Abstract
Multiple APOBEC3 proteins are expressed in HIV-1 target cells, but their individual contributions to viral suppression when expressed at endogenous levels remain largely unknown. We used an HIV NL4-3 mutant that selectively counteracts APOBEC3G (A3G) but not APOBEC3F (A3F) to dissect the relative contribution of A3F to the inhibition of HIV-1 replication in primary human lymphocytes (peripheral blood mononuclear cells [PBMCs]). This HIV Vif mutant replicated similarly to wild-type virus in PBMCs, suggesting that the effect of A3F on HIV restriction in these cells is limited. The different A3F variants found in PMBC donors displayed either comparable activity or less activity than wild-type A3F. Lastly, the endogenous A3F mRNA and protein expression levels in PBMCs were considerably lower than those of A3G. Our results suggest that A3F neutralization is dispensable for HIV-1 replication in primary human T-lymphocytes.
APOBEC3F (A3F) is a member of the APOBEC3 cytidine deaminase family which, if left unchecked, are potent restriction factors of retroviral replication. In the absence of a functional HIV-1 Vif protein, A3 molecules are copackaged into egressing virions and exert their mutagenic activity by modifying cytidines to uracils in the viral minus strand during reverse transcription in the next round of infection (4, 10, 27). HIV-1 circumvents the restriction posed by APOBEC3s by mediating their proteasome-dependent degradation through HIV-1 Vif (4, 11, 20). Circulating viral populations harbor Vif molecules that differ widely in their capacity to limit APOBEC3 activity (1, 19). In addition, proviruses with G-to-A mutations in either GG or GA dinucleotide contexts have been detected in chronically infected patients and long-term nonprogressors (7, 8, 15, 24). On a population level, the high degree of functional variation of Vif may be exploited by HIV-1, possibly allowing the appearance of beneficial mutations that increase fitness and/or drug resistance (13, 18). The Vif protein contains several functional motifs, some of which were shown to be responsible for selective activity against A3G (40YRHHY44), A3F (14DRMR17), or both (55VXIPLX4-5LXΦX2YWXL72 and 144SLQ146) (4, 6, 12, 25, 26) (Fig. 1 A). We took advantage of the differential APOBEC3-specific neutralization activity associated with Vif mutations (19, 22) to probe for the relative contribution of A3F to HIV-1 restriction in human primary lymphocytes. Indeed, one of the naturally circulating Vif mutants previously described contains a substitution (W11R) (Fig. 1A) adjacent to the 14DRMR17 A3F binding motif which prevents it from rescuing viral infectivity of a ΔVif virus in the presence of A3F (19). When NL4-3 encoding an arginine at position 11 of Vif (HIV 11R) was produced in HEK 293T cells in the presence of A3F, it failed to mediate A3F degradation and, consequently, showed a lack of infectivity comparable to that of negative-control viruses with a defective BC box (SLQ144-146AAA) (HIV ΔSLQ) (Fig. 1B). This defect selectively influenced the activity against A3F, since the infectivity of HIV 11R produced in the presence of A3G was comparable to that of HIV WT virus (Fig. 1B). None of the viruses counteracted the inhibitory activity of A3B, confirming its previously reported resistance to HIV-1 Vif (3). We also showed that the efficiency with which the different Vifs rescue viral infectivity in the presence of A3 correlates well with their ability to degrade these molecules (Fig. 1C). Taken together, the results of these overexpression experiments demonstrate that HIV-1 Vif mutant 11R behaves like wild-type (WT) HIV in the presence of A3G and A3B but that its infectivity in the presence of A3F resembles that of the Vif-defective mutant HIV ΔSLQ (Fig. 1B and 2 A).
FIG. 1.
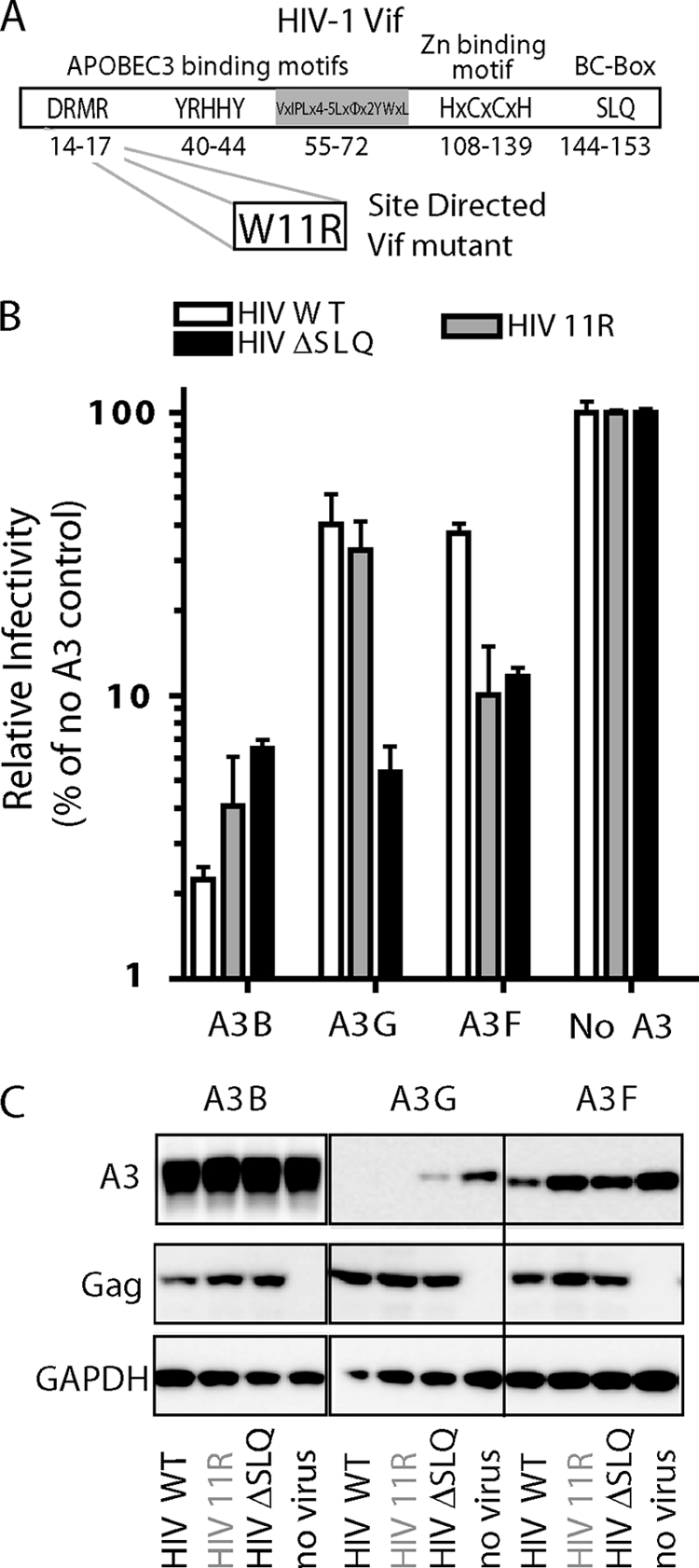
(A) Schematic representation of HIV-1 Vif domain organization, including the domains that provide selective A3G neutralization activity. (B) Infectivity of WT, W11R mutant, and SLQ144-146AAA Vif full-length NL4-3 viruses (HIV WT, HIV 11R, and HIV ΔSLQ, respectively) produced in the presence or absence of A3F, A3G, and A3B, measured as β-galactosidase expression levels in TZM-bl cells. Data are shown relative to the infectivity of viruses produced without any APOBEC3. Error bars show standard deviations. (C) Levels of degradation of A3F, A3G, and A3B achieved by the different Vif variants in the context of the full-length virus. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
FIG. 2.
(A) Schematic representation of the anti-APOBEC3 activities of WT HIV, HIV 11R, and HIV ΔSLQ. The symbol ⊥ indicates inhibition. (B and C) Replication kinetics of the full-length HIV-1 NL4-3 W11R variant (HIV 11R) compared to those of WT virus and negative control HIV ΔSLQ in MT2 and MT4 T-cell lines (B) and PBMCs (C). Infections were carried out using 20 ng p24 equivalents for each virus in PHA-stimulated PBMCs from 5 anonymous donors. CA-p24 production was measured every 2 to 4 days for 12 to 15 days. The results shown are representative of two independent replication experiments.
We next tested the replication capacity of the HIV 11R virus under more physiologically relevant conditions, by infecting phytohemagglutinin (PHA)-stimulated peripheral blood mononuclear cells (PBMCs) from five anonymous HIV-1-negative blood donors. Contrary to our expectations based on the results of the experiments shown in Fig. 1B and C, in which A3 molecules were overexpressed, the replication capacity of the HIV 11R virus was comparable to that of WT HIV in cells derived from all donors (Fig. 2C). The replication of HIV ΔSLQ, however, was 5 to 20 times lower than that of WT HIV, suggesting that the endogenous expression levels of Vif-sensitive APOBEC3 proteins such as A3G exert a substantial inhibition of viruses lacking a functional BC box in Vif. Interestingly, PBMCs from donor 5 and, to a smaller extent, from donor 4 allow some replication of HIV ΔSLQ, which may indicate donor variation in APOBEC3 expression (A3G) (see Fig. 4). Of note, the low level of p24 production observed for the Vif-defective viruses likely represents viral output from the first round of infection. Interestingly, when T-cell lines such as MT2 (“nonpermissive,” i.e., high levels of A3G and A3F expression) (Fig. 2A and B; also see Fig. 4) and MT4 (“permissive,” i.e., low levels of A3G and A3F expression) (Fig. 2A and B; also see Fig. 4) were used in replication experiments with the same viruses, we observed that HIV 11R replicated in the permissive MT4 but not in the nonpermissive MT2 cell line. Thus, viral replication of HIV 11R in the nonpermissive T cell-line MT2 recapitulated well the phenotypes observed in overexpression experiments (Fig. 1B), while its replication in PBMCs was more comparable to that observed in the permissive MT4 cell line (compare Fig. 2B and C).
FIG. 4.

Expression of APOBEC3G and APOBEC3F. (A to C) Endogenous transcript expression levels of A3B, A3G, and A3F in PHA-IL-2-stimulated PBMCs, as well as in MT2 and MT4 T-cell lines. These cells were used for the HIV-1 replication experiments described in the legend to Fig. 2B and C. mRNA quantification was performed using TaqMan gene expression assays (Applied Biosystems) specific for human A3B (Hs00358981_m1), A3F (Hs00736570_m1), and A3G (Hs00222415_m1). Data are expressed as the number of molecules per 10 ng of reverse-transcribed RNA, with error bars representing the standard deviations from the means. The results shown represent two independent experiments. (D) Endogenous expression levels of A3G and A3F proteins in the same PBMCs and T-cell lines used for the experiments whose results are shown in panel A.
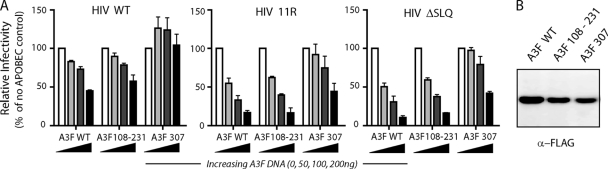
This discrepancy between the restriction of HIV 11R in MT2 cells and its robust replication in PBMCs (Fig. 2C) could be due to the presence of less active A3F variants caused by single-nucleotide polymorphisms (SNPs), reminiscent of A3H haplotypes (5, 14, 21), or to insufficient A3F expression in PBMCs. In order to test these hypotheses, we cloned A3F from our PBMC donors into the PCR cloning vector pSC-A (StrataClone; Stratagene). Sequencing of the A3F clones revealed three different A3F haplotypes. Donors 1, 2, and 3 encoded A3F variants with two SNPs leading to A108S and V231I substitutions (A3F 108-231). The A3F of donor 5 was instead characterized by a Y307C substitution (A3F 307). Donor 4 harbored only wild-type A3F transcripts (A3F WT [A108, V231, and Y307]). An in-depth database analysis of A3F SNPs in human populations revealed that A3F variants A108S and V231I are frequent: A108S reaches over 40% allele frequency (highest in European [54%] and lowest in West African populations [29%]), and V231I reaches 64% (highest in East Asian populations [74%] and lowest in West African populations [15%]). In contrast, SNP Y307C in A3F is rare, with an allele frequency of 3% (highest in West Africans [6%] and none in the other populations tested) (sources, NCBI SNP database and Ensembl). These donor-derived A3F cDNAs were cloned as N-terminal-FLAG fusions in an expression vector (pTR600) (5) and tested for restriction activity against the wild type and HIV-1 Vif mutants. The inhibitory efficacy of the A3F 108-231 variant was comparable to that of A3F WT when targeting WT HIV (A3F 108-231 had 45% infectivity, and A3F WT had 57% infectivity), HIV 11R (both had 17% infectivity), and HIV ΔSLQ (A3F 108-231 had 11% infectivity, and A3F WT had 16%) (all values reflect 200 ng of the A3F plasmid) (Fig. 3 A). Interestingly, A3F 307 was significantly less active than A3F WT or A3F 108-231 (no reduction of infectivity for WT HIV, 44% infectivity for HIV 11R, and 42% infectivity for HIV ΔSLQ) (Fig. 3A), despite having an expression level comparable to that of A3F 108-231 upon transfection into HEK 293T cells (Fig. 3B). These results together show that A3F is polymorphic in sequence, with at least three different haplotypes, one of which (Y307C) is impaired in restricting WT HIV, HIV 11R, and HIV ΔSLQ viruses. The results of these overexpression experiments indicate that the presence of the natural alternative A3F allele A108S-V231I is not the underlying cause for the unrestricted replication of HIV 11R in the primary cells from donors 1, 2, and 3. Only donor 5 carries the A3F Y307 variant, and the phenotype of these cells is somewhat atypical: very little overall A3-mediated restriction is observed, resembling the replication curves observed in the permissive MT4 T-cell line (compare Fig. 2C and B). Thus, A3F SNPs are probably not the underlying reason for the widespread lack of HIV 11R restriction observed in primary PBMCs.
FIG. 3.
(A) Measurement of the restriction activities of the different A3F variants (A3F WT, 108-231, and 307) on the infectivity of HIV WT, HIV 11R, and HIV ΔSLQ full-length viruses. Viruses were produced in the absence or presence of increasing amounts (50 ng, 100 ng, and 200 ng) of the A3F plasmids, and infectivity measured as β-galactosidase expression in TZM-bl cells. Data are shown relative to the infectivity of viruses produced without any A3F. The averages ± standard deviations of the results of two independent experiments are shown. (B) Protein expression levels of the different A3F haplotypes upon transfection of 100 ng of each plasmid into HEK293T cells. A3 proteins were detected with anti-FLAG antibody.
Next, we determined A3F expression in PBMCs (donors 1 to 5) and T-cell lines (MT2 and MT4). For comparison, we also included A3G and A3B in the quantification. Total cellular RNA was extracted from primary cells, incubated for 48 h with PHA-interleukin-2 (IL-2), and from the MT2/MT4 cell lines and was reverse transcribed with an iScript cDNA synthesis kit (Bio-Rad). The abundance of A3F, A3G, and A3B transcripts was quantified by quantitative PCR (qPCR) using TaqMan assays as described by Koning et al. (9). We extrapolated the absolute number of molecules per 10 ng of total RNA from standard curves obtained by serial dilutions of plasmids encoding the relevant A3 molecule.
The A3 expression levels varied considerably among PBMCs of the five donors (Fig. 4). Taken as a whole, A3F and A3B were expressed in significantly smaller amounts than A3G (approximately 4- to 5-fold and 10- to 20-fold lower, respectively; Fig. 4A to C). Importantly, the three different A3F variants in the PBMCs were expressed at low levels that were comparable to expression levels in MT4 cells (Fig. 4C). The observation that A3F is expressed at lower levels in PBMCs is in agreement with the qPCR data recently reported by two different laboratories (9, 17). We also measured A3F and A3G protein expression in PBMCs and T-cell lines (no reliable antibody is currently available for A3B endogenous expression). Briefly, the same number of PBMCs from the 5 donors, as well as MT2 and MT4 cells, were lysed, and proteins were separated on 10% polyacrylamide gels (Invitrogen), transferred, and probed with antibodies against A3F (no. 7304; ImmunoDiagnostics) or A3G (ApoC17; NIH AIDS Research and Reference Reagent Program). While the use of two different antibodies prevented us from comparing their relative expression levels, the results in Fig. 4D show that the protein expression levels in each donor closely followed their respective mRNA levels. Interestingly, A3G protein expression in PBMCs varied considerably among donors and was significantly lower in donors 4 and 5, which may explain the low levels of replication of HIV ΔSLQ in their respective PBMCs (Fig. 2C). Of note, MT4 cells expressed considerably less A3F and A3G than MT2 cells, whereas PBMCs from the five donors expressed as little A3F as the permissive MT4 cell line. It is conceivable, therefore, that in order to restrict the replication of the HIV 11R Vif mutant, A3F expression needs to reach levels comparable to those detected in MT2 cells (Fig. 4D).
Taken together, our data suggest that A3F neutralization is dispensable for spreading HIV-1 infection in primary lymphocytes, given that the HIV-1 W11R Vif mutant, which retains activity against A3G but not A3F, replicates efficiently in human PBMCs. The induction of A3 molecules by type I interferons, as well as cell type-specific A3 expression (macrophages and dendritic cells), has been reported (9, 16, 23), and viruses egressing from such cells with high A3F expression levels could infect PBMCs and edit HIV proviruses in a GA dinucleotide context. Alternatively, other APOBEC3 molecules, such as A3B and A3H, could introduce G-to-A mutations in this GA dinucleotide context (2, 5). Our current understanding of the repertoire of APOBEC3 molecules restricting HIV-1 in its natural target cells may be incomplete, and future studies are needed to elucidate their individual roles.
Acknowledgments
We thank I. Chen for critically reading the manuscript and the members of the Simon Laboratory for insightful discussions.
This work was supported by NIH grants R01 AI064001 (V.S.) and R21 AI073213 (L.C.F.M) and the Max Kade Foundation (A.K.). V.S. is a Sinsheimer Scholar (Alexandrine and Alexander L. Sinsheimer Fund).
Footnotes
Published ahead of print on 30 June 2010.
REFERENCES
- 1.Alexander, L., M. J. Aquino-DeJesus, M. Chan, and W. A. Andiman. 2002. Inhibition of human immunodeficiency virus type 1 (HIV-1) replication by a two-amino-acid insertion in HIV-1 Vif from a nonprogressing mother and child. J. Virol. 76:10533-10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, and B. R. Cullen. 2007. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology 364:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doehle, B. P., A. Schafer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281-288. [DOI] [PubMed] [Google Scholar]
- 4.Goila-Gaur, R., and K. Strebel. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harari, A., M. Ooms, L. C. Mulder, and V. Simon. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He, Z., W. Zhang, G. Chen, R. Xu, and X. F. Yu. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381:1000-1011. [DOI] [PubMed] [Google Scholar]
- 7.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75:7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieffer, T. L., P. Kwon, R. E. Nettles, Y. Han, S. C. Ray, and R. F. Siliciano. 2005. G→A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J. Virol. 79:1975-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koning, F. A., E. N. Newman, E. Y. Kim, K. J. Kunstman, S. M. Wolinsky, and M. H. Malim. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 11.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 12.Mehle, A., H. Wilson, C. Zhang, A. J. Brazier, M. McPike, E. Pery, and D. Gabuzda. 2007. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J. Virol. 81:13235-13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulder, L. C., A. Harari, and V. Simon. 2008. Cytidine deamination induced HIV-1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 105:5501-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.OhAinle, M., J. A. Kerns, M. M. Li, H. S. Malik, and M. Emerman. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4:249-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pace, C., J. Keller, D. Nolan, I. James, S. Gaudieri, C. Moore, and S. Mallal. 2006. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J. Virol. 80:9259-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng, G., K. J. Lei, W. Jin, T. Greenwell-Wild, and S. M. Wahl. 2006. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 203:41-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Refsland, E. W., M. D. Stenglein, K. Shindo, J. S. Albin, W. L. Brown, and R. S. Harris. 22 March 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 18.Sadler, H. A., M. D. Stenglein, R. S. Harris, and L. M. Mansky. 2010. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J. Virol. 84:7396-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, J. L., W. Bu, R. C. Burdick, and V. K. Pathak. 2009. Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends Pharmacol. Sci. 30:638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan, L., P. T. Sarkis, T. Wang, C. Tian, and X. F. Yu. 2009. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 23:279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian, C., X. Yu, W. Zhang, T. Wang, R. Xu, and X. F. Yu. 2006. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 80:3112-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapp, S., N. R. Derby, R. Singer, A. Shaw, V. G. Williams, S. G. Turville, J. W. Bess, Jr., J. D. Lifson, and M. Robbiani. 2009. Double-stranded RNA analog poly(I:C) inhibits human immunodeficiency virus amplification in dendritic cells via type I interferon-mediated activation of APOBEC3G. J. Virol. 83:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei, M., H. Xing, K. Hong, H. Huang, H. Tang, G. Qin, and Y. Shao. 2004. Biased G-to-A hypermutation in HIV-1 proviral DNA from a long-term non-progressor. AIDS 18:1863-1865. [DOI] [PubMed] [Google Scholar]
- 25.Xiao, Z., E. Ehrlich, K. Luo, Y. Xiong, and X. F. Yu. 2007. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. FASEB J. 21:217-222. [DOI] [PubMed] [Google Scholar]
- 26.Xiao, Z., E. Ehrlich, Y. Yu, K. Luo, T. Wang, C. Tian, and X. F. Yu. 2006. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349:290-299. [DOI] [PubMed] [Google Scholar]
- 27.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]