individuals and determined the frequency of dual infection among cohorts in Cameroon, Kenya, Tanzania, South Africa, the United States, and elsewhere (6, 8, 9, 16, 17, 21, 26; B. Chohan, A. Piantadosi, L. Lavreys, and J. Overbaugh, presented at AIDS Vaccine 2007, Seattle, WA; P. Hawkins, O. Manigart, C. Kraft, E. Chomba, J. Mulenga, C. Derdeyn, S. A. Allen, and E. Hunter, presented at AIDS Vaccine 2007, Seattle, WA). Most of these studies were conducted with high-risk commercial sex worker (CSW) cohorts or cohorts of men who have sex with men (MSM); however, our longitudinal study of a normal-risk cohort of HIV-positive (HIV+) individuals in Yaoundé, Cameroon, found nearly 16% of study subjects to be dually infected within 36 months of follow-up (21). This analysis was generally consistent with a cross-sectional survey of normal-risk HIV+ Tanzanians, which estimated the frequency of dual infection at 9% (9). Moreover, two studies conducted on CSW cohorts in East Africa have determined the rate of reinfection per year to be roughly half that of primary infection, while a cohort of subtype B-infected MSM in the United States found reinfection to occur at a rate comparable to that of primary infection (3, 18, 26). It is apparent from these studies that dual HIV-1 infection is not an exceptional occurrence across Africa or in any setting where exposure to the virus is relatively frequent.
Limited data exist regarding the effects of a dual HIV-1 infection on the host. In terms of disease pathogenesis, small studies of dual infection by subtypes B and C in the United States and South Africa, respectively, have demonstrated a positive correlation between dual infection, increased viral load, and a more rapid disease progression, though this topic has not been thoroughly investigated (7, 8). To date, two studies have examined whether neutralizing antibodies (nAbs) generated during single HIV-1 infection correlate with protection from dual infection; however, the results have been conflicting (1, 25). In one study of dually subtype-B-infected subjects, nAbs to heterologous laboratory virus isolates were lacking in these subjects prior to dual infection compared to findings for control subjects; however, a study of Kenyan women dually infected by various subtypes contradicted this data, finding that prior to dual infection, relatively broad and potent nAb responses to heterologous HIV-1 viruses were present (1, 25).
Although dual infections provide a unique opportunity to investigate how the host immune response is affected when challenged by diverse HIV-1 antigens, which has significant implications for vaccine design, no studies have been conducted regarding the effect of dual HIV-1 infection on host immunity, particularly with regard to any impact on the induction of potent nAbs. Focusing for the first time on the effect of a dual infection on the nAb response, changes in the breadth and potency of humoral anti-HIV-1 immunity exhibited by pairs of sequential plasma samples obtained from 4 Cameroonian subjects shown previously to be dually infected were analyzed in the present study (21). These changes were compared to those exhibited by a control group of pairs of sequential plasma samples obtained from 23 singly infected subjects of the same HIV-positive cohort, matched for study duration and change in CD4 T-cell count (ΔCD4) (21).
MATERIALS AND METHODS
Study subject data.
Blood samples were collected in Yaoundé, Cameroon, at 3- to 6-month intervals (calendar years 2001 to 2004) from a cohort of 64 HIV+, chronically infected individuals. In addition, blood was collected from certain subjects 50 to 60 months after the initial sample. All cohort study subjects were antiretroviral drug naïve, and the time since seroconversion was unknown.
In addition, a blood sample collected in calendar year 2006 from subject 06ARC064, who was not part of this cohort but was part of a separate cohort of chronically infected HIV+ individuals on antiretroviral therapy (ARV) in Cameroon, was also used as a control for antiretroviral activity in plasma against neutralization of HIV-1 pseudotyped with murine leukemia virus (MLV) Env. This individual, referred to as 06ARC064, was on Triomune, a triple combination therapy comprised of stavudine (d4T), lamivudine (3TC), and nevirapine (NVP) which is a first-line antiretroviral therapy administered to HIV patients in Cameroon.
In the present study, 4 dually infected and 23 singly infected subjects from the Yaoundé cohort were analyzed (21). Dual or single infection status was previously determined using a mixed-time-point heteroduplex assay (HDA), in which a gag fragment (HxB2 location, 1577 to 2040) was amplified by nested reverse transcriptase PCR (RT-PCR) from RNA isolated from the initial blood sample obtained from each study subject and from each sequentially obtained blood sample (22). In this assay, sequential pairs of amplicons are mixed, denatured, and rapidly cooled so as to generate various types of DNA duplexes, which can be differentiated by their mobility in a urea-acrylamide gel. Discernible heteroduplexes—slowly migrating duplexes consisting of one strand of amplicon from the initial sample and one strand from the sequential sample—indicate ≥5% genetic discordance between samples in the gag fragment, and therefore, their formation reveals dual infection (22). Thus, dual infection status was determined based on the presence of heteroduplexes upon mixed-time-point HDA, while single infection status was determined by the absence of heteroduplexes in all mixed-time-point HDAs performed for each sequential sample pair. Notably, the primers used for the HDA are capable of amplifying HIV-1 type N and O strains but likely not HIV-2 sequences, as determined using the primalign tool on the HIV sequence database (http://www.hiv.lanl.gov/content/sequence/QUICK_ALIGN/QuickAlign.html) (10).
Selection of the 4 subjects analyzed in the present study from the 10 dually infected subjects found in our previous cohort study was based on their identification as dually infected at an interim study time point to allow the analysis of blood samples obtained at least 12 months after dual infection in order to give any potential anti-HIV-1 immune response sufficient time to mature. The HIV-1 strains infecting the 4 dually infected subjects were classified previously in the C1C2 region of env as follows: CMNYU107, a CRF01_AE/CRF36_cpx URF and a subtype G virus; CMNYU129, a CRF02_AG virus and a subtype F2 virus; CMNYU6518, two CRF01_AE/CRF36_cpx URFs possessing intersubsubtype (∼12%) genetic distance from one another; and CMNYU6544, a CRF02_AG/F2 URF and CRF02_AG virus (20) (Table 1). The times elapsed between plasma samples for subjects CMNYU107 and CMNYU129 were 36 and 33 months, respectively, with samples being obtained 21 and 9 months, respectively, before dual infection and 24 and 15 months, respectively, after dual infection. For subjects CMNYU6518 and CMNYU6544, the times elapsed between plasma samples were 60 and 53 months, respectively, with samples being obtained 3 and 6 months, respectively, before and 57 and 47 months, respectively, after the dual infections were identified. Given the significant difference in study duration between these two groups, changes in neutralization capacities of each group were compared to those exhibited by a “short-duration” or “long-duration” control group of sequential plasma samples obtained from 23 singly infected subjects of the same HIV-positive cohort (21). There were no significant differences in the study duration or changes in CD4 T-cell count (ΔCD4) over the study period between these control groups and their respective dually infected groups. Notably, there were no significant differences in the ΔCD4 value between the two dually infected groups or between the control groups. Viral loads were determined for the sequential samples obtained from the dually infected subjects and for the 23 singly infected controls by the Versant HIV RNA 3.0 assay (bDNA, Siemens, IL), as recommended by the manufacturer (see Table 4).
TABLE 1.
Study subject dataa
| Subject ID | Duration of study (mo) | Subtype(s)b | CD4 count (cells/mm3) |
ΔCD4 (cells/mm3) | |
|---|---|---|---|---|---|
| Initial sample | Final sample | ||||
| Dual infections | |||||
| CMNYU107 | 36 (15 since dual infection) | 01_AE-36_cpx URF/G | 526 | 184 | −342 |
| CMNYU129 | 33 (24 since dual infection) | F2/CRF02_AG | 603 | 337 | −266 |
| CMNYU6518 | 60 (57 since dual infection) | 01_AE-36_cpx URF/01_AE-36_cpx URF | 345 | 389 | 44 |
| CMNYU6544 | 53 (47 since dual infection) | CRF02_AG-F2 URF/CRF02_AG | 479 | 289 | −190 |
| Singly infected controls | |||||
| CMNYU40 | 24 | NDc | 624 | 719 | 95 |
| CMNYU110d | 30 | 02_AG | 393 | 220 | −173 |
| CMNYU113d | 27 | ND | 964 | 355 | −609 |
| CMNYU122 | 36 | 02_AG | 334 | 208 | −126 |
| CMNYU128d | 36 | 02_AG | 389 | 230 | −159 |
| CMNYU133 | 21 | F2 | 923 | 592 | −331 |
| CMNYU153d | 30 | H | 300 | 279 | −21 |
| CMNYU157 | 21 | ND | 373 | 306 | −67 |
| CMNYU161 | 30 | ND | 408 | 185 | −223 |
| CMNYU164d | 27 | 02_AG | 865 | 746 | −119 |
| CMNYU6489 | 30 | A | 421 | 126 | −295 |
| CMNYU6490 | 21 | CRF11_cpx | 338 | 209 | −129 |
| CMNYU6495 | 24 | ND | 347 | 231 | −116 |
| CMNYU6500 | 27 | A | 371 | 274 | −97 |
| CMNYU166 | 76 | ND | 534 | 313 | −221 |
| CMNYU179 | 77 | 02_AG | 515 | 404 | −111 |
| CMNYU6504 | 65 | ND | 516 | 86 | −430 |
| CMNYU6537 | 51 | 02_AG-A1 URF | 725 | 329 | −396 |
| CMNYU6540 | 53 | ND | 1180 | 520 | −660 |
| CMNYU6542d | 51 | 02_AG-F2 URF | 1206 | 599 | −607 |
| CMNYU6553 | 52 | ND | 503 | 239 | −264 |
| CMNYU6556 | 52 | 02_AG-F2-A1 URF | 428 | 174 | −254 |
| CMNYU6561 | 51 | ND | 482 | 475 | −7 |
Subjects shown in boldface comprise the long-duration control group.
Subtyping was done for C1C2 region of env.
ND, not done.
Used as controls for IC50 analysis.
TABLE 4.
Viral load data
| Subject ID | Viral load (copies/ml) |
||
|---|---|---|---|
| Initial sample | Final sample | Change | |
| Dual infections | |||
| CMNYU107 | 12,877 | 13,354 | 477 |
| CMNYU129 | 4,202 | 2,850 | −1,352 |
| CMNYU6518 | 2,885 | 50 | −2,835 |
| CMNYU6544 | 1,6,847 | 50 | −16,797 |
| Singly infected controls | |||
| CMNYU40 | 100 | 200 | 100 |
| CMNYU110 | 437 | 135 | −302 |
| CMNYU113 | 2,682 | 2,327 | −355 |
| CMNYU122 | 909 | 389 | −520 |
| CMNYU128 | 14,262 | 144,543 | 130,281 |
| CMNYU133 | 3,925 | 1,749 | −2,176 |
| CMNYU153 | 2,454 | 8,421 | 5,967 |
| CMNYU157 | 432 | 156 | −276 |
| CMNYU161 | 30,311 | 1,347 | −28,964 |
| CMNYU164 | 211 | 345 | 134 |
| CMNYU6489 | 10,955 | 3,405 | −7,550 |
| CMNYU6490 | 2,274 | 3,893 | 1,619 |
| CMNYU6495 | 1,856 | 210 | −1,646 |
| CMNYU6500 | 103,880 | 279,271 | 175,391 |
| CMNYU166 | 102 | 147 | 45 |
| CMNYU179 | 4,775 | 770 | −4,005 |
| CMNYU6504 | 1,607 | 2,930 | 1,323 |
| CMNYU6537 | 5,074 | 39,451 | 34,377 |
| CMNYU6540 | 2,490 | 292 | −2,198 |
| CMNYU6542 | 244 | 2,344 | 2,100 |
| CMNYU6553 | 675 | 12,031 | 11,356 |
| CMNYU6556 | 589 | 2,244 | 1,655 |
| CMNYU6561 | 6,338 | 1,291 | −5,047 |
Virus isolates and HIV-1 pseudotyped with MLV Env.
Seven primary isolates heterologous to the viruses infecting the cohort study subjects were used, including isolates SF162 and Bx08 (subtype B) and the non-B primary Cameroonian isolates 102 (CRF02_AG), 104 (subsubtype F2), 122 (CRF02_AG), 161 (CRF02_AG), and 6491 (subtype G), which were randomly selected from frozen virus stocks. All virus stocks were prepared by infecting phytohemagglutinin (PHA)-activated human peripheral blood mononuclear cells (PBMCs), as described previously (31). Briefly, frozen PBMCs from HIV-1-negative blood donors were thawed, stimulated with PHA (3 μg/ml; Difco, Detroit, MI) for 3 days, centrifuged, and infected with 1 ml of virus-infected culture supernatant. After a 1 h of exposure of the cells to the virus, the volume of the cell suspension was adjusted to a concentration of 2 × 106 cells/ml and the culture was maintained in RPMI 1640 medium with 10% fetal bovine serum (FBS) and interleukin-2 (20 U/ml; Boehringer Mannheim Biochemicals, Indianapolis, IN) at the same cell concentration. The concentration of p24 in the infected culture supernatant was checked every 3 to 4 days by a noncommercial enzyme-linked immunosorbent assay (14). The infected culture supernatant was collected when the concentration of p24 was >100 ng/ml.
MLV Env was obtained through the AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH, donated by Nathaniel Landau and Dan Littman as SV-A-MLV-env. This (SV-A-MLV-env) was used to generate the HIV-1 pseudotyped with MLV Env as described below.
Cells.
CCR5-GHOST cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% glutamine, 2% penicillin plus streptomycin, Geneticin (200 μg/ml), hygromycin (25 μg/ml), and puromycin (1 μg/ml) (GHOST-DMEM). TZM-bl cells were maintained in DMEM supplemented with 10% fetal bovine serum, 1% glutamine, and 2% penicillin plus streptomycin (TZM-DMEM). The cultures were maintained at 37°C in a 5%-CO2 humidified incubator.
Virus titration and GHOST cell neutralization assay.
GHOST cells were seeded in 24-well plates at 6 × 104 cells/well/0.5 ml. Forty-eight hours later, medium was removed and cells were infected with undiluted virus stocks (110 μl/well). DEAE-dextran was added to each well at a final concentration of 8 μg/ml. The virus was left to adsorb overnight. Cell monolayers were then washed with sterile phosphate-buffered saline (PBS), and 1 ml/well of medium, as described above, was added. Four days postinfection (p.i.), cells were washed with PBS, resuspended in 300 μl of 1 mM EDTA, and fixed in formaldehyde at a final concentration of 2%. Cells were analyzed for green fluorescent protein (GFP) expression with a FACScan flow cytometer (Becton Dickenson, San Jose, CA). Titers yielding ∼500 infected (GFP+) cells/well were used for neutralization assays.
Neutralization assays were performed as previously described, using a fixed dilution of each virus stock (2). Plasma to be tested, obtained by Ficoll-Hypaque gradient centrifugation of whole blood, was heat inactivated at 56°C for 30 min and diluted with GHOST-DMEM. Diluted plasma and diluted virus were mixed in equal volumes (120 μl each) and incubated for 1 h at 37°C; 110 μl of each virus-plasma mixture was added to duplicate wells, 8 μg/ml DEAE-dextran was added to each well, and cells were incubated overnight. Subsequent washes, incubations, harvest, and readout procedures were performed as described above.
Production and neutralization assay of HIV-1 pseudotyped with MLV Env.
293T cells were cotransfected with the pNL4-3Δenv and SV-A-MLV-env plasmids using Fugene 6, according to the manufacturer's protocol (Roche, Manheim, Germany) (15). After 48 h, supernatant was removed from cells, spun at 3,000 rpm for 10 min, brought to an FBS concentration of 20%, and stored at −80°C until use. HIV-1 pseudotyped with MLV Env was titrated and tested in neutralization assays with TZM-bl cells expressing CD4, CCR5, and CXCR4 as previously described (30). Diluted heat-inactivated plasma and 100 50% tissue culture infective doses (TCID50s) of pseudotyped HIV-1 were mixed in equal volumes (50 μl each) and incubated for 1 h at 37°C; 10,000 cells in 100 μl of TZM-DMEM containing 25 μg/ml DEAE-dextran was added to each well, and cells were incubated for 48 h. Luminescence was measured using the Bright Glo reagent (Bright-Glo luciferase assay system; Promega, Madison, WI) on a Lumimark Plus system microplate reader (Bio-Rad Laboratories, Hercules, CA).
Statistical analysis and calculation of percent neutralization, plasma dilution capable of neutralizing 50% of virus (IC50) titer, and breadth and potency scores.
Neutralization experiments were repeated a minimum of 3 times. The mean number of GFP-positive cells for each set of replicates was divided by the mean number of GFP-positive cells in the appropriate replicates of virus-only control wells in order to determine percent neutralization. Statistical significance was determined using an unpaired two-tailed Student t test. Mean IC50s were determined from plasma titrations using the previously described method of plotting the mean titers with which greater than 50% and less than 50% neutralization were achieved and using the plotted line's equation to interpolate the titer at which 50% neutralization would be achieved (5). Significance of overall pairwise neutralization comparisons was determined using Fisher's exact test.
Breadth and potency score calculations were adapted from the method of Blish et al. (1). Briefly, a median IC50 was determined for each virus. For breadth, the plasma/virus combinations for which the IC50 was above the median IC50 for that virus were scored as 1, and those below were scored as 0. The potency score was obtained by dividing the IC50 of a given plasma/virus combination by the median virus IC50. The scores for all 6 viruses tested were added to obtain an overall potency and breadth score for each plasma sample. The average change in breadth and potency scores for the dually infected plasma pairs was compared to that of the singly infected controls using an unpaired one-tailed Student t test.
RESULTS
Percent neutralization at 1:80 dilution.
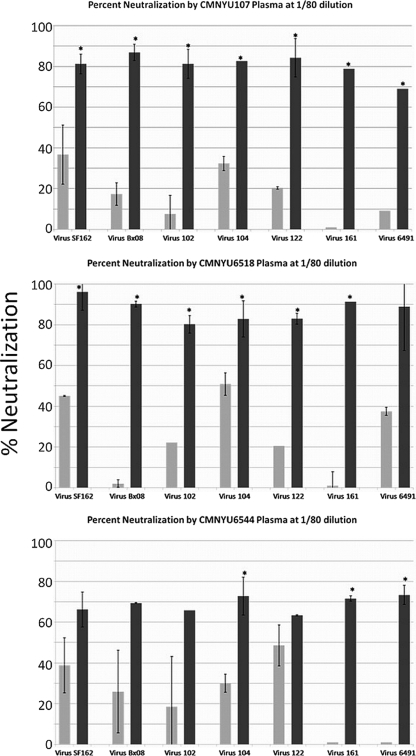
Initially, the neutralization capacities of the sequential plasma samples at a final dilution in medium of 1:80 were assayed against 7 heterologous HIV-1 viruses, including the tier 1, subtype B viruses SF162 and Bx08, and 5 primary isolates, 102, 104, 122, 161, and 6491, obtained from infected subjects in Cameroon. The isolates were randomly selected from our frozen Cameroonian virus stocks. This analysis found that plasma samples obtained from 3 of 4 dually infected subjects after the identification of dual infection (CMNYU107, CMNYU6518, and CMNYU6544) exhibited significantly increased (P < 0.05) neutralization capacities against at least 3 of the 5 heterologous Cameroonian viruses compared to the neutralization capacities of the initial plasma samples against the same viruses, which were generally below 50% (Fig. 1 and Table 2). Furthermore, 2 of 4 dually infected plasma samples (CMNYU107 and CMNYU6518) exhibited a significant increase in neutralization relative to results for the initial samples of both tier 1 viruses and of 4 or more Cameroonian isolates. The remaining dually infected subject, CMNYU129, did not exhibit any significant changes in neutralization against any of the viruses tested. Among the dually infected samples exhibiting increased neutralization, the average increase in percent neutralization against the tier 1 viruses was 9.8-fold (range, 1.7- to 45-fold), while against the Cameroonian isolates, the average increase, at 23.9-fold, tended to be greater (range, 1.3- to 91.4-fold). The significant increases resulted in average neutralization capacities of the sequential plasma samples from the dually infected subjects of 74% (range, 63% to 96% neutralization).
FIG. 1.
Neutralization of heterologous virus at 1:80 dilution. Dually infected plasma exhibited significant increases in neutralization. Light- and dark-gray bars represent plasma samples obtained before and after the dual infection, respectively. Mean values with standard deviations are shown. *, significant change, P < 0.05.
TABLE 2.
Mean percent neutralization of heterologous viruses by singly or dually infected control plasma samplesa
| Plasma sample | % neutralization of virus |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SF162 | Bx08 | 102 | 104 | 122 | 161 | 6491 | |||||||
| Singly infected | |||||||||||||
| CMNYU40 | |||||||||||||
| (1) | 0 | 0 | 0 | 38 | 0 | 0 | 0 | ||||||
| (2) | 12 | 11 | 0 | 21 | 0 | 0 | |||||||
| CMNYU110 | |||||||||||||
| (1) | 77 | 38 | 0 | 30 | 0 | 35 | 0 | ||||||
| (2) | 83 | 45 | 6 | 21 | 0 | 24 | 0 | ||||||
| CMNYU113 | |||||||||||||
| (1) | 40 | 17 | 2 | 33 | 0 | 0 | 20 | ||||||
| (2) | 16 | 23 | 0 | 0 | 0 | 0 | 16 | ||||||
| CMNYU122 | |||||||||||||
| (1) | 0 | 0 | 0 | 9 | ND | 0 | 0 | ||||||
| (2) | 51* | 0 | 0 | 33 | ND | 0 | 0 | ||||||
| CMNYU128 | |||||||||||||
| (1) | 97 | 75 | 62 | 64 | 73 | 8 | 5 | ||||||
| (2) | 98 | 76 | 41 | 70 | 60 | 39 | 10 | ||||||
| CMNYU133 | |||||||||||||
| (1) | 73 | 57 | 25 | 32 | 5 | 19 | 0 | ||||||
| (2) | 47 | 43 | 39 | 31 | 28 | 23 | 0 | ||||||
| CMNYU153 | |||||||||||||
| (1) | 82 | 52 | 2 | 43 | 0 | 2 | 2 | ||||||
| (2) | 89 | 55 | 31 | 30 | 10 | 24 | 31 | ||||||
| CMNYU157 | |||||||||||||
| (1) | 20 | 0 | 0 | 30 | 0 | 2 | 36 | ||||||
| (2) | 8 | 0 | 0 | 12 | 0 | 0 | 22 | ||||||
| CMNYU161 | |||||||||||||
| (1) | 0 | 22 | 0 | 13 | 0 | ND | 0 | ||||||
| (2) | 0 | 12 | 0 | 22 | 0 | ND | 13 | ||||||
| CMNYU164 | |||||||||||||
| (1) | 43 | 29 | 30 | 31 | 6 | 18 | 30 | ||||||
| (2) | 62 | 46 | 26 | 15 | 9 | 19 | 26 | ||||||
| CMNYU166 | |||||||||||||
| (1) | 0 | 35 | 0 | 28 | 0 | 0 | 0 | ||||||
| (2) | 40 | 36 | 0 | 40 | 0 | 0 | 0 | ||||||
| CMNYU179 | |||||||||||||
| (1) | 41 | 46 | 0 | 28 | 0 | 4 | 0 | ||||||
| (2) | 81* | 67 | 0 | 19 | 0 | 0 | 0 | ||||||
| CMNYU6489 | |||||||||||||
| (1) | 90 | 79 | 0 | 32 | 0 | 0 | 25 | ||||||
| (2) | 13 | 23 | 15 | 34 | 0 | 7 | 4 | ||||||
| CMNYU6490 | |||||||||||||
| (1) | 0 | 58 | 0 | 48 | 0 | 0 | 0 | ||||||
| (2) | 0 | 50 | 0 | 41 | 0 | 0 | 0 | ||||||
| CMNYU6495 | |||||||||||||
| (1) | 0 | 2 | 0 | 0 | 0 | 0 | 20 | ||||||
| 0 | 26 | 0 | 28 | 0 | 0 | 0 | |||||||
| CMNYU6500 | |||||||||||||
| (1) | 11 | 12 | 0 | 26 | 0 | 0 | 21 | ||||||
| (2) | 25 | 42 | 0 | 21 | 11 | 7 | 20 | ||||||
| CMNYU6504 | |||||||||||||
| (1) | 0 | 22 | 3 | 37 | 0 | 15 | 0 | ||||||
| (2) | 0 | 32 | 0 | 8 | 0 | 0 | 0 | ||||||
| CMNYU6537 | |||||||||||||
| (1) | 48 | 34 | 0 | 60 | 0 | 37 | 0 | ||||||
| (2) | 54 | 43 | 0 | 35 | 0 | 16 | 20 | ||||||
| CMNYU6540 | |||||||||||||
| (1) | 0 | 0 | 0 | 11 | 0 | 0 | 3 | ||||||
| (2) | 0 | 49 | 0 | 25 | 0 | 3 | 37 | ||||||
| CMNYU6542 | |||||||||||||
| (1) | 65 | 55 | 44 | 12 | 5 | 0 | 0 | ||||||
| (2) | 71 | 31 | 50 | 19 | 0 | 0 | 0 | ||||||
| CMNYU6553 | |||||||||||||
| (1) | 70 | 50 | 0 | 20 | 0 | 0 | 19 | ||||||
| (2) | 86 | 56 | 16 | 35 | 0 | 11 | 2 | ||||||
| CMNYU6556 | |||||||||||||
| (1) | 39 | 2 | 0 | 71 | 0 | 15 | 13 | ||||||
| (2) | 93* | 79* | 0 | 69 | 6 | 43 | 0 | ||||||
| CMNYU6561 | |||||||||||||
| (1) | 51 | 64 | 0 | 16 | 0 | 0 | 0 | ||||||
| (2) | 64 | 63 | 0 | 14 | 0 | 0 | 0 | ||||||
| Dually infected | |||||||||||||
| CMNYU107 | |||||||||||||
| (1) | 37 | 17 | 8 | 32 | 20 | 0 | 9 | ||||||
| (2) | 81* | 87* | 81* | 83* | 84* | 79* | 69* | ||||||
| CMNYU129 | |||||||||||||
| (1) | 78 | 36 | 0 | 39 | 53 | 15 | 0 | ||||||
| (2) | 58 | 32 | 9 | 32 | 41 | 0 | 8 | ||||||
| CMNYU6518 | |||||||||||||
| (1) | 45 | 2 | 22 | 51 | 21 | 0 | 38 | ||||||
| (2) | 96* | 90* | 80* | 83* | 83* | 91* | 89 | ||||||
| CMNYU6544 | |||||||||||||
| (1) | 39 | 26 | 19 | 30 | 49 | 0 | 0 | ||||||
| (2) | 66 | 69 | 66* | 73 | 63* | 72 | 73* | ||||||
For single infections, (1), 1st plasma sample; (2), sequential plasma sample. For dual infections, (1), 1st plasma sample; (2) sequential plasma obtained after dual infection. *, significant increase, P < 0.05. Boldface indicates >60% neutralization. Underlining indicates 50% to 60% neutralization. Plasmas were assayed at a 1:80 dilution. ND, not done.
The initial samples obtained from the control subjects exhibited neutralization of the 7 viruses tested that was generally below 50% and not significantly different from those of the initial samples obtained from the dually infected subjects (P = 0.15), indicating that dual infection had occurred in the presence of nAbs that were no less robust than those in the subjects who did not become dually infected (1). However, the sequential singly infected control plasma samples exhibited neutralization capacities that were in striking contrast to those of the dually infected samples. None of the 23 sequential samples exhibited a significant increase in their capacity to neutralize any of the 5 Cameroonian isolates (Table 2). One control subject, CMNYU6556, exhibited a significant increase in neutralization of both tier 1 viruses, while two additional control subjects, CMNYU122 and CMNYU179, exhibited significant increases in the neutralization of SF162 only. As such, examining the neutralization of each heterologous virus by each plasma sample in pairwise combinations, significant increases in neutralization capacity were found for 16 of 28 dually infected plasma/virus pairs, while only 4 of 159 such pairs comprising the singly infected control plasma panel exhibited significant increases in neutralization, yielding a highly significant result overall regarding changes in neutralization of heterologous virus over time for the dually infected subjects versus the singly infected control subjects (P < 0.0001; Table 2). Comparing the short-duration and long-duration dually infected groups to their respective control groups, significant increases were found for 7 of 14 dually infected plasma/virus pairs of the short-duration group, with only 1 of 96 such pairs of the short-duration control group exhibiting a significant increase (P < 0.0001). Similarly, significant increases were found for 9 of 14 dually infected plasma/virus pairs of the long-duration group, with only 3 of 63 such pairs of the long-duration control group exhibiting a significant increase (P < 0.0001).
IC50 titers.
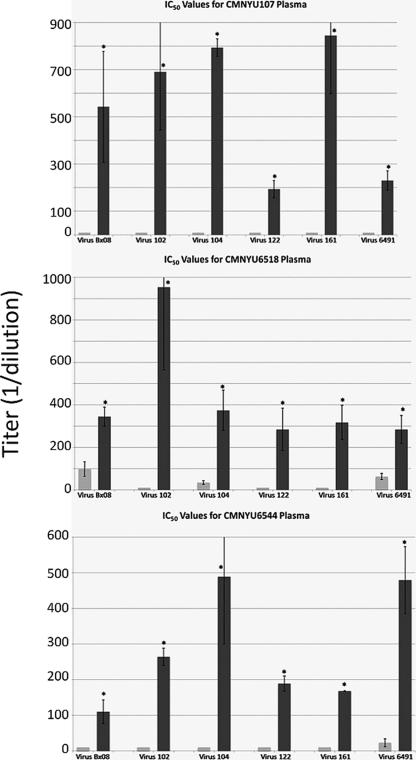
To verify the specificity of neutralization and to compare the difference in the magnitude of neutralization for each plasma pair, neutralization assays using 6 of the 7 heterologous viruses (omitting SF162, a highly neutralization-sensitive laboratory strain) were repeated using 2-fold serial dilutions of plasma (1:20 to 1:1,280) obtained from all 4 dually infected subjects and 6 control subjects randomly selected from the 23 control subjects included previously. IC50s (plasma titer at which 50% of virus is neutralized) were determined using the titration curves obtained for each sample. As predicted by the initial analysis at a 1:80 dilution, the plasma samples obtained from 3 of the 4 dually infected subjects after the dual infection was identified (CMNYU107, CMNYU6518, and CMNYU6544) exhibited significant increases (P < 0.05) in IC50 titers relative to those of the initial samples, in this case against all 6 viruses tested (Fig. 2 and Table 3). This was in complete contrast to the IC50 titers of the control plasma pairs, which exhibited no significant changes in magnitude over the study period (Table 3). Against the tier 1 virus Bx08, the average increase in IC50 titer for the dually infected plasma was 22.9-fold (range, 3.5- to 54.3-fold), while against the Cameroonian isolates, there was an even more potent average increase in IC50 titer of 38.1-fold (range, 7.6- to 95.4-fold). Again examining the neutralization of each heterologous virus by each plasma sample in pairwise combinations, the IC50 titers for 18 of 24 dually infected plasma/virus pairs showed significant increases, while none of the 36 combinations for the singly infected controls exhibited a significant increase (P < 0.0001). Comparing the short-duration and long-duration dually infected groups to their respective control groups, significant increases were found for 6 of 12 dually infected plasma/virus pairs of the short-duration group, with none of the 30 plasma/virus pairs of the short-duration control group exhibiting a significant increase (P = 0.002). Similarly, significant increases were found for all 12 dually infected plasma/virus pairs of the long-duration group, while none of the 6 plasma/virus pairs of the long-duration group in the control group exhibited a significant increase in neutralization (P = 0.031).
FIG. 2.
Neutralization of heterologous virus by serially diluted plasma. Dually infected plasma exhibited significant increases in IC50 titers. Light- and dark-gray bars represent plasma samples obtained before and after the dual infection, respectively. Mean values with standard deviations are shown. *, significant change, P < 0.05.
TABLE 3.
Mean IC50s of singly infected control or dually infected plasma samples against heterologous virusesa
| Plasma sample | IC50 against virus |
||||||
|---|---|---|---|---|---|---|---|
| Bx08 | 102 | 104 | 122 | 161 | 6491 | MLV | |
| Singly infected | |||||||
| CMNYU110 | |||||||
| (1) | 10 | 10 | 10 | 10 | 36 | 141 | |
| (2) | 10 | 10 | 10 | 10 | 10 | 10 | |
| CMNYU133 | |||||||
| (1) | 58 | 25 | 10 | 10 | 10 | 10 | |
| (2) | 10 | 10 | 10 | 10 | 10 | 10 | |
| CMNYU128 | |||||||
| (1) | 547 | 110 | 60 | 239 | 10 | 10 | |
| (2) | 882 | 75 | 50 | 126 | 20 | 10 | |
| CMNYU153 | |||||||
| (1) | 25 | 10 | 10 | 10 | 10 | 10 | |
| (2) | 41 | 10 | 10 | 10 | 10 | 10 | |
| CMNYU164 | |||||||
| (1) | 10 | 10 | 10 | 10 | 10 | 10 | |
| (2) | 10 | 10 | 10 | 10 | 10 | 10 | |
| CMNYU6542 | |||||||
| (1) | 44 | 10 | 10 | 10 | 10 | 10 | |
| (2) | 10 | 10 | 10 | 10 | 20 | 10 | |
| Dually infected | |||||||
| CMNYU107 | |||||||
| (1) | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| (2) | 543* | 691* | 794* | 194* | 845* | 231* | 10 |
| CMNYU129 | |||||||
| (1) | 10 | 10 | 10 | 10 | 10 | 10 | ND |
| (2) | 10 | 10 | 10 | 10 | 10 | 10 | ND |
| CMNYU6518 | |||||||
| (1) | 98 | 10 | 35 | 10 | 10 | 63 | 10 |
| (2) | 345* | 954* | 375* | 285* | 318* | 479* | 10 |
| CMNYU6544 | |||||||
| (1) | 10 | 10 | 10 | 10 | 10 | 23 | 25 |
| (2) | 110* | 264* | 489* | 189* | 168* | 284* | 10 |
(1), 1st plasma sample; (2), sequential plasma sample. For dual infections, the sequential sample was obtained after dual infection. *, significant increase, P < 0.05. MLV. neutralization of HIV-1 pseudotyped with MLV Env. ND, not done. Boldface indicates IC50 titer of >100. Underlining indicates IC50 titer of 50 to 100. A value of 10, i.e., half the minimum dilution tested, indicates 50% neutralization was not achieved at any dilution.
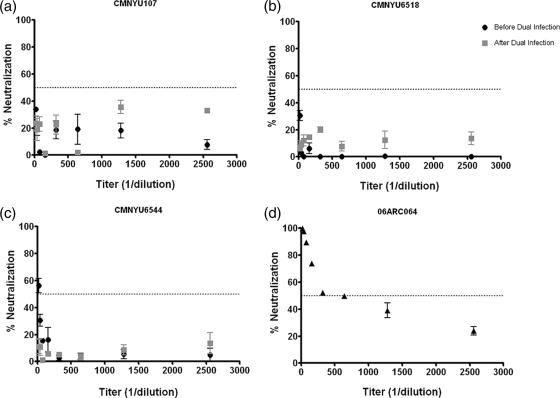
As a specificity control for HIV-1 NAbs and to confirm that these subjects had not been exposed to antiretroviral therapy, the sequential samples obtained from the dually infected subjects CMNYU107, CMNYU6518, and CMNYU6544, as well as plasma obtained from subject 06ARC064, who is on ARV, were tested for their ability to neutralize HIV-1 pseudotyped with MLV Env using 2-fold serial dilutions of plasma (1:20 to 1:2,560). HIV-1 pseudotyped with MLV Env would be susceptible to the action of antiretroviral drugs present in plasma but not to anti-HIV-Nabs. This analysis found that the plasma obtained from the subject 06ARC064 was able to strongly neutralize the MLV Env control virus, exhibiting an IC50 titer of 903 (range, 519 to 1,278). In contrast, the plasma obtained from each dually infected subject before and after dual infection generally could not neutralize HIV-1 pseudotyped with MLV Env, except for the initial CMNYU6544 sample, which exhibited a mean IC50 titer of 25 (range, 17 to 32) (Fig. 3). Therefore, there were no significant differences in neutralization potency against HIV-1 pseudotyped with MLV Env between samples obtained before and after the dual infections; however, the IC50 titer for subject 06ARC064 against this control virus was significantly higher than those for the dually infected subjects at either time point tested (P = 0.0003) (Table 3 and Fig. 3).
FIG. 3.
Neutralization patterns of HIV-1 pseudotyped with MLV Env by serially diluted plasma. Subjects CMNYU107, CMNYU6518, and CMNYU6544 are dually infected with HIV-1 and are ARV naive. Subject 06ARC064 is on ARV treatment. Horizontal dotted line indicates 50% neutralization.
Breadth and potency scores.
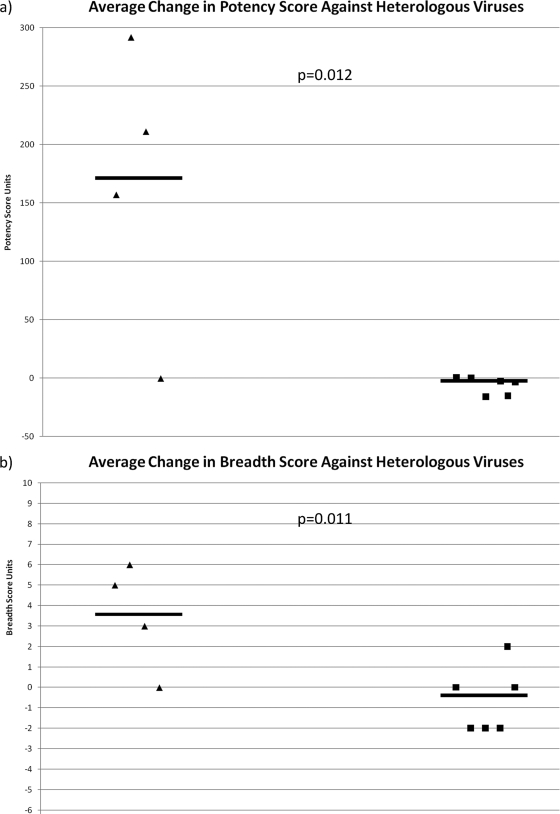
In order to further characterize this enhancement of anti-HIV-1 immunity, breadth and potency scores were calculated from the IC50s according to the method of Blish et al. for each plasma sample (1). For the dually infected cases and singly infected controls, the average change in breadth and potency for each plasma pair against all 6 viruses was determined and compared. In terms of neutralization potency, the average change in potency score among the 4 dually infected subjects from the initial plasma sample to the final one was an increase of 165 points (range, −0.16 to 292 points), while for the 6 control subjects, there was an average decrease of 6.11 points (range, −15.8 to 0.45 points), demonstrating that the dually infected subjects exhibited a significantly potent nAb response after the dual infection compared to any such changes over a comparable time period among the singly infected controls (P = 0.012) (Fig. 4a). Furthermore, the average change in the breadth score for the dually infected subjects was an increase of 3.5 points (range, 0 to 6 points), while the average change in the breadth score among the control subjects was a decrease of 0.6 points (range, −2 to 2 points), indicating a significant increase in breadth for the dually infected subjects compared to any broadening of the nAb response among the controls over the study period (P = 0.011) (Fig. 4b).
FIG. 4.
Average change in potency and breadth of neutralization. Dually infected and singly infected samples are represented by triangles and squares, respectively. Horizontal lines indicate mean values.
Viral load analysis.
Viral loads were measured using the Versant HIV RNA 3.0 assay, which is known to efficiently detect different group M subtype URFs and CRFs, in order to determine if nAb enhancement might have been attributable to higher viral loads in the dually infected subjects than in the singly infected controls (4, 19, 24, 27). While the mean viral loads of the initial samples obtained from the broadly neutralizing dually infected subjects were higher than those of the initial samples from the controls (n = 23), this difference was not statistically significant (P = 0.08). In the final samples obtained from the control subjects and the broadly neutralizing dually infected subjects, the mean viral loads were also not significantly different statistically (P = 0.47). The average change in viral load for the control group in the final sample was 13,535 copies/ml, while that of the dually infected group was a decrease of 6,385 copies/ml. These values were not significantly different (P = 0.19). Furthermore, there were no significant differences in the initial, final, or change in viral load between the short-duration case and control groups (P = 0.33, P = 0.33, and P = 1, respectively) or between the long-duration case and control groups (P = 0.22, P = 0.15, and P = 0.14, respectively) (Table 4).
DISCUSSION
The occurrence of dual HIV-1 infection provides a unique opportunity to investigate the effect on the immune response of exposure to multiple viral variants. Specifically, these infections provide the best model for examining and comparing the effect of infection by a single HIV-1 strain to that of infection by multiple, genetically divergent variants on the host immune system in generating nAbs and to study whether such antibodies exhibit differences in their potency and breadth. By testing of the neutralization capacities of pairs of sequential plasma samples obtained before and after dual infection, it was shown in the present study that both the potency and breadth of the humoral anti-HIV-1 immune response are significantly enhanced after a dual infection. These results were in clear contrast to data obtained for control subjects matched for study period duration, whose sequential plasma samples exhibited no significant increases in their magnitudes of neutralization against the heterologous isolates tested. Furthermore, the lack of neutralization potency by plasma from the dually infected subjects against HIV-1 pseudotyped with MLV Env and the ability of the last-time-point plasma samples from these subjects to neutralize the different HIV-1 primary isolates constitute evidence that the neutralization exhibited by the dually infected subjects is due to antibodies directed specifically at HIV-1 envelopes in the primary HIV-1 isolates tested and also confirm that these dually infected patients were not exposed to antiretroviral therapy. The data presented herein establish a strong correlation between repeated infection by genetically distant HIV-1 viruses and the broadening and strengthening of the anti-HIV-1 nAb response, relative to the response generated by a single infection. Importantly, since the viral loads for the broadly neutralizing, dually infected subjects compared to those for the control subjects were not significantly different in either the first or final samples obtained from these subjects, nor was there a significant difference in the change in their viral loads over time, it does not appear that viral loads were responsible for the observed nAb enhancement. Notably, the dually infected subjects experienced an overall decrease in viral loads, with subjects CMNYU6518 and CMNYU6544 possessing undetectable viral loads in their final, broadly neutralizing plasma samples, while the control subjects experienced an overall increase. None of these subjects were known to have been exposed to antiretroviral drugs; therefore, it is possible that the broad nAbs detected in the dually infected subjects may have had a potent, autologous neutralizing effect by this point in the study, though this remains to be confirmed through future analysis. This potential correlation between broad nAbs and reduced viral loads is in contradiction to the direct correlation between these variables observed in previous studies; however, these studies were not conducted using dually infected subjects, and therefore, it is difficult to draw conclusions regarding this apparent discrepancy (4, 19, 24, 27).
Using these dual HIV-1 infections as an in vivo model, our results, for the first time, provide proof-of-principle that repeated exposure of the immune system to highly diverse HIV-1 antigens can significantly improve anti-HIV-1 immunity. These data support the use of vaccine constructs that carry epitopes of divergent HIV-1 variants rather than of single strains, and they imply that polyvalent vaccination schemes may be preferable to monovalent ones. Indeed, recent vaccine studies support this hypothesis, suggesting that vaccines based on multiple strains do induce superior nAbs (11, 23, 28-30). Zolla-Pazner et al. demonstrated that immunization of rabbits with V3 fusion proteins from HIV-1 clades A, B, and C induces anti-HIV-1 antibodies with better potency and breadth than immunization of rabbits with V3 fusion proteins from a single clade (30). Furthermore, in a study evaluating the induction of HIV-1-specific immune responses in adults immunized with either monovalent or polyvalent HIV-1 env formulations, it was demonstrated that plasma from individuals who received the polyvalent formulations exhibited higher titers of broad nAb responses (28).
It is not clear from the present study of these few subjects how genetically distant HIV-1 antigens must be to cause the observed effect on host immunity. Studies of expanded dually and singly infected cohorts that include more subjects who may be dually infected with a wider array of genetically divergent viruses will shed light on this issue and further inform vaccine design as to the required genetic distance of vaccine immunogens. Available sequence information on the present subjects suggests that the enhancement of the nAb response can occur after dual infection by viruses that possess only subsubtype distances from one another, as shown by subject CMNYU6518, who was dually infected by unrelated strains of the same URF and exhibited significant enhancement of the immune response. Yet subject CMNYU129 was dually infected by 2 distinct HIV-1 subtypes (CRF02_AG and F2) and thus might have been expected to exhibit this same effect but did not. It is not known why this subject failed to exhibit an enhancement of the immune response, though various virus and host factors may be accountable. It may have been that this subject's immune system was incapable of generating such a robust response due to general polymorphisms or concurrent disease; also, insufficient time may have elapsed since the dual infection, since this subject's final sample was obtained 24 months after the dual infection, 2 to 3 years less than the period between the dual infection and the final samples obtained from two of the subjects exhibiting broadly neutralizing nAbs. In this vein, since it was not known how long these individuals had been infected with HIV-1 prior to dual infection, it is possible that the results presented herein may have been skewed if the dually infected subjects with potent nAb responses were infected for a much longer period of time prior to dual infection than the control group. However, given the size of the control group, this is highly unlikely, although in future larger studies, this variable should be controlled for. Furthermore, differences in viral fitness may have inhibited the growth of one of the infecting strains and thus failed to stimulate an enhanced immune response, and it is also possible that specific timing between the initial and dual infections, not experienced by subject CMNYU129, is critical to generating this stimulatory effect. Detailed investigation of these topics will be reported elsewhere.
It remains to be seen if dual infection in these subjects has resulted in differential autologous immune control of each dually infecting virus. Future study of autologous neutralization in these and similar subjects will shed light on this question, which may be of interest for the design of therapeutic HIV-1 vaccines, i.e., investigating if challenge with vaccine antigens that are genetically divergent from an HIV-1 strain already infecting a patient would produce a beneficial effect in controlling the established infection. Such studies will also address whether individuals exhibiting broad and potent neutralization as a result of dual infection harbor fewer recombinant viruses than those not exhibiting this effect. If such were the case, this would suggest that dual infection induces nAb responses that may control virus evolution, improving our understanding of disease progression and the relationships between infection, protection, and humoral immunity.
Various studies of HIV-positive individuals, conducted globally, have found that dual infection is not exceptional, thus demonstrating that the immune response generated by a single infection is insufficient to prevent a 2nd infection (6, 8, 9, 16, 17, 21, 26; B. Chohan et al., AIDS Vaccine 2007; Hawkins et al., AIDS Vaccine 2007). Similarly, most trial vaccines have failed to generate a protective anti-HIV-1 response, due at least in part to the extreme genetic diversity of the HIV pandemic, and have sought to overcome this roadblock by including immunogens based on epitopes conserved among the HIV-1 clades or by tailoring vaccines to the strains circulating in specific regions of endemicity (12). Ironically, it may be that this apparent obstacle can be surmounted by exploiting HIV-1 diversity. Vaccination schemes consisting of polyvalent, genetically divergent immunogens may yield an immune response of extremely heterologous and potent anti-HIV-1 nAbs, and importantly, it may be that the heterologous HIV-1 strains comprising such a vaccine need not all be derived from those circulating among a vaccine target population, alleviating the need for regionally tailored vaccines.
Acknowledgments
We are grateful to the individuals who have donated their blood for these studies. We acknowledge the continued support of the Ministry of Public Health, Cameroon.
This work was supported by grant no. AI47053 from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), the Fogarty International Center (no. TW01409), and the Center for AIDS Research (no. AI027742-17) and funds from the Department of Veterans Affairs (Merit Review Award and the Research Enhancement Program).
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Blish, C. A., O. C. Dogan, N. R. Derby, M. A. Nguyen, B. Chohan, B. A. Richardson, and J. Overbaugh. 2008. HIV-1 superinfection occurs despite relatively robust neutralizing antibody responses. J. Virol. 82:12094-12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chohan, B., L. Lavreys, S. M. Rainwater, and J. Overbaugh. 2005. Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype. J. Virol. 79:10701-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doria-Rose, N. A., R. M. Klein, M. G. Daniels, S. O'Dell, M. Nason, A. Lapedes, T. Bhattacharya, S. A. Migueles, R. T. Wyatt, B. T. Korber, J. R. Mascola, and M. Connors. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenyo, E. M., A. Heath, S. Dispinseri, H. Holmes, P. Lusso, S. Zolla-Pazner, H. Donners, L. Heyndrickx, J. Alcami, V. Bongertz, C. Jassoy, M. Malnati, D. Montefiori, C. Moog, L. Morris, S. Osmanov, V. Polonis, Q. Sattentau, H. Schuitemaker, R. Sutthent, T. Wrin, and G. Scarlatti. 2009. International network for comparison of HIV neutralization assays: the NeutNet report. PLoS One 4:e4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerhardt, M., D. Mloka, S. Tovanabutra, E. Sanders-Buell, O. Hoffmann, L. Maboko, D. Mmbando, D. L. Birx, F. E. McCutchan, and M. Hoelscher. 2005. In-depth, longitudinal analysis of viral quasispecies from an individual triply infected with late-stage human immunodeficiency virus type 1, using a multiple PCR primer approach. J. Virol. 79:8249-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottlieb, G. S., D. C. Nickle, M. A. Jensen, K. G. Wong, J. Grobler, F. Li, S. L. Liu, C. Rademeyer, G. H. Learn, S. S. Karim, C. Williamson, L. Corey, J. B. Margolick, and J. I. Mullins. 2004. Dual HIV-1 infection associated with rapid disease progression. Lancet 363:619-622. [DOI] [PubMed] [Google Scholar]
- 8.Grobler, J., C. M. Gray, C. Rademeyer, C. Seoighe, G. Ramjee, S. A. Karim, L. Morris, and C. Williamson. 2004. Incidence of HIV-1 dual infection and its association with increased viral load set point in a cohort of HIV-1 subtype C-infected female sex workers. J. Infect. Dis. 190:1355-1359. [DOI] [PubMed] [Google Scholar]
- 9.Herbinger, K. H., M. Gerhardt, S. Piyasirisilp, D. Mloka, M. A. Arroyo, O. Hoffmann, L. Maboko, D. L. Birx, D. Mmbando, F. E. McCutchan, and M. Hoelscher. 2006. Frequency of HIV type 1 dual infection and HIV diversity: analysis of low- and high-risk populations in Mbeya Region, Tanzania. AIDS Res. Hum. Retroviruses 22:599-606. [DOI] [PubMed] [Google Scholar]
- 10.Heyndrickx, L., W. Janssens, L. Zekeng, R. Musonda, S. Anagonou, G. Van der Auwera, S. Coppens, K. Vereecken, K. De Witte, R. Van Rampelbergh, M. Kahindo, L. Morison, F. E. McCutchan, J. K. Carr, J. Albert, M. Essex, J. Goudsmit, B. Asjo, M. Salminen, A. Buve, Study Group on Heterogeneity of HIV Epidemics in African Cities, and G. van Der Groen. 2000. Simplified strategy for detection of recombinant human immunodeficiency virus type 1 group M isolates by gag/env heteroduplex mobility assay. J. Virol. 74:363-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurwitz, J. L., K. S. Slobod, T. D. Lockey, S. Wang, T. H. Chou, and S. Lu. 2005. Application of the polyvalent approach to HIV-1 vaccine development. Curr. Drug Targets Infect. Disord. 5:143-156. [DOI] [PubMed] [Google Scholar]
- 12.Johnston, M. I., and A. S. Fauci. 2008. An HIV vaccine—challenges and prospects. N. Engl. J. Med. 359:888-890. [DOI] [PubMed] [Google Scholar]
- 13.Korber, B., et al. (ed.). 2004. HIV immunology and sequence databases, Los Alamos, NM. Los Alamos National Laboratory, Los Alamos, NM. www.hiv.lanl.gov.
- 14.Laal, S., S. Burda, S. Sharpe, and S. Zolla-Pazner. 1993. A rapid, automated microtiter assay for measuring neutralization of HIV-1. AIDS Res. Hum. Retroviruses 9:781-785. [DOI] [PubMed] [Google Scholar]
- 15.Landau, N. R., K. A. Page, and D. R. Littman. 1991. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J. Virol. 65:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manigart, O., V. Courgnaud, O. Sanou, D. Valea, N. Nagot, N. Meda, E. Delaporte, M. Peeters, and P. Van de Perre. 2004. HIV-1 superinfections in a cohort of commercial sex workers in Burkina Faso as assessed by an autologous heteroduplex mobility procedure. AIDS 18:1645-1651. [DOI] [PubMed] [Google Scholar]
- 17.McCutchan, F. E., M. Hoelscher, S. Tovanabutra, S. Piyasirisilp, E. Sanders-Buell, G. Ramos, L. Jagodzinski, V. Polonis, L. Maboko, D. Mmbando, O. Hoffmann, G. Riedner, F. von Sonnenburg, M. Robb, and D. L. Birx. 2005. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J. Virol. 79:11693-11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piantadosi, A., B. Chohan, V. Chohan, R. S. McClelland, and J. Overbaugh. 2007. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 3:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piantadosi, A., D. Panteleeff, C. A. Blish, J. M. Baeten, W. Jaoko, R. S. McClelland, and J. Overbaugh. 2009. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J. Virol. 83:10269-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell, R. L., L. Lezeau, T. Kinge, and P. Nyambi. 2010. Longitudinal quasispecies analysis of viral variants in HIV-1 dually-infected individuals highlights the importance of sequence identity in viral recombination. AIDS Res. Hum. Retroviruses 26:253-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell, R. L., M. M. Urbanski, S. Burda, T. Kinge, and P. N. Nyambi. 2009. High frequency of HIV-1 dual infections among HIV-positive individuals in Cameroon, West Central Africa. J. Acquir. Immune Defic. Syndr. 50:84-92. [DOI] [PubMed] [Google Scholar]
- 22.Powell, R. L., M. M. Urbanski, and P. N. Nyambi. 2008. A heteroduplex assay for the rapid detection of dual human immunodeficiency virus type 1 infections. J. Virol. Methods 149:20-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakhatskyy, P., S. Wang, C. Zhang, T. H. Chou, M. Kishko, and S. Lu. 2008. Immunogenicity and protection efficacy of subunit-based smallpox vaccines using variola major antigens. Virology 371:98-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sather, D. N., J. Armann, L. K. Ching, A. Mavrantoni, G. Sellhorn, Z. Caldwell, X. Yu, B. Wood, S. Self, S. Kalams, and L. Stamatatos. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, D. M., M. C. Strain, S. D. Frost, S. K. Pillai, J. K. Wong, T. Wrin, Y. Liu, C. J. Petropolous, E. S. Daar, S. J. Little, and D. D. Richman. 2006. Lack of neutralizing antibody response to HIV-1 predisposes to superinfection. Virology 355:1-5. [DOI] [PubMed] [Google Scholar]
- 26.Smith, D. M., J. K. Wong, G. K. Hightower, C. C. Ignacio, K. K. Koelsch, E. S. Daar, D. D. Richman, and S. J. Little. 2004. Incidence of HIV superinfection following primary infection. JAMA 292:1177-1178. [DOI] [PubMed] [Google Scholar]
- 27.Swanson, P., C. de Mendoza, Y. Joshi, A. Golden, R. L. Hodinka, V. Soriano, S. G. Devare, and J. Hackett, Jr. 2005. Impact of human immunodeficiency virus type 1 (HIV-1) genetic diversity on performance of four commercial viral load assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v1.5, VERSANT HIV-1 RNA 3.0, and NucliSens HIV-1 QT. J. Clin. Microbiol. 43:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, S., J. S. Kennedy, K. West, D. C. Montefiori, S. Coley, J. Lawrence, S. Shen, S. Green, A. L. Rothman, F. A. Ennis, J. Arthos, R. Pal, P. Markham, and S. Lu. 2008. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 26:3947-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, S., R. Pal, J. R. Mascola, T. H. Chou, I. Mboudjeka, S. Shen, Q. Liu, S. Whitney, T. Keen, B. C. Nair, V. S. Kalyanaraman, P. Markham, and S. Lu. 2006. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology 350:34-47. [DOI] [PubMed] [Google Scholar]
- 30.Zolla-Pazner, S., S. S. Cohen, C. Krachmarov, S. Wang, A. Pinter, and S. Lu. 2008. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology 372:233-246. [DOI] [PubMed] [Google Scholar]
- 31.Zolla-Pazner, S., and S. Sharpe. 1995. A resting cell assay for improved detection of antibody-mediated neutralization of HIV type 1 primary isolates. AIDS Res. Hum. Retroviruses 11:1449-1458. [DOI] [PubMed] [Google Scholar]