Abstract
Nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) are important components of multidrug therapy for HIV-1. Understanding the effect of NNRTI-resistant mutants on virus replication and reverse transcriptase (RT) function is valuable for the development of extended-spectrum NNRTIs. We measured the fitness of six NNRTI-resistant mutants, the K103N, V106A, Y181C, G190A, G190S, and P236L viruses, using a flow cytometry-based cell culture assay. K103N and Y181C viruses had fitness similar to that of the wild type while V106A, G190A, G190S, and P236L viruses had reduced fitness. We also determined the biochemical correlates of fitness by measuring the RNase H and polymerization activities of recombinant mutant RTs and virion-associated RTs. The RNase H activities of recombinant and virion-associated RTs correlated with the relative fitness for each mutant. K103N and Y181C mutants had normal RNase H activity; V106A, G190A, and G190S mutants had moderate reductions in activity; and the P236L mutant had substantially reduced activity. With the exception of the P236L mutant, reduced fitness correlates with low virion-associated polymerization efficiency and reduced RT content. Reduced polymerase function in virions derived from low RT content rather than an intrinsic polymerization defect in each RT protein. In conclusion, severe defects in RNase H activity alone, exemplified by the P236L mutant, appear sufficient to cause a substantial reduction in fitness. For the other NNRTI mutants, reductions in RT content decreased both polymerization and RNase H activity in virions. RNase H reduction was compounded by intrinsic RNase H defects in the mutant RTs.
HIV-1 reverse transcriptase (RT) is a multifunctional enzyme that converts the single-stranded RNA viral genome into double-stranded DNA. It has RNA- and DNA-dependent DNA polymerase activities, which copy RNA or DNA templates, respectively, into double-stranded DNA, as well as RNase H activity that cleaves genomic RNA still hybridized to DNA (40). Although both activities are essential for HIV-1 replication, some studies have shown that polymerization and RNase H activities are not rigorously coupled (11, 12, 27, 36). RT is a heterodimer comprised of two subunits, a 66-kDa (p66) subunit and a 51-kDa (p51) subunit (20). The p66 subunit has both the polymerase and RNase H domains, which make contact simultaneously with the RNA genome (12). The p51 subunit is an N-terminal cleavage product of p66 which lacks the RNase H domain and serves as a structural scaffold for the p66 subunit (2).
Because of its essential role in the virus life cycle, RT is an important antiretroviral target for the treatment of HIV-1. Nonnucleoside RT inhibitors (NNRTIs) are one of three classes of antiretroviral RT inhibitors that are commonly used in treatment. They are a group of structurally diverse small hydrophobic compounds, which can specifically bind and inhibit RT (reviewed in reference 34). One limitation of NNRTIs is that they face a low genetic barrier to resistance. Previous studies of viral replication fitness have shown that NNRTI resistance mutations impact replication fitness and that the relative fitness of the viruses with these mutations is associated with their prevalence in vivo, independent of the level of resistance that they confer (4, 19, 38). Mutants such as the G190S, V106A, and P236L viruses, which have lower prevalence in clinical isolates, are less fit than are mutants such as the K103N and Y181C viruses, which are similar in fitness to drug-sensitive isolates. The relative fitness of these mutants in cell culture correlates with the relative RNase H activities of the purified mutant RTs, which display RNase H deficiencies but no defects in DNA polymerization with DNA or RNA primers (reviewed in reference 14). Therefore, decreased RNase H activity is one apparent mechanism of reduced replication fitness of viruses with NNRTI resistance mutations.
One study has shown that amino acid substitutions at G190 of HIV-1 RT severely decreased virion-associated mature RT protein content and RT activity, which correlated with impaired viral replication capacity (25). We hypothesize that in addition to reduced RNase H activity, reduced RT content of NNRTI-resistant mutants in virions, which incidentally reduces RNase H, could contribute to reduced replication fitness in cell culture. We therefore determined the effects of the K103N, Y181C, P236L, G190S, G190A, and V106A NNRTI resistance mutants on the polymerization and RNase H activities of both purified recombinant protein and virion-associated RT activity and content.
MATERIALS AND METHODS
Cell culture.
The 293 cell line (ATCC, Manassas, VA) was grown in Dulbecco modified Eagle medium (Cellgro, Herndon, VA) with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 U/ml). The PM1 cell line is derived from HUT 78 and is permissive for both macrophage-tropic and lymphocyte-tropic strains of HIV-1. It was obtained from Marvin Reitz through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID) (31), and was cultured in RPMI medium with l-glutamine (2 mM) (Cellgro, Herndon, VA), supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 U/ml).
Antibodies.
The following antibody was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: the HIV-1 RT monoclonal antibody 8C4 was obtained from D. Helland and A. M. Szilvay and recognizes both subunits of RT (35); p24 monoclonal antibody 183-H12-5C was obtained from Bruce Chesebro and Kathy Wehrly (8, 37); polyclonal HIV-1 integrase antiserum recognizing epitopes mapping to amino acids 1 to 16 was obtained from Duane P. Grandgenett (6).
Site-directed mutagenesis.
Each of the 6 NNRTI drug resistance mutations K103N, V106A, Y181C, G190A, G190S, and P236L was introduced into pRHA1 (19) by using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). A molecular clone of HIV-1, pNL4-3, was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Malcolm Martin. The same mutants were introduced into pRHAXX (16). The pAT1 and pAT2 plasmids are derived from pNL4-3 and have the mouse Thy1.1 or Thy1.2 genes in place of HIV-1 nef, respectively (17). The NNRTI drug-resistant mutations were subcloned from pRHA1 into pNL4-3 by using ApaI and AgeI restriction sites and from pRHAXX into pAT2 by using XmaI and XbaI restriction sites as previously described (17, 38).
Generation of NNRTI-resistant virus stocks.
A human cell line, 293, was transiently transfected with wild-type (WT) NL4-3, the NNRTI drug-resistant mutants of NL4-3, or the NNRTI drug-resistant mutants of AT2 by using Lipofectin (Superfect; Qiagen, Santa Clarita, CA). After 72 h, clarified supernatants were harvested and the p24 capsid protein of each stock was measured using enzyme-linked immunosorbent assay (ELISA) (Perkin-Elmer, Wellesley, MA). Wild-type and mutant stocks in the pNL4-3 background were used for Western blot analysis to measure the relative RT content and for virion-associated RT activity. Wild-type AT1 and mutant AT2 stocks were used for virus growth competition assays.
Virus growth competition assay.
The multiple-cycle growth competition assays were performed by the method developed by our research group using flow cytometry to quantify the relative amounts of reference and test virus as previously described (17). Briefly, we produced separate virus stocks by transfecting 293 cells with wild-type pAT1 or mutant pAT2. PM1 cells were coinfected with the wild type and each NNRTI mutant stock. On days 3, 4, 5, and 6 postinfection, half the culture was removed and replaced with fresh medium; the numbers of wild-type- and mutant-infected cells were determined by fluorescence-activated cell sorting (FACS) analysis after the cells were stained with antibodies specific to Thy1.1 and Thy1.2 (Becton Dickinson, San Jose, CA). The relative fitness 1 + s was calculated using procedures from the website http://bis.urmc.rochester.edu/vFitness/FitnessTwo.aspx, where s is the fitness coefficient and 1 + s is calculated using the following formula: exp{(1/Δt) ln[Tm(t2)Tw(t1)/Tw(t2)Tm(t1)]}, where Tm(t2) is the number of mutant-infected cells at day 6 postinfection, Tw(t1) is the number of wild-type-infected cells at day 3 postinfection, Tw(t2) is the number of wild-type-infected cells at day 6 postinfection, and Tm(t1) is the number of mutant-infected cells at day 3 postinfection (42).
Construction of expression plasmids for NNRTI-resistant mutants.
The full-length wild-type and mutant RT sequences were amplified by PCR using pRHA and the following primers: GATATACATATGCCCATTAGTCCTATTGAG (forward) and GCAAGCTTCTATAGTACTTTCCTGATTCC (reverse). These primers introduced an NdeI restriction site (underlined) with an initiation ATG codon (forward primer) and a HindIII restriction site with a terminating TAG codon (reverse primer). Amplified products were cloned into the expression vector pET21a(+) (Novagen) (24) using the NdeI and HindIII restriction sites to create pET21ap66 wild type and mutant. Similarly, the wild-type and mutant RT sequences spanning the p51 subunit of RT were amplified from pRHA using the following primers: GATATACATATGCCCATTAGTCCTATTGAG (forward) and CATAAGCTTGTTAGAAAGTTTCTGCTCCTAT (reverse). Amplified products were cloned into the expression vector pET-28a(+), which contains an N-terminal hexahistidine tag, by using the NdeI and HindIII restriction sites. After sequencing to confirm the presence of the mutation and the absence of spurious mutations, the expression plasmids were transformed into Escherichia coli BL21(DE3) cells. The Autoinduction System 1 was used to induce overnight cultures (Novagen/EMD, Darmstadt, Germany). The presence of each RT subunit was determined by protein SDS-PAGE analysis using 5 μl of culture pellets.
Protein purification.
Wild-type and mutant p66 and p51 heterodimeric recombinant RTs were purified from induced bacterial cultures by using chromatography. Since the p51 subunit contained the 6×His tag, p66 and p51 cultures were mixed prior to chromatography to ensure that p66/p51 heterodimers were purified. The following columns were used with the AKTAprime Plus system (Amersham/GE, Piscataway, NJ) to purify p66/p51 heterodimer: Q-Sepharose (Amersham/GE, Piscataway, NJ) and Talon Ni metal affinity resin (BD Biosciences, Palo Alto, CA). All eluted fractions from the Q-Sepharose column were run over the Talon column. The eluted fractions from the Talon column that had the highest total amount of protein were used for separation of p51 monomer from p66/p51 heterodimer, which was performed using a Sesource S column as previously described (24). The eluted fractions with equal relative amounts of p51 and p66 were determined using SDS-PAGE and chosen to measure DNA polymerization and RNase H activities.
Pre-steady-state kinetic measurements with purified recombinant RTs.
Pre-steady-state and steady-state kinetic measurements of polymerization were performed as previously described (38). Briefly, single nucleotide incorporation was measured using a 5′-radiolabeled DNA primer annealed to a 40-mer DNA template. Steady-state assays of single nucleotide incorporation were performed on a DNA-DNA primer template over a range of nucleotide concentrations. Pre-steady-state burst reactions were performed in a KinTek rapid quench-flow machine using the same primer template. Relative amounts of mutant and wild-type RTs were normalized based on active RT concentration.
DNA polymerization and RNase H assays of recombinant RTs.
Specific DNA polymerization activity of each RT preparation was measured by using a poly(rA)/oligo(dT) template/primer and an [α-32P]dTTP substrate (4). A unit was defined as the amount of enzyme required to incorporate 1 nmol of dTTP into nucleic acid product in 10 min at 37°C. Equivalent numbers of units of specific polymerization activity were added to both the polymerization rate and RNase H assay mixtures. DNA 3′-end-directed and RNA 5′-end-directed RNase H activities were measured as previously described (38).
To determine the rate of RNA-dependent DNA polymerization activity of each RT, the polymerization activities were tested using a 5′-radiolabeled primer binding site (PBS) oligomer DNA primer annealed to the RNA template D199, +1 to +199 of the pNL4-3 genomic RNA, which included the PBS. The primer/template (15 nM/25 nM) was prebound with purified RT normalized by relative specific activities of polymerization in reaction buffer containing 25 mM Tris-HCl, pH 8.0, 25 mM NaCl, and 0.5 mM EDTA and dithiothreitol (DTT) at 37°C for 2 min. The reactions were initiated by adding 6 mM MgCl2 and 500 μM deoxynucleoside triphosphates (dNTPs) and quenched at various times with an equal volume of 2× DNA loading buffer (Ambion). Extension products were analyzed by SDS-PAGE and quantified by phosphorimaging.
Purified 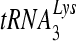 from human placenta (Bio S&T, Canada) was 5′ end labeled by [γ-32P]ATP as previously described (43) and was annealed to D199 at a primer/template (P/T) ratio of 1:2. After binding with purified RT, the reactions were initiated by adding 6 mM MgCl2 and 500 μM dNTPs in a reaction buffer containing 25 mM Tris-HCl, pH 8.0, 25 mM NaCl, and 0.5 mM EDTA and DTT. The reactions were quenched at various times with an equal volume of 2× DNA loading buffer (Ambion). Extension products were analyzed by SDS-PAGE.
from human placenta (Bio S&T, Canada) was 5′ end labeled by [γ-32P]ATP as previously described (43) and was annealed to D199 at a primer/template (P/T) ratio of 1:2. After binding with purified RT, the reactions were initiated by adding 6 mM MgCl2 and 500 μM dNTPs in a reaction buffer containing 25 mM Tris-HCl, pH 8.0, 25 mM NaCl, and 0.5 mM EDTA and DTT. The reactions were quenched at various times with an equal volume of 2× DNA loading buffer (Ambion). Extension products were analyzed by SDS-PAGE.
Virion-associated RT polymerization and RNase activity.
Supernatant solutions from 293 cells transfected with wild type or NNRTI-resistant mutants were pelleted by centrifugation at 19,500 rpm for 1 h at 4°C. Virus pellets containing 100 ng of p24 capsid protein were resuspended in 10 μl PBS and were lysed by adding 10 μl 0.3% Igepal CA-630 (Sigma-Aldrich, Castle Hill, NSW, Australia) (39). The specific activities of DNA polymerization and the 5′-end-directed RNase H of RT in the viral lysates were measured as previously described (4, 38, 39).
Quantification of RT content in virions using Western blotting.
Supernatant solutions from 293 cells transfected with wild type or NNRTI-resistant mutants were pelleted by centrifugation at 19,500 rpm for 1 h at 4°C. Virus pellets containing 200 ng of p24 antigen for mutants and different amounts (5 to 100 ng) for wild type were resuspended with 15 μl NuPAGE 2× sample buffer (Invitrogen) and fractionated by electrophoresis accordingly to the NuPAGE manufacturer's instructions. The proteins were separated using a 4 to 12% Bis-Tris gel with 1× MOPS (morpholinepropanesulfonic acid) running buffer (Invitrogen), transferred to nitrocellulose membranes, blocked with 5% milk in Tris-buffered saline (TBS) with 0.05% Tween 20 (TBST) overnight, and bound with primary antibody (anti-RT [8C4]), anti-IN, or anti-p24) and secondary antibody conjugated to horseradish peroxidase (HRP; Bio-Rad). Viral proteins were visualized using SuperSignal West Femto chemiluminescence substrate (Thermo Scientific) and quantified with one-dimensional (1D) image analysis software (Kodak Digital Science). Purified RT preparations, described below, were used to verify that RT was recognized by the RT monoclonal antibody 8C4. gag processing was also visualized by Western blotting using the method described above, except that an antibody specific to p24 was used and the amount of virus loaded on the gel was increased to 800 ng in order to see the processing intermediates.
RESULTS
V106A, G190S, and P236L viruses have reduced fitness.
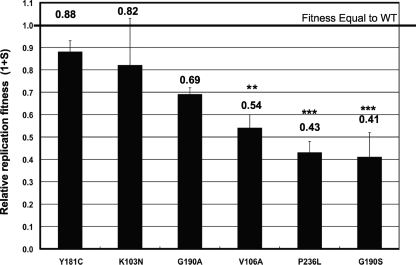
Using a flow cytometry-based growth competition assay developed by our research group (17), we measured the relative replication fitness of the NNRTI-resistant K103N, V106A, Y181C, G190A, G190S, and P236L mutant viruses. The 1 + s fitness values of K103N (0.82 ± 0.22) and Y181C (0.88 ± 0.05) viruses were not significantly different from those of the wild type. However, the 1 + s fitness values of G190A (0.69 ± 0.03), V106A (0.54 ± 0.05), P236L (0.43 ± 0.05), and G190S (0.41 ± 0.11) viruses were all reduced (Fig. 1). The rank order of fitness of these mutant viruses was as follows: wild type (WT) ≥ Y181C ≥ K103N > G190A > V106A > P236L ≥ G190S. This order was confirmed by direct competition of the mutants against each other in the NL4-3 construct by using a sequencing-based method.
FIG. 1.
Relative replication fitness of NNRTI-resistant mutants. Each mutant virus was competed against the wild type in PM1 cells. A fitness value of 1.0 indicates fitness equal to that of the wild type. Each bar represents the mean 1 + s value and the standard deviation from 6 to 11 independent infections. Mutants that had a mean 1 + s value that was statistically different from that of the K103N mutant are indicated with asterisks (**, P < 0.005; ***, P < 0.0001).
Preparation of purified recombinant RT using His-tagged p51.
Previous work characterizing the biochemical effects of NNRTI resistance mutations on RT function has revealed defects in DNA polymerization by using 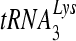 as a primer and deficiencies in the different modes of RNase H function (4, 19, 38). In those studies, the p66 and p51 subunits were both His tagged and purified separately with a Ni affinity column. The individual subunits were mixed and incubated on ice to allow for p66/p51 heterodimer formation. However, this method was inefficient in the amount of heterodimer that was obtained and did not prevent the formation of p66/p66 homodimer. Another method of purification, which was chosen for the current analyses, tagged only the p51 subunit (24). Individual bacterial cultures expressing either p66 or tagged p51 subunits were mixed before column purification. Only RT molecules containing p51 were purified by column chromatography: either single p51 subunits, p51/p51 homodimers, or p66/p51 heterodimers. Fractions of purified protein with equal molar amounts of p66 and p51 were chosen for study.
as a primer and deficiencies in the different modes of RNase H function (4, 19, 38). In those studies, the p66 and p51 subunits were both His tagged and purified separately with a Ni affinity column. The individual subunits were mixed and incubated on ice to allow for p66/p51 heterodimer formation. However, this method was inefficient in the amount of heterodimer that was obtained and did not prevent the formation of p66/p66 homodimer. Another method of purification, which was chosen for the current analyses, tagged only the p51 subunit (24). Individual bacterial cultures expressing either p66 or tagged p51 subunits were mixed before column purification. Only RT molecules containing p51 were purified by column chromatography: either single p51 subunits, p51/p51 homodimers, or p66/p51 heterodimers. Fractions of purified protein with equal molar amounts of p66 and p51 were chosen for study.
NNRTI-resistant heterodimer RTs do not affect pre-steady-state rates of polymerization.
We wanted to confirm that the pre-steady-state polymerization rates of wild-type and mutant RTs purified by the p51-His tag method were similar to the previously reported values (13, 38). Therefore, we performed pre-steady-state kinetic analysis using a DNA-DNA primer/template on a select set of RTs: wild type and G190S and P236L mutants. The percent active sites were 80% for wild type and 65% for G190S and P236L mutants. This is higher than the percent active sites obtained using the p66- and p51-His tag method, which averaged 10 to 18%. The rate of pre-steady-state polymerization, kpol (rate of nucleotide incorporation per second), was also similar to previous results: 31.35 ± 3.43 for wild type, 45.43 ± 8.89 for the G190S mutant, and 36.38 ± 11.81 for the P236L mutant.
NNRTI-resistant heterodimer RTs are not altered in polymerization from a tRNA3Lys primer bound to the primer binding site (PBS).
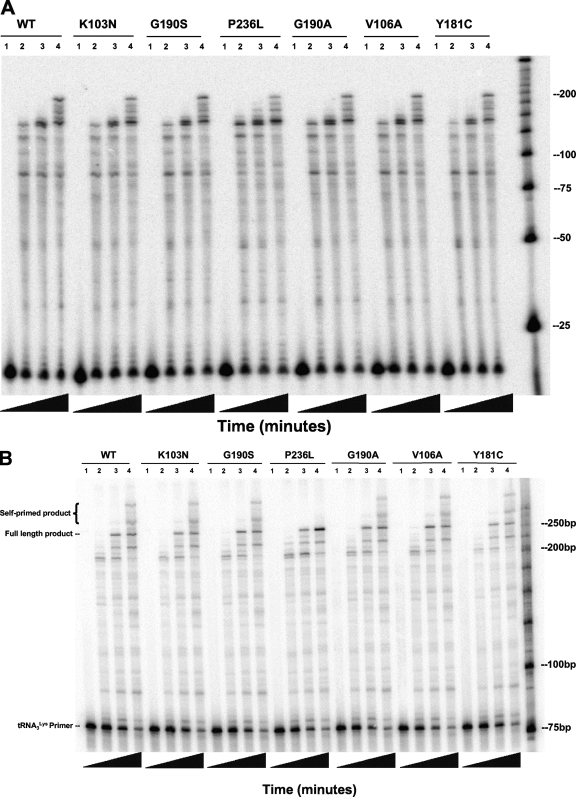
We also wanted to confirm earlier results showing the impact of NNRTI-resistant mutations on steady-state DNA polymerization from the PBS using either a DNA or a 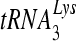 primer (38). We measured the synthesis of DNA from a DNA primer bound to the template D199, which contains +1 to +199 of the HIV-1 genomic sequence containing the PBS, U5, and R regions. The amount of each mutant enzyme added was normalized, based on specific activity of polymerization. All 6 mutants, the K103N, V106A, Y181C, G190S, G190S, and P236L enzymes, had levels of synthesis similar to that of the wild type (Fig. 2A), confirming previous results. We next measured the synthesis of DNA from a
primer (38). We measured the synthesis of DNA from a DNA primer bound to the template D199, which contains +1 to +199 of the HIV-1 genomic sequence containing the PBS, U5, and R regions. The amount of each mutant enzyme added was normalized, based on specific activity of polymerization. All 6 mutants, the K103N, V106A, Y181C, G190S, G190S, and P236L enzymes, had levels of synthesis similar to that of the wild type (Fig. 2A), confirming previous results. We next measured the synthesis of DNA from a 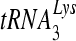 primer annealed to the same template, D199. Contrasting with previously published observations in which the G190A and G190S mutant RTs exhibited relatively slower synthesis from a tRNA primer, synthesis of DNA from the tRNA primer was the same for all 6 mutants compared to that for the wild type, including the G190A and G190S mutants (Fig. 2B).
primer annealed to the same template, D199. Contrasting with previously published observations in which the G190A and G190S mutant RTs exhibited relatively slower synthesis from a tRNA primer, synthesis of DNA from the tRNA primer was the same for all 6 mutants compared to that for the wild type, including the G190A and G190S mutants (Fig. 2B).
FIG. 2.
RNA-dependent DNA polymerization activity of purified wild-type and mutant RTs. (A) Representative polyacrylamide gel of the RNA-dependent DNA polymerization activity of wild-type and mutant RTs using a DNA primer. The substrate used was made by annealing a 5′-end-labeled PBS DNA primer (26 nt) to RNA template D199 (RNA containing +1 to +199 of the NL4-3 HIV-1 genomic sequence). Reactions were allowed to proceed for the indicated lengths of time: lanes 1, 0 s; lanes 2, 15 s; lanes 3, 1 min; lanes 4, 4 min. Nucleotide markers are indicated on the right. (B) Representative polyacrylamide gel of the RNA-dependent DNA polymerization activity of wild-type and mutant RTs using 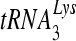 as primer. The substrate used was made by annealing a 5′-end-labeled human
as primer. The substrate used was made by annealing a 5′-end-labeled human 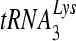 primer to D199. Reactions were allowed to proceed for the indicated lengths of time: lanes 1, 0 s; lanes 2, 1 min; lanes 3, 4 min; lanes 4, 16 min. Reactions for both experiments were started with 500 μM dNTP and 6 mM MgCl2. Equal amounts of specific activity units of RT as measured by poly(rA)/oligo(dT) template/primer synthesis were used per reaction.
primer to D199. Reactions were allowed to proceed for the indicated lengths of time: lanes 1, 0 s; lanes 2, 1 min; lanes 3, 4 min; lanes 4, 16 min. Reactions for both experiments were started with 500 μM dNTP and 6 mM MgCl2. Equal amounts of specific activity units of RT as measured by poly(rA)/oligo(dT) template/primer synthesis were used per reaction.
RNase H activity of NNRTI-resistant recombinant RTs correlates with fitness.
Previously published studies have shown that NNRTI-resistant mutants with reduced replication in cell culture have reduced 5′-end-directed and 3′-end-directed RNase H activity (4, 19, 38). Briefly, the RT bound in an orientation for DNA synthesis makes rapid primary cuts about 18 nucleotides (nt) in from the DNA 3′ end and slower secondary cuts about 9 nt in. The RT also binds RNA segments annealed to DNA and makes rapid primary cuts about 18 nt in from the RNA 5′ end and slower cuts about 9 nt in. The secondary cuts are not obligatorily sequential to the primary cuts on a particular substrate; the secondary cut rate is just lower (41). The 3′-end-directed cuts generate the RNA segments for the 5′-end-directed cut. The two cutting modes are thought to effectively clear away genomic RNA in preparation for synthesis of the second DNA strand.
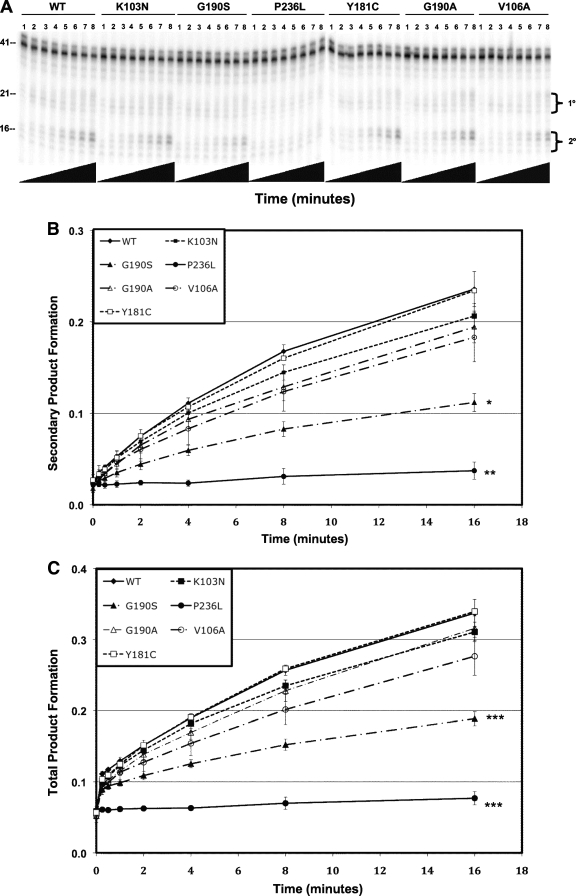
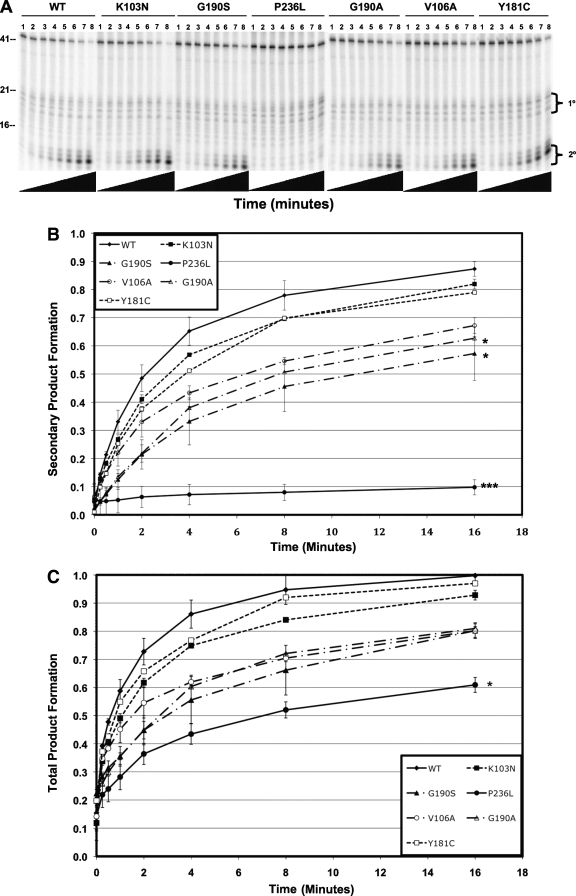
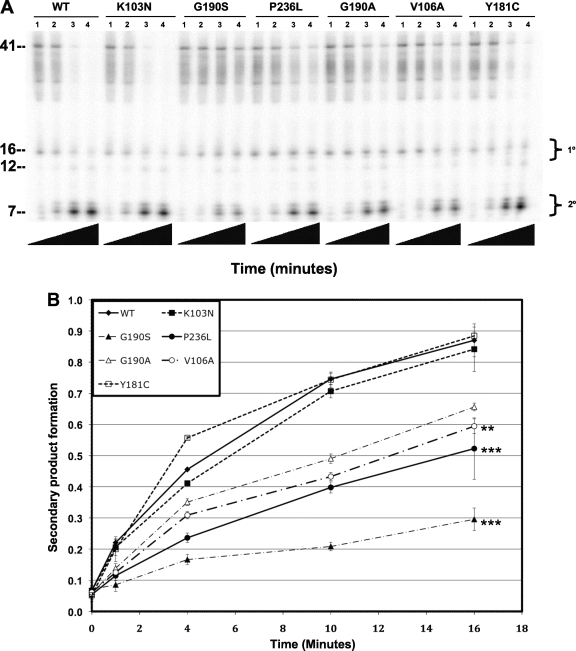
We measured the 3′-end-directed RNase H activity of each NNRTI mutant recombinant RT compared to that of the wild type (Fig. 3A, B, and C). We found that the RNase H activities of the K103N and Y181C mutants were similar to those of the wild type. The activities of the G190A, G190S, and V106A mutants were decreased relative to that of the wild type, and the activity of the P236L mutant was substantially lower than those of any of the other RTs tested. Less than 10% of the starting substrate was converted to secondary cleavage product after 16 min for P236L RT compared to 85% for wild-type RT. The RNase H defect of P236L RT was confirmed with a second protein preparation (data not shown). A similar result was seen for 5′-end-directed RNase H activity (Fig. 4A, B, and C). Notably, whereas the P236L RT is profoundly defective in 3′-end-directed primary and secondary cleavages and in 5′-end-directed secondary cleavage, it exhibits only a moderate defect in 5′-end-directed primary cleavage. The relative RNase H activity hierarchy for purified protein matched the relative fitness hierarchy for the viruses: WT ≥ Y181C ≥ K103N > G190A ≥ V106A ≥ G190S > P236L.
FIG. 3.
Polymerase-dependent RNase H activity of purified wild-type and mutant RTs. (A) Representative polyacrylamide gel of DNA 3′-end-directed RNase H activity. The substrate used was made by annealing a 26-nt-long DNA oligonucleotide primer to a 5′-end-labeled 41-nt-long RNA such that the 3′ end of the DNA primer was recessed relative to the RNA 5′ end. Reactions were allowed to proceed for the indicated lengths of time: lanes 1, 0 s; lanes 2, 15 s; lanes 3, 30 s; lanes 4, 1 min; lanes 5, 2 min; lanes 6, 4 min; lanes 7, 8 min; lanes 8, 16 min. Nucleotide size markers are indicated on the left. (B) Rate of secondary product formed. (C) Rate of total product formed. Secondary cleavage products were quantified by phosphorimaging and plotted on the y axis versus time on the x axis. Duplicate RNase H reactions were performed in duplicate using a single virus stock for each mutant. Results represent the means and standard deviations of 4 values. Mutants that had a mean rate of product formation at 16 min that was statistically different from that of the K103N mutant are indicated with asterisks (*, P < 0.01; **, P < 0.005; ***, P < 0.0001).
FIG. 4.
Polymerase-independent RNase H activity of purified wild-type and mutant RTs. (A) Representative polyacrylamide gel of RNA 5′-end-directed RNase H activity. The substrate used was made by annealing a 5′-end-labeled 41-nt RNA to a 77-nt DNA primer such that the 5′ end of the RNA was recessed relative to the DNA. The reactions were allowed to proceed for the indicated lengths of time: lanes 1, 0 s; lanes 2, 15 s; lanes 3, 30 s; lanes 4, 1 min; lanes 5, 2 min; lanes 6, 4 min; lanes 7, 8 min; lanes 8, 16 min. Nucleotide size markers are indicated on the left. (B) Rate of secondary product formed. (C) Rate of total product formed. Secondary cleavage products were quantified by phosphorimaging and plotted on the y axis versus time on the x axis. Duplicate RNase H reactions were performed in duplicate using a single virus stock for each mutant. Results represent the means and standard deviations of 4 values. Mutants that had a mean rate of product formation at 16 min that was statistically different from that of the K103N mutant are indicated with asterisks (*, P < 0.01; **, P < 0.005; ***, P < 0.0001).
RT protein content of virions correlates with virion-associated polymerization.
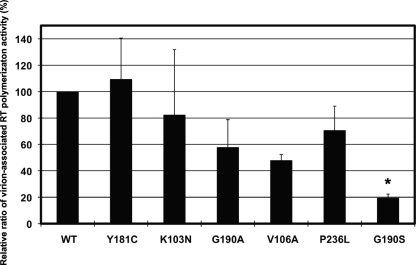
Previous studies have shown that the level of RT in the virion correlates with replication fitness (15, 25, 27). Clearly, the concentration of RT could also influence fitness. In order to determine whether the RNase H activity defect of NNRTI-resistant mutants is primarily responsible for the fitness defect in cell culture, we measured the virion-associated specific polymerase activity and RT content in virions. Unlike the polymerization activity of purified, recombinant RTs, the relative polymerization activity in virion lysates was reduced for viruses with RTs bearing the V106A and G190S mutations compared to that of the wild type (P < 0.01) (Fig. 5). Even though V106A, P236L, and G190S mutants had significantly reduced fitness compared to that of the K103N mutant, the G190S mutant was the only one that had significantly reduced polymerization activity (P < 0.01). The relative polymerization activity of the P236L mutant was similar to those of the wild type and the K103N mutant. Since the relative virion polymerization activity of the P236L mutant was much higher than expected from the poor fitness of the P236L virus, based on the good correlation of polymerization activities and fitness for the other NNRTI-resistant viruses, we hypothesized that the RT content of the P236L virus is higher than that of the other mutant viruses.
FIG. 5.
Virion-associated RT polymerization activity of wild-type and mutant virus. Virus stocks were prepared by transfecting 293 cells with either wild-type or mutant plasmid DNA. Virus pellets were prepared with 100 ng of capsid protein. The bar graph shows the virion-associated RT activity of lysed virus pellet extracts with a poly(rA)/oligo(dT) template/primer and an [α-32P]dTTP substrate. Three replicate polymerization reactions were performed with a single virus stock. The y axis shows the relative ratio of RT polymerization activity for each mutant compared to that of the wild type as a percentage. The wild-type value was set at 100%. The mutant that had a mean difference in the relative ratio compared with that of the K103N mutant is indicated with an asterisk (*, P < 0.01).
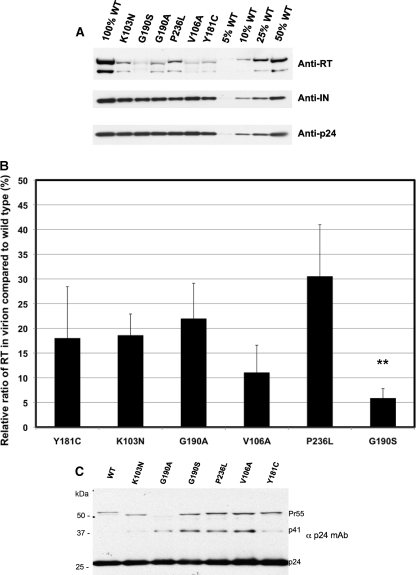
The protein contents of wild-type, K103N, Y181C, P236L, G190A/S, and V106A virus particles were assessed by Western blotting using the virus supernatant solutions prepared from transfected 293 cells. Different amounts based on p24 antigen of wild-type virus were used to create a linear standard curve (R2 = 0.98). Mutant viral stocks were normalized by p24 antigen and were analyzed using antibodies specific to RT, integrase, or capsid protein (p24) (Fig. 6A). All mutants had reduced RT contents (Fig. 6B), and content correlated with relative fitness (compare Fig. 1 with Fig. 6B), except for the P236L virus, which had more RT than did other mutants with similar fitness such as V106A and G190S viruses.
FIG. 6.
Western blot showing the relative amounts of RT in wild-type and mutant virions. (A) Representative Western blot probed with antibodies that recognize HIV-1 RT p66 and p51 subunits, integrase, or gag capsid p24 protein. Virus stocks were prepared by transfecting 293 cells with either wild-type or mutant plasmid DNA. Mutant virus pellets were prepared with an equal amount (200 ng) of capsid protein. Different amounts of p24 capsid protein were pelleted for the wild type: 10 ng (5%), 20 ng (10%), 50 ng (25%), and 100 ng (50%). These amounts were used to calculate a standard curve. (B) Relative amount of RT for each mutant compared to that of the wild type in percent on the y axis. The bars represent the means and standard deviations of triplicate Western blots. The mutant that had a mean amount of RT that was statistically different from that of the K103N mutant is indicated with asterisks (**, P < 0.005). (C) Relative amounts of gag processing intermediates. Equal amounts of p24 capsid protein were loaded for each virus (800 ng). The molecular masses in kilodaltons of the markers are shown on the left side.
Since both integrase and RT are cleaved from the Pr160Gag-Pol polyprotein, we measured the virion-associated integrase content. There were no significant differences in the amount of integrase for any of the mutants tested (Fig. 6A), indicating that the amount of Pr160Gag-Pol polyprotein incorporated into the virions was the same. We also looked at the amount of GagPr55 processing using an antibody specific to capsid, p24 (Fig. 6C). The ratios of GagPr55 to GagPr41 to p24 for K103N and Y181C mutants were similar to that of the wild type, suggesting that Gag cleavage was normal. The ratio of GagPr55 to p24 was more than four times higher for G190S, P236L, and V106A mutants than for the wild type, and the ratio of GagPr41 to p24 was 10 to 20 times higher for G190A, G190S, P236L, and V106A mutants, suggesting that these mutations affect the Gag processing after viral particle release. These data suggest that reduced levels of polyprotein processing result in reduced levels of p66 and p51 subunits of RT for mutants with reduced RT contents.
Virion-associated RNase H activity of NNRTI resistance mutations correlates with fitness.
We also measured the 5′-end-directed RNase H activities of wild-type and mutant viral lysates (Fig. 7A and B). K103N and Y181C mutants had levels of 5′-end-directed RNase H activity similar to that of the wild type. The RNase H activities of G190A, V106A, P236L, and G190S mutants were all reduced relative to the wild type with the G190S mutant having the lowest level of activity. The 5′-end-directed RNase H activity hierarchy was WT ≥ Y181C ≥ K103N > G190A > V106A > P236L > G190S, which is very similar to the replication fitness hierarchy (Fig. 1). These observations show that virion-associated RNase H activity of NNRTI-resistant mutants correlates better with fitness than do polymerization activity and RT content.
FIG. 7.
Relative virion-associated RNase H activities of wild-type and mutant viruses. Virus stocks were prepared by transfecting 293 cells with either wild-type or mutant plasmid DNA. Virus pellets were prepared with 100 ng of capsid protein. (A) Representative polyacrylamide gel of RNA 5′-end-directed RNase H activity using the lysed pellet. The substrate used was made by annealing a 5′-end-labeled 41-nt RNA to a 77-nt DNA primer such that the 5′ end of the RNA was recessed relative to the DNA. The reactions were allowed to proceed for the indicated lengths of time: lanes 1, 0 s; lanes 2, 1 min; lanes 3, 10 min; lanes 4, 16 min. Nucleotide size markers are indicated on the left. (B) Rate of secondary product formed. Secondary cleavage products were quantified by phosphorimaging and plotted on the y axis versus time on the x axis. Duplicate RNase H reactions were performed in duplicate using a single virus stock for each mutant. Results represent the means and standard deviations of 4 values. Mutants that had a mean rate of product formation at 16 min that was statistically different from that of the K103N mutant are indicated with asterisks (**, P < 0.005; ***, P < 0.0001, t test).
DISCUSSION
We measured the relative fitness of NNRTI-resistant viruses compared to the wild type using a flow cytometry-based cell culture assay (17) and then looked for biochemical correlates of reduced fitness for the V106A, G190A, G190S, and P236L mutant RT strains. Previous studies from our research group using a sequenced-based fitness assay are consistent with the fitness hierarchy of the panel of 6 mutants that we tested in this study: WT ≥ Y181C ≥ K103N > G190A > V106A > P236L ≥ G190S (4, 19, 38).
Previous biochemical studies of recombinant RTs revealed that RNase H activity correlates with fitness and that tested NNRTI-resistant RTs have normal polymerization activity (4, 13, 14, 19, 38). However, these studies were performed with recombinant RTs in which the p66 and p51 subunits were both His tagged and mixed after separate column chromatography purification. This method produced less active protein and had a smaller amount of p66/p51 heterodimers than that for the method that we used to purify the RT for the current studies. Here, recombinant heterodimer RT purified by mixing the His-tagged p51 and untagged p66 bacterial lysates before column chromatography resulted in four to eight times more active enzyme at the same protein concentration. Since fractions with equal molar amounts of p66 and p51 were chosen for study, the amount of p66/p51 heterodimer versus p51 homodimer is almost certainly larger as well.
In this study we show that the pre-steady-state and steady-state polymerization activities of recombinant NNRTI-resistant mutant RTs are similar to what we previously published, except that there was no difference in 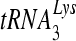 primer extension. Previous studies showed that G190A and G190S mutants had reduced rates of polymerization from a tRNA primer despite having normal rates of polymerization for a DNA PBS primer (38). In this study the rate of polymerization from the tRNA primer was similar to that of the wild type for all mutant RTs tested. These discordant results are likely explained by the purification method, since the older preparation had a much lower proportion of active p66/p51 heterodimer. Catalytically inactive RT or alternative RT structural forms may have produced the observed result. Specifically, the packaging and annealing of tRNA into virions involve a complex set of interactions between the tRNA, lysyl-tRNA synthetase, and Gag and Gag/Pol polyproteins (7, 32). It is known that
primer extension. Previous studies showed that G190A and G190S mutants had reduced rates of polymerization from a tRNA primer despite having normal rates of polymerization for a DNA PBS primer (38). In this study the rate of polymerization from the tRNA primer was similar to that of the wild type for all mutant RTs tested. These discordant results are likely explained by the purification method, since the older preparation had a much lower proportion of active p66/p51 heterodimer. Catalytically inactive RT or alternative RT structural forms may have produced the observed result. Specifically, the packaging and annealing of tRNA into virions involve a complex set of interactions between the tRNA, lysyl-tRNA synthetase, and Gag and Gag/Pol polyproteins (7, 32). It is known that 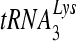 can stimulate polymerization by p66 homodimers, induce conformational changes, and increase proteolytic cleavage of the RNase H domain (3). In addition the initiation from a tRNA primer involves complex enzyme and substrate interactions that change as nucleotides are added (28-30). Therefore, heterodimers and homodimers of RT may have different kinetic properties that are mutant specific when a tRNA primer is used. Although previously published methods for purifying RT, which did not control for the formation of p66 and p51 homodimers, may have been adequate for measuring most kinetic parameters, they were apparently not adequate specifically for tRNA initiation.
can stimulate polymerization by p66 homodimers, induce conformational changes, and increase proteolytic cleavage of the RNase H domain (3). In addition the initiation from a tRNA primer involves complex enzyme and substrate interactions that change as nucleotides are added (28-30). Therefore, heterodimers and homodimers of RT may have different kinetic properties that are mutant specific when a tRNA primer is used. Although previously published methods for purifying RT, which did not control for the formation of p66 and p51 homodimers, may have been adequate for measuring most kinetic parameters, they were apparently not adequate specifically for tRNA initiation.
Consistent with our previous work, the RNase H activities of V106A, G190A, G190S, and P236L recombinant RTs were reduced relative to those of wild-type, K103N, and Y181C RTs. The relative level of RNase H activities of recombinant proteins correlated with fitness of the corresponding virus. The P236L mutant RT was consistently more defective in RNase H than indicated in previous studies as confirmed with independent protein preparations. The effect of the P236L mutation on RNase H activity is consistent with known structural information on RT. P236 is located at the base of the thumb in the p51 subunit, which supports the RNase H domain of p66 and could therefore influence domain activity (26, 33).
During reverse transcription the minus-strand strong-stop DNA (−SSDNA) that is synthesized from the tRNA primer strand transfers to the 3′ end of the RNA genome to complete minus-strand synthesis (5, 21). Studies of tRNA initiation and minus-strand transfer in vitro have shown that the −SSDNA can fold back to create a self-primed primer/template (P/T) (22). When the template that we used for the tRNA initiation studies, D199, is copied, it produces −SSDNA, which self-primes. The extension of this self-primed P/T is dependent on the removal of the original RNA template, D199, by RT RNase H activity (Fig. 2B). Consistent with previous studies, P236L, which has greatly reduced RNase H activity, did not form the extended self-primed product because it cannot degrade the RNA template.
To confirm that defects in RNase H activity of recombinant RTs are responsible for the reduced fitness of NNRTI-resistant mutant viruses, we measured the virion-associated polymerization and RNase H activities. Virion-associated polymerization activity also correlated with fitness, with the exception of the virus with P236L RT. This mutant had polymerization activity that was similar to that of the K103N mutant, a fit mutant, with a polymerization level that was 70% of the wild-type level. Since the polymerization activity of recombinant protein was normal for each mutant, we hypothesized that the absolute amount of RT in the mutant variants must be reduced in order for the virion-associated RT activity to correlate with fitness. When we quantified the RT content in the virions, this was indeed the case. However, the P236L mutant had 30% of the amount of RT in the virion whereas the G190S and V106A mutants had 6% and 11%, respectively. This explains why the polymerization activity of the P236L mutant was higher than expected based on its relative fitness.
In contrast, the virion-associated RNase H activity uniformly correlated with recombinant protein activity and with fitness. K103N and Y181C RT viruses, which replicated at a rate similar to that of the wild type in cell culture, had virion-associated RNase H activity equivalent to that of the wild type. The remaining mutant viruses had reduced RNase H activity, although the RNase H activity of the P236L mutant was slightly higher than that of the G190S mutant. This is most likely explained by the increased amount of RT in P236L viruses. However, the RNase H activities of the unfit NNRTI mutant viruses were ≤50% of that of the wild type, indicating that RNase H is greatly impaired and that this is a likely explanation for reduced fitness.
HIV-1 p66/p51 heterodimer RT formation in virions requires the activity of protease such that mature RT is formed by the proteolytic cleavage of the Gag-Pol polyprotein (Pr160Gag-Pol) during virion maturation (reviewed in reference 23). The order and timing of Gag-Pol precursor processing are well established, with cleavage shown to occur in a sequential manner. Studies of the fitness, RT content, and Gag-Pol processing of G190 mutant viruses showed that reduced RT content apparently resulted from reduced Gag and Gag-Pol processing due to reduced Gag-Pol incorporation into virions (25). Our data clearly show that diminished RT content does not completely correlate with Gag processing of NNRTI-resistant mutants. P236L virus had RT levels similar to those of Y181C and K103N viruses despite having defective Gag processing. In addition the mutants in our study had normal amounts of integrase, indicating normal Gag-Pol incorporation into virions. Studies of NNRTI mutations showed that some mutants such as the K103N, Y181C, and G190A mutants increased heterodimer stability (18). This suggests that the P236L mutation promotes RT stability, resulting in an overall increase in RT content despite reduced proteolytic cleavage. However, it is not clear whether increased stability of heterodimers results in proteolytic stability of RT, since mutations in the protease cleavage site which prevent cleavage of p66 into p51 result in reduced amounts of RT in the virion (1). Our results suggest that NNRTI resistance mutants do not affect Gag-Pol incorporation and that all NNRTI resistance mutants affect proteolytic stability to different extents, resulting in different amounts of RT in the virion.
What have we learned here about the mechanisms that reduce fitness of NNRTI-resistant viruses? The effects of the P236L mutation and those of reduced RT content have an obvious commonality. The P236L mutation severely impairs 3′-end-dependent RNase H primary and secondary cleavages. With the P236L mutation in RT, as minus-strand DNA is made, almost none of the template cleavages that that would normally accompany synthesis can actually occur. Since cleavages during synthesis produce the RNA oligomer substrates for later 5′-end-directed cleavage, the defects in polymerization-associated cleavage alone would be sufficient to greatly reduce RNA template degradation. Additionally, although P236L RT exhibits significant 5′-end-directed primary cleavage activity, it is defective for 5′-end secondary cleavage activity. The latter defect would additionally reduce whatever 5′-end cleavage activity can occur at the infrequent cleavage sites generated during synthesis.
The corresponding effect of RT content reduction is that overall RNase H activity is also impaired. Details of the mechanism, however, are different from the effects of mutations that impair RNase H in each RT. There are normally 50 to 70 RT molecules per virion (for a review, see reference 9). Since only one RT is needed for primer extension, the likely reason for the excess is that the additional RTs are employed to carry out 5′-end-directed RNase H activity. Cuts made during synthesis should produce hundreds of RNA oligomers that likely require many RTs for efficient degradation. Depletion of RT is unlikely to affect polymerization or polymerization-dependent RNase H. However, it should reduce the rate at which the resultant RNA oligomers are degraded. While this is clearly different mechanistically from the effects of the RNase H defect in P236L RT, the impact on reverse transcription should be similar. That is, the long-lived RNA oligomers would hinder strand transfers and second-strand RNA synthesis, slowing viral replication.
The effect of the reduction of DNA polymerase active sites caused by RT depletion is difficult to predict and cannot be approached experimentally by the techniques used here, since polymerization and RNase H are reduced concomitantly. One view is that since only one RT is needed for polymerization, the effects should be minimal. However, reduction of polymerization activity in virions has previously been reported to decrease substantially the rate of viral replication (27).
Another possibility is that maintaining the natural activity ratio between the polymerization and RNase H, as occurs when RT content is reduced, is less disruptive of reverse transcription than is the great imbalance caused by the P236L mutation. This again would be hard to verify experimentally.
Finally, the differences in characteristics of the 3′-end-directed RNase H and the 5′-end-directed RNase H activities of the P236L mutant RT are noteworthy. While the positioning of the RT for these two RNase H functions is determined by two different structures, the orientations of the RT with respect to the RNA and DNA strands are much the same (10). Nevertheless, the 3′-end-directed primary cleavages are strongly suppressed by the mutation while the 5′-end-directed cleavages are not. This suggests that a difference in protein-nucleic acid contacts between one binding mode and the other is capable of activating the RNase H. Understanding the structural basis of this RNase H regulation could have therapeutic potential.
In conclusion, our current results indicate that the reduction of fitness in viruses with NNRTI resistance mutations is based on either of two possible mechanisms. We had previously reported that the mutant RTs shared the property that they are deficient in RNase H activity. We showed here that the P236L HIV-1 has a normal content of RT but that the RT is severely deficient in RNase H function. We propose that the observed fitness defect derives from the inability of the P236L RT to effectively degrade the viral RNA to allow strand transfer and second-strand DNA synthesis. While the defect in cleavage most severely affects polymerization-dependent RNase H activity, this activity creates the substrate for polymerization-independent activity, so that overall RNase H activity is impaired. Viruses with the G190S, G190A, and V106A RTs have substantially reduced RT content. This situation also reduces the amount of RT available for 5′-end-directed RNase H activity and would also impair proper degradation of the viral RNA. The problem is further exacerbated by moderate intrinsic defects in the RNase H of the mutant RTs. While the correlation between poor fitness and RNase H deficiency is unmistakable, the effect of lowered polymerization activity in those mutants with low RT content still needs to be evaluated.
Acknowledgments
We acknowledge the technical support of Dongge Li, Kora Fox, and Xiufang Liu.
This work was supported by NIH grant R01 AI-41387 and the University of Rochester Developmental Center for AIDS Research (NIH P30AI078498).
Footnotes
Published ahead of print on 30 June 2010.
REFERENCES
- 1.Abram, M. E., and M. A. Parniak. 2005. Virion instability of human immunodeficiency virus type 1 reverse transcriptase (RT) mutated in the protease cleavage site between RT p51 and the RT RNase H domain. J. Virol. 79:11952-11961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amacker, M., and U. Hubscher. 1998. Chimeric HIV-1 and feline immunodeficiency virus reverse transcriptases: critical role of the p51 subunit in the structural integrity of heterodimeric lentiviral DNA polymerases. J. Mol. Biol. 278:757-765. [DOI] [PubMed] [Google Scholar]
- 3.Andreola, M. L., G. A. Nevinsky, P. J. Barr, L. Sarih-Cottin, B. Bordier, M. Fournier, S. Litvak, and L. Tarrago-Litvak. 1992. Interaction of tRNALys with the p66/p66 form of HIV-1 reverse transcriptase stimulates DNA polymerase and ribonuclease H activities. J. Biol. Chem. 267:19356-19362. [PubMed] [Google Scholar]
- 4.Archer, R. H., C. Dykes, P. Gerondelis, A. Lloyd, P. Fay, R. C. Reichman, R. A. Bambara, and L. M. Demeter. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arts, E. J., and M. A. Wainberg. 1996. Human immunodeficiency virus type 1 reverse transcriptase and early events in reverse transcription. Adv. Virus Res. 46:97-163. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. U. S. A. 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cen, S., M. Niu, and L. Kleiman. 2004. The connection domain in reverse transcriptase facilitates the in vivo annealing of tRNALys3 to HIV-1 genomic RNA. Retrovirology 1:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin, J. M., and S. H. Hughes (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 10.DeStefano, J. J. 1995. The orientation of binding of human immunodeficiency virus reverse transcriptase on nucleic acid hybrids. Nucleic Acids Res. 23:3901-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeStefano, J. J., R. G. Buiser, L. M. Mallaber, T. W. Myers, R. A. Bambara, and P. J. Fay. 1991. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J. Biol. Chem. 266:7423-7431. [PubMed] [Google Scholar]
- 12.Ding, J., K. Das, Y. Hsiou, S. G. Sarafianos, A. D. Clark, Jr., A. Jacobo-Molina, C. Tantillo, S. H. Hughes, and E. Arnold. 1998. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution. J. Mol. Biol. 284:1095-1111. [DOI] [PubMed] [Google Scholar]
- 13.Domaoal, R. A., R. A. Bambara, and L. M. Demeter. 2006. HIV-1 reverse transcriptase mutants resistant to nonnucleoside reverse transcriptase inhibitors do not adversely affect DNA synthesis: pre-steady-state and steady-state kinetic studies. J. Acquir. Immune Defic. Syndr. 42:405-411. [DOI] [PubMed] [Google Scholar]
- 14.Domaoal, R. A., and L. M. Demeter. 2004. Structural and biochemical effects of human immunodeficiency virus mutants resistant to non-nucleoside reverse transcriptase inhibitors. Int. J. Biochem. Cell Biol. 36:1735-1751. [DOI] [PubMed] [Google Scholar]
- 15.Dunn, L. L., M. J. McWilliams, K. Das, E. Arnold, and S. H. Hughes. 2009. Mutations in the thumb allow HIV-1 RT to be cleaved by protease in virions. J. Virol. 83:12336-12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dykes, C., K. Fox, A. Lloyd, M. Chiulli, E. Morse, and L. M. Demeter. 2001. Impact of clinical reverse transcriptase sequences on the replication capacity of HIV-1 drug-resistant mutants. Virology 285:193-203. [DOI] [PubMed] [Google Scholar]
- 17.Dykes, C., J. Wang, X. Jin, V. Planelles, D. S. An, A. Tallo, Y. Huang, H. Wu, and L. M. Demeter. 2006. Evaluation of a multiple-cycle, recombinant virus, growth competition assay that uses flow cytometry to measure replication efficiency of human immunodeficiency virus type 1 in cell culture. J. Clin. Microbiol. 44:1930-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figueiredo, A., S. Zelina, N. Sluis-Cremer, and G. Tachedjian. 2008. Impact of residues in the nonnucleoside reverse transcriptase inhibitor binding pocket on HIV-1 reverse transcriptase heterodimer stability. Curr. HIV Res. 6:130-137. [DOI] [PubMed] [Google Scholar]
- 19.Gerondelis, P., R. H. Archer, C. Palaniappan, R. C. Reichman, P. J. Fay, R. A. Bambara, and L. M. Demeter. 1999. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end- and DNA 3′-end-directed RNase H activities. J. Virol. 73:5803-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff, S. P. 1990. Retroviral reverse transcriptase: synthesis, structure, and function. J. Acquir. Immune Defic. Syndr. 3:817-831. [PubMed] [Google Scholar]
- 21.Gotte, M., X. Li, and M. A. Wainberg. 1999. HIV-1 reverse transcription: a brief overview focused on structure-function relationships among molecules involved in initiation of the reaction. Arch. Biochem. Biophys. 365:199-210. [DOI] [PubMed] [Google Scholar]
- 22.Guo, J. H., L. E. Henderson, J. Bess, B. Kane, and J. G. Levin. 1997. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 71:5178-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, M., G. Tachedjian, and J. Mak. 2005. The packaging and maturation of the HIV-1 Pol proteins. Curr. HIV Res. 3:73-85. [DOI] [PubMed] [Google Scholar]
- 24.Hou, E. W., R. Prasad, W. A. Beard, and S. H. Wilson. 2004. High-level expression and purification of untagged and histidine-tagged HIV-1 reverse transcriptase. Protein Expr. Purif. 34:75-86. [DOI] [PubMed] [Google Scholar]
- 25.Huang, W., A. Gamarnik, K. Limoli, C. J. Petropoulos, and J. M. Whitcomb. 2003. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J. Virol. 77:1512-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobo-Molina, A., A. D. Clark, Jr., R. L. Williams, R. G. Nanni, P. Clark, A. L. Ferris, S. H. Hughes, and E. Arnold. 1991. Crystals of a ternary complex of human immunodeficiency virus type 1 reverse transcriptase with a monoclonal antibody Fab fragment and double-stranded DNA diffract x-rays to 3.5-A resolution. Proc. Natl. Acad. Sci. U. S. A. 88:10895-10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanchy, J. M., C. Ehresmann, S. F. Le Grice, B. Ehresmann, and R. Marquet. 1996. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J. 15:7178-7187. [PMC free article] [PubMed] [Google Scholar]
- 29.Lanchy, J. M., C. Isel, C. Ehresmann, R. Marquet, and B. Ehresmann. 1996. Structural and functional evidence that initiation and elongation of HIV-1 reverse transcription are distinct processes. Biochimie 78:1087-1096. [DOI] [PubMed] [Google Scholar]
- 30.Lanchy, J. M., C. Isel, G. Keith, S. F. Le Grice, C. Ehresmann, B. Ehresmann, and R. Marquet. 2000. Dynamics of the HIV-1 reverse transcription complex during initiation of DNA synthesis. J. Biol. Chem. 275:12306-12312. [DOI] [PubMed] [Google Scholar]
- 31.Lusso, P., F. Cocchi, C. Balotta, P. D. Markham, A. Louie, P. Farci, R. Pal, R. C. Gallo, and M. S. Reitz, Jr. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saadatmand, J., F. Guo, S. Cen, M. Niu, and L. Kleiman. 2008. Interactions of reverse transcriptase sequences in Pol with Gag and LysRS in the HIV-1 tRNALys3 packaging/annealing complex. Virology 380:109-117. [DOI] [PubMed] [Google Scholar]
- 33.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sluis-Cremer, N., and G. Tachedjian. 2008. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Res. 134:147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szilvay, A. M., S. Nornes, I. R. Haugan, L. Olsen, V. R. Prasad, C. Endresen, S. P. Goff, and D. E. Helland. 1992. Epitope mapping of HIV-1 reverse transcriptase with monoclonal antibodies that inhibit polymerase and RNase H activities. J. Acquir. Immune Defic. Syndr. 5:647-657. [PubMed] [Google Scholar]
- 36.Telesnitsky, A., and S. P. Goff. 1993. Two defective forms of reverse transcriptase can complement to restore retroviral infectivity. EMBO J. 12:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toohey, K., K. Wehrly, J. Nishio, S. Perryman, and B. Chesebro. 1995. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology 213:70-79. [DOI] [PubMed] [Google Scholar]
- 38.Wang, J., C. Dykes, R. A. Domaoal, C. E. Koval, R. A. Bambara, and L. M. Demeter. 2006. The HIV-1 reverse transcriptase mutants G190S and G190A, which confer resistance to non-nucleoside reverse transcriptase inhibitors, demonstrate reductions in RNase H activity and DNA synthesis from tRNA(Lys, 3) that correlate with reductions in replication efficiency. Virology 348:462-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wapling, J., K. L. Moore, S. Sonza, J. Mak, and G. Tachedjian. 2005. Mutations that abrogate human immunodeficiency virus type 1 reverse transcriptase dimerization affect maturation of the reverse transcriptase heterodimer. J. Virol. 79:10247-10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitcomb, J. M., and S. H. Hughes. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8:275-306. [DOI] [PubMed] [Google Scholar]
- 41.Wisniewski, M., M. Balakrishnan, C. Palaniappan, P. J. Fay, and R. A. Bambara. 2000. The sequential mechanism of HIV reverse transcriptase RNase H. J. Biol. Chem. 275:37664-37671. [DOI] [PubMed] [Google Scholar]
- 42.Wu, H., Y. Huang, C. Dykes, D. Liu, J. Ma, A. S. Perelson, and L. M. Demeter. 2006. Modeling and estimation of replication fitness of human immunodeficiency virus type 1 in vitro experiments by using a growth competition assay. J. Virol. 80:2380-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, T., J. Guo, J. Bess, L. E. Henderson, and J. G. Levin. 1999. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J. Virol. 73:4794-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]