Abstract
The hepatitis C virus (HCV) nonstructural protein NS5A is critical for viral genome replication and is thought to interact directly with both the RNA-dependent RNA polymerase, NS5B, and viral RNA. NS5A consists of three domains which have, as yet, undefined roles in viral replication and assembly. In order to define the regions that mediate the interaction with RNA, specifically the HCV 3′ untranslated region (UTR) positive-strand RNA, constructs of different domain combinations were cloned, bacterially expressed, and purified to homogeneity. Each of these purified proteins was probed for its ability to interact with the 3′ UTR RNA using filter binding and gel electrophoretic mobility shift assays, revealing differences in their RNA binding efficiencies and affinities. A specific interaction between domains I and II of NS5A and the 3′ UTR RNA was identified, suggesting that these are the RNA binding domains of NS5A. Domain III showed low in vitro RNA binding capacity. Filter binding and competition analyses identified differences between NS5A and NS5B in their specificities for defined regions of the 3′ UTR. The preference of NS5A, in contrast to NS5B, for the polypyrimidine tract highlights an aspect of 3′ UTR RNA recognition by NS5A which may play a role in the control or enhancement of HCV genome replication.
Hepatitis C virus (HCV) is a human pathogen which chronically infects nearly 3% of the world's population (36, 37). Persistent infection, in 80% of cases, leads to chronic hepatitis which can progress to liver cirrhosis and, in the worst cases, hepatocellular carcinoma (37). Current therapies lack specificity and efficacy due largely to an incomplete understanding of the complex molecular mechanisms of virus infectivity, RNA replication, and assembly (4, 36). HCV is a member of the Flaviviridae family of enveloped viruses (30), with a positive-sense RNA genome of ∼9.6 kb consisting of a single open reading frame (ORF) that encodes 10 structural and nonstructural viral proteins (3, 16, 25). Cap-independent translation of the ORF (29) yields a large polyprotein of approximately 3,000 amino acid residues that is cleaved co- and posttranslationally by host and viral proteases into 10 mature virus proteins; these cleavage products are ordered from the amino to the carboxy terminus as follows: core (C), envelope proteins 1 and 2 (E1 and E2), p7, nonstructural protein 2 (NS2), NS3, NS4A, NS4B, NS5A, and NS5B (3, 16, 25). At the flanking ends of the genome are two highly conserved untranslated regions (UTRs). The 5′ UTR is highly structured and consists of the internal ribosome entry site (IRES), which is important for the initiation of cap-independent translation of the polyprotein (29). The 3′ UTR consists of a short genotype-specific variable region, a tract of variable length comprising solely pyrimidine residues (predominantly U), and a conserved 98-nucleotide sequence, known as the X region, containing three stem-loops (13, 23) (Fig. 1A). The 3′ UTR is the initiation site for the synthesis of the negative-strand RNA during viral replication (13) and is involved in translational regulation.
FIG. 1.
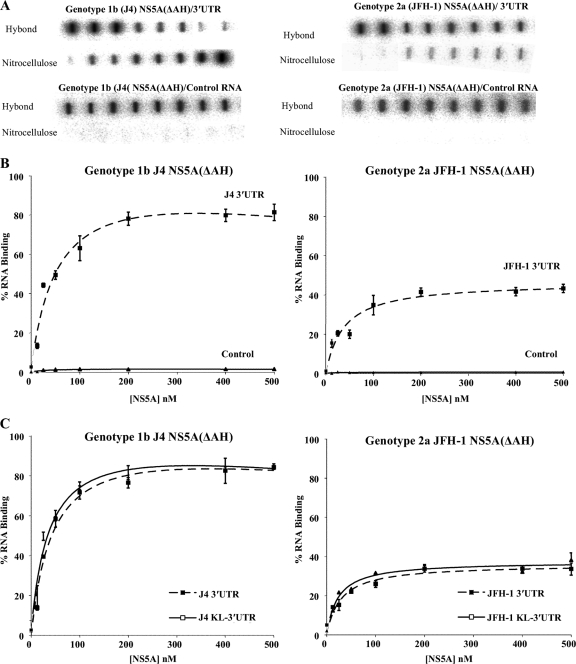
The HCV 3′ UTR RNA. (A) The positive-strand 3′ UTR consists of three distinct regions, i.e., a short genotype-specific variable region, a polypyrimidine tract [poly(U/UC)] of variable length, and a conserved 98-nucleotide sequence known as the X region containing three stable stem-loops. The predicted structure of the genotype 1b 3′ UTR is shown. (B) Left panel, the integrities of in vitro-transcribed radiolabeled full-length 3′ UTR RNAs of genotypes 1b (nucleotides 9375 to 9595) and 2a (nucleotides 9443 to 9678) and the poly(U/UC) (nucleotides 9406 to 9497) and X region (nucleotides 9498 to 9595) of genotype 1b are shown on denaturing polyacrylamide gels. Right panel, the integrities of in vitro-transcribed radiolabeled RNAs comprising the 3′-terminal NS5B-coding region plus the 3′ UTR RNAs of genotypes 1b (nucleotides 9136 to 9595) and 2a (nucleotides 9204 to 9678) (KL-3′ UTR) are shown on denaturing polyacrylamide gels.
HCV RNA replication occurs on membranous structures derived from the endoplasmic reticulum (ER) in a complex that includes host cell factors as well as viral nonstructural proteins, including NS5B, the RNA-dependent RNA polymerase (RdRp) which replicates the viral genome in vivo and in vitro (2, 25, 30). Initiation of the synthesis of the negative-strand RNA is thought to occur upon recognition and specific binding of the NS5B polymerase to the 3′ UTR of the genomic RNA (2, 16, 26). This replication activity and template specificity of NS5B in vivo are dependent, however, on the presence of the other nonstructural proteins, such as the proteases NS2 and NS3, which are required for polyprotein processing and helicase activity, and the multifunctional protein NS5A (16).
NS5A is a proline-rich phosphoprotein that is absolutely required for viral replication and is also involved in virus particle assembly (9, 10, 20, 22, 35). Its specific function in the latter process is, however, still unknown. NS5A is membrane associated due to the presence of an N-terminal amphipathic helix that serves as a membrane anchor allowing association with ER-derived membranes (Fig. 2) (24, 27). The cytoplasmic portion of NS5A is organized into three domains that are separated by low-complexity sequences (Fig. 2A) (20). The X-ray crystal structure of domain I has revealed that it is a zinc binding domain which forms a homodimer with contacts at the N-terminal ends of the molecules; the resultant large, basic groove at the dimeric interface has been proposed to be involved in RNA binding during viral replication (17, 33). NS5A has also been shown to interact with uridylate and guanylate-rich RNA and to bind to the 3′ ends of the HCV positive- and negative-strand RNAs (8). These observations suggest that NS5A may specifically interact with the large U/G stretches in the IRES of the 5′ UTR, implying a role in HCV translation and genome multiplication, while its interactions with the polypyrimidine tract of the 3′ UTR suggest that NS5A may affect the efficiency of RNA synthesis by NS5B (8, 28, 32). The reported interactions with both flanking regions of the HCV genome imply that NS5A may play a role in the switch between translation and replication that must occur during the viral life cycle (8).
FIG. 2.
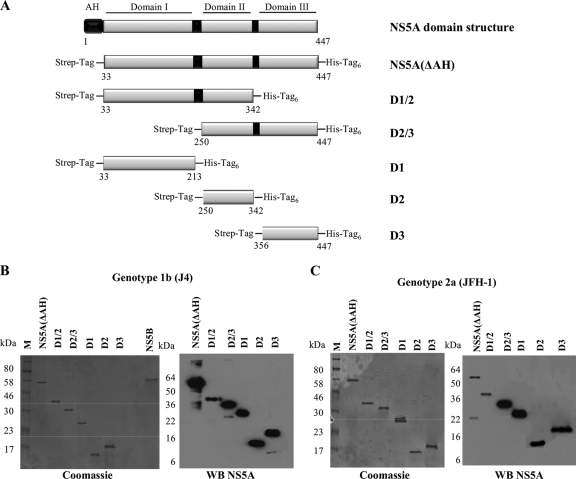
Domain structure and expression of HCV NS5A. (A) Schematic diagram of the functional domains of NS5A and design of the constructs used in the study (genotype 1b NS5A protein numbering). The N-terminal amphipathic helix of NS5A (black box) is responsible for the interaction of NS5A with membranes. NS5A is organized into three domains that are separated by low-complexity sequences, indicated by black boxes. The NS5A constructs used all lacked the N-terminal amphipathic helix and were designed to include an N-terminal Strep tag and a C-terminal hexahistidine tag. (B and C) SDS-PAGE and Western blot analysis of the NS5A(ΔAH) and NS5A domain constructs purified by nickel affinity and Streptactin tag affinity chromatography. Coomassie brilliant blue-stained gels and Western blots (WB) using anti-NS5A antibodies for NS5A proteins of genotype 1b strain J4 (B) and genotype 2a strain JFH-1 (C) are shown.
Among HCV genotypes, domains II and III are less well conserved than domain I (34). By mutational analysis, domain II, along with domain I, has been attributed to the replicase activity of NS5A (12). Contrastingly, domain III has been shown to be dispensable for RNA replication, and large heterologous insertions and deletions in this region can be tolerated, maintaining RNA replication (34). It has been shown, however, that these insertions and deletions within domain III do have an impact on virus particle assembly, highlighting the critical role of domain III NS5A in the viral life cycle (1, 10). Recent nuclear magnetic resonance (NMR) studies of domains II and III of NS5A revealed that they both adopt a natively unfolded state (6, 14, 15). The high degree of disorder and flexibility observed in these domains may contribute to the promiscuity of NS5A, which has been shown to interact with a variety of biological partners essential for NS5A function and virus persistence (11, 18, 19, 21, 31). In addition, regions within domains I and II of NS5A interact with NS5B, stimulating the in vitro activity of the polymerase and supporting the hypothesis that NS5A has a role in the modulation of RNA replication (28, 32).
In this study, we have investigated in detail the RNA binding properties of NS5A. We have mapped the RNA binding regions of NS5A using bacterially expressed deletion constructs of NS5A and have assayed their binding affinity for HCV positive-strand 3′ UTR RNA. In addition, we provide evidence that the RNA binding activity of NS5A is specific and that NS5A interacts preferentially with the polypyrimidine region of the 3′ UTR.
MATERIALS AND METHODS
DNA manipulations.
PCR was used to amplify the sequence corresponding to full-length NS5A, lacking the amphipathic helix at the N terminus and thus termed NS5A(ΔAH) here, from the HCV genotype 1b clone J4 (GenBank accession number AF054247) and the HCV genotype 2a infectious clone JFH-1 (GenBank accession number AB047639). The PCR products were digested with XmaI and NotI and introduced into the expression vector pET52b to create N-terminal Strep-tagged and C-terminal decahistidine-tagged proteins of genotype 1b NS5A(ΔAH) (amino acids [aa] 33 to 447) and genotype 2a NS5A(ΔAH) (aa 35 to 466), respectively. Appropriate primers were designed to create the following domain constructs of NS5A(ΔAH) from both genotypes (amino acid selections and nomenclature are in parentheses): genotype 1b domain I (J4 D1, aa 33 to 213), domain II (J4 D2, aa 250 to 342), domain III (J4 D3, aa 356 to 447), domains I and II (J4 D1/2, aa 33 to 342), and domains II and III (J4 D2/3, aa 250 to 447); genotype 2a domain I (JFH-1 D1, aa 35 to 215), domain II (JFH-1 D2, aa 248 to 341), domain III (JFH-1 D3, aa 353 to 466), domains I and II (JFH-1 D1/2, aa 35 to 341), and domains II and III (JFH-1 D2/3, aa 248 to 466). For the production of RNA probes by in vitro transcription, PCR was used to amplify sequences corresponding to either the 3′-terminal NS5B-coding region and 3′ UTR of genotypes 1b (nucleotides 9136 to 9595) and 2a (nucleotides 9204 to 9678) (KL-3′ UTR) or the 3′ UTR alone of genotype 1b (nucleotides 9375 to 9595) or 2a (nucleotides 9443 to 9678). In addition, the polypyrimidine tract [poly(U/UC); nucleotides 9406 to 9497] and X region (nucleotides 9498 to 9595) of genotype 1b were amplified. PCR products were cloned into pBluescript II SK+ and linearized with XbaI prior to in vitro transcription. All plasmid constructs were verified by sequencing. Primer sequences are available on request.
Expression and purification of recombinant NS5A proteins.
NS5A(ΔAH) plasmids were transformed into Escherichia coli BL21(DE3) pLysS STAR cells. Cultures were grown in Luria-Bertani (LB) medium supplemented with 50 μg/μl ampicillin, 1% (wt/vol) glucose, and 0.5 M NaCl. The cells were grown at 37°C to an optical density at 600 nm (OD600) of 0.5 to 0.7 and then cooled to 4°C for 30 min prior to induction with IPTG (isopropyl-β-d-thiogalactopyranoside) (0.5 mM) for 4 h. at 20°C. The cells were harvested by centrifugation at 7,000 rpm for 10 min. The NS5A(ΔAH) protein or its derivatives were purified by sequential Ni2+ affinity and Strep tag affinity purifications. Briefly, cell pellets were suspended in 20 ml NS5A lysis buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 10 mM 2-mercaptoethanol, 2 mM cysteine, 10 mM imidazole) supplemented with 2 μg/μl DNase and EDTA-free protease inhibitor cocktail tablets (Roche). The cell suspension was lysed by sonication on ice at an amplitude of 10 μm for six pulses of 20 s separated by 20 s and the extract clarified by centrifugation at 16,000 × g for 30 min at 4°C. The supernatant was then applied to Ni2+-charged chelating Sepharose (Chelating Sepharose Fast Flow [Amersham Biosciences]; the resin was charged with Ni2+ prior to use, according to the manufacturer's instructions), and the proteins were allowed to bind in batch by rotation at 4°C for 1 h. The mixture was centrifuged at 2,000 × g for 2 min and the flowthrough sample recovered. The resin was washed four times with 10 volumes of NS5A lysis buffer supplemented with 50 mM imidazole. Purified NS5A protein was eluted in 5-ml fractions with NS5A lysis buffer supplemented with 0.3 M imidazole. These fractions were assessed by SDS-PAGE analysis for NS5A and pooled before dialysis against 100 mM Tris-HCl (pH 8.0)-150 mM NaCl overnight at 4°C. The second step of purification was performed using a Streptactin purification kit (Novagen) at 4°C. Briefly, the dialyzed protein sample was applied to a 1-ml Streptactin Superflow agarose column (Novagen) designed for gravity flow, which was equilibrated with two column volumes of wash buffer (100 mM Tris-HCl [pH 8.0], 150 mM NaCl). The column was loaded with the protein sample, after which the column was washed five times with wash buffer. The protein was then eluted in 2-ml fractions from the resin with elution buffer (100 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2.5 mM desthiobiotin [Novagen]). The fractions were assessed by SDS-PAGE analysis for NS5A. The fractions containing the majority of NS5A were pooled and dialyzed against binding buffer (40 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 2 mM dithiothreitol [DTT]) overnight at 4°C. Protein concentrations were measured by the Bradford assay method (Bio-Rad).
Expression and purification of NS5B.
NS5B of genotype 1b HC-J4 strain (GenBank accession number AF054250) lacking the C-terminal membrane anchor sequence was expressed in E. coli BL21(DE3) cells and purified as described previously (26).
In vitro transcription reactions and labeling of RNA probes.
Transcription reaction mixtures (20 μl) contained 40 mM Tris-HCl (pH 8.0); 6 mM MgCl2; 10 mM DTT; 2 mM spermidine; 500 μM (each) ATP, CTP, and GTP; 12 μM UTP; 50 μCi [α-32P]UTP; 40 units of RNaseOUT RNase (Invitrogen); 40 units of T7 RNA polymerase according to the manufacturer's instructions (Roche); and 1 μg appropriate DNA template (linearized with XbaI). Reaction mixtures were incubated for 1.5 h at 37°C. RQ1 DNase (Promega) was added to remove the DNA template. The transcripts were purified using NucAway spin columns (Ambion) before extraction with acidified phenol-chloroform and ethanol precipitation. Purified RNA transcripts were analyzed on an 8% denaturing polyacrylamide gel (SequaGel; National Diagnostics) in Tris-borate-EDTA (TBE) buffer. RNA concentrations were determined by absorbance at 260 nm.
RNA filter binding assays.
Radiolabeled RNA transcripts and NS5A proteins were diluted in binding buffer (40 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 10 mM DTT, 50 μg/ml bovine serum albumin [BSA], 10 μg/ml yeast tRNA [Ambion]) and preincubated separately for 10 min at 4°C. The binding reaction was initiated by mixing 1 nM radiolabeled RNA and NS5A proteins (0 to 500 nM) in a 20-μl final volume at 4°C for 30 min. Membranes were presoaked in binding buffer supplemented with 5% (vol/vol) glycerol and assembled from bottom to top as follows in a slot-blot apparatus (Bio-Rad): filter paper, nitrocellulose (Schleicher & Schuell) to trap soluble protein-RNA complexes, and Hybond-N nylon (Amersham Biosciences) to bind free RNA molecules. After assembly, 20 μl of each binding reaction mixture was applied to each slot and filtered through the membranes. Each slot was washed with 0.5 ml of binding buffer and air dried, and quantification of radioactivity was performed using an image plate, BAS 1000 Bioimager (Fuji), and Aida Image Analyser v4.22 software. As a negative control, an aptamer selected against the foot-and-mouth disease virus (FMDV) 3D RNA-dependent RNA polymerase was used (sequence, 5′-GGGAAAGGAUCCACAUCUACGAAUUCGGCUCAAAAAUAGUCCGCACCAUACAUUCACUGCAGACUUGACGAAGCUU-3′). This aptamer is predicted by M-Fold analysis (38) to adopt a hairpin loop structure. Fitting was performed using GraphPad Prism 5 software (GraphPad Software). In each case, the data were fitted to the hyperbolic equation R = Rmax × R/(Kd + [P]), where R is the percentage of bound RNA, Rmax is the maximal percentage of RNA competent for binding, [P] is the concentration of NS5A, and Kd is the apparent dissociation constant.
EMSA.
For electrophoretic mobility shift assays (EMSA), 1 nM radiolabeled RNA was diluted in binding buffer (40 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 2 mM DTT, 50 μg/ml BSA, 10 μg/ml yeast tRNA [Ambion]), incubated with 200 nM purified NS5A protein construct or NS5B protein in binding buffer in a final reaction volume of 20 μl, and incubated at 4°C for 30 min. Five microliters of 50% (vol/vol) glycerol was added to each reaction mixture before loading onto a 7.5% native polyacrylamide (acrylamide-bisacrylamide, 37.5:1) gel to separate the protein-bound and free RNA molecules. Electrophoresis was carried out in 0.5× Tris-borate-EDTA (TBE) buffer at 150 V for 2 h and visualized by autoradiography.
RESULTS
NS5A binds with high specificity to HCV 3′ UTR RNA.
NS5A is an essential component of the HCV replication complex and has been shown to bind RNA; however, the domains of NS5A that are responsible for RNA binding in vitro have not been identified. To address this question, full-length HCV NS5A (lacking the N-terminal amphipathic helix) from both genotypes 1b and 2a, individual domains, or combinations of domains were expressed in E. coli and purified to high homogeneity (Fig. 2B). The 3′ UTR is a 221-nucleotide highly structured RNA molecule that corresponds to the positive-strand HCV RNA sequence and functions as the initiation site for negative-strand HCV RNA synthesis. Radiolabeled 3′ UTR RNA was produced by in vitro transcription (Fig. 1B) and used in a filter binding assay to determine the binding activity of purified NS5A(ΔAH), genotypes 1b and 2a. Increasing concentrations of NS5A(ΔAH) (0 to 500 nM) were mixed with radiolabeled 3′ UTR RNA of the corresponding HCV genotype. As shown in Fig. 3A and B, for both genotype 1b and 2a NS5A(ΔAH), the fraction of RNA bound to protein increased as a function of NS5A concentration. The data were fitted to a hyperbolic function to yield similar Kd values of 55 ± 10 nM and 57 ± 7 nM for the cognate binding partners from genotypes 1b and 2a, respectively. For genotype 1b, the highest concentration of NS5A(ΔAH) assayed bound 82% ± 4% of the input genotype 1b 3′ UTR RNA, compared to 42% ± 4.1% of genotype 2a 3′ UTR RNA bound by genotype 2a NS5A(ΔAH) (Fig. 3A and B). These values are comparable to the previously published value of 80 ± 10 nM (7) for the affinity of the NS5A-3′ UTR interaction with a maximal binding of 80% ± 4%. The small differences in affinity can be accounted for by variations in filter binding assay conditions. Negative-control radiolabeled RNA corresponding to an aptamer raised against the FMDV 3D polymerase was used in the filter binding assay. This aptamer is predicted to adopt significant secondary and tertiary structure (data not shown). In contrast to the interaction with HCV 3′ UTR RNA, NS5A(ΔAH) of both genotypes bound less than 2% of this aptamer (Fig. 3A and B). These data demonstrate that the interaction between NS5A and the HCV 3′ UTR is specific and of high affinity. Both NS5A(ΔAH) proteins of the two genotypes exhibit similar affinities of interaction; however, differences in the efficiencies of these interactions were observed (Table 1).
FIG. 3.
Filter binding analysis of the interaction between NS5A(ΔAH) and the HCV 3′ UTR RNA. (A) The indicated proteins were incubated with radiolabeled RNA (1 nM), either 3′ UTR (upper panel) or control FMDV 3C aptamer (lower panel), before application to a slot blot apparatus, filtering through nitrocellulose and Hybond-N membranes, and visualization by phosphorimaging. From left to right, the slots contained 0, 12.5, 25, 50, 100, 200, 400, and 500 nM protein. (B) The percentage of RNA bound to the nitrocellulose membrane was quantified and plotted as a function of the NS5A concentration. The data were fitted to a hyperbolic equation. Experiments were performed in triplicate, and the means and standard errors are plotted. (C) NS5A(ΔAH) (0 to 500 nM) was incubated with the corresponding genotype radiolabeled RNA (1 nM), corresponding to either the 3′ UTR or the 3′ terminal NS5B coding region plus 3′ UTR (KL-3′ UTR). Experiments were performed in triplicate, and the means and standard errors are plotted.
TABLE 1.
Binding of 3′ UTR or KL-3′ UTR RNA to NS5A(ΔAH) protein constructs of genotypes 1b and 2a as evaluated by filter binding assaya
| Genotype (clone) and binding partners | Kd, nM (mean ± SE) | Endpoint, % (mean ± SE) |
|---|---|---|
| 1b (J4) | ||
| NS5A(ΔAH) + J4 3′ UTR | 55 ± 1.0 | 85 ± 1.8 |
| NS5A(ΔAH) + J4 KL-3′ UTR | 59 ± 3.0 | 86 ± 2.7 |
| NS5A(ΔAH) + control RNA | NDb | 1.8 ± 0.63 |
| NS5A(ΔAH) + JFH-1 3′ UTR | 85 ± 3.1 | 73 ± 3.1 |
| 2a (JFH-1) | ||
| NS5A(ΔAH) + JFH-1 3′ UTR | 57 ± 0.7 | 37 ± 6.2 |
| NS5A(ΔAH) + JFH-1 KL-3′ UTR | 54 ± 6.0 | 35 ± 7.3 |
| NS5A(ΔAH) + control RNA | ND | 1.6 ± 0.76 |
| NS5A(ΔAH) + J4 3′ UTR | 37 ± 6.7 | 46 ± 3.9 |
The binding affinity and percentage of the total RNA bound to the NS5A proteins for interactions between 3′ UTR RNA and control FMDV 3C aptamer RNA are detailed. Results from comparative experiments to measure the genotype specificity of the NS5A(ΔAH)-3′ UTR interaction are also shown.
ND, not determinable. The low affinity of NS5A(ΔAH) for the control aptamer RNA meant that it was not possible to calculate the Kd from the binding curve.
The 3′-terminal sequence of the NS5B-coding region contains three conserved RNA stem-loop elements; the second of these (SL3-2) has been reported to form a kissing-loop interaction with the second stem-loop (SL2) in the X region of the 3′ UTR (5), an interaction shown to be essential for RNA replication. To investigate whether this kissing-loop interaction influenced the binding of NS5A to the 3′ UTR region, a radiolabeled RNA comprising both the 3′-terminal sequence of the NS5B-coding region and the complete 3′ UTR (termed KL-3′ UTR) was in vitro transcribed and used in filter binding assays with NS5A(ΔAH) of genotypes 1b and 2a. With increasing concentrations of NS5A(ΔAH) (0 to 500 nM), the fraction of protein-RNA complexes formed increased in the case of both genotypes (Fig. 3C). The data were fitted to a hyperbolic equation, yielding Kd values of 59 ± 3 nM and 54 ± 6 nM for the cognate binding partners from genotype 1b or 2a, respectively. These values were essentially indistinguishable from the corresponding Kd values for the interaction of NS5A(ΔAH) with 3′UTR RNA (61 ± 3 nM and 58 ± 6 nM for genotypes 1b and 2a, respectively). In addition, the efficiencies of the interactions in the presence and absence of the NS5B stem-loop elements were also very similar, implying that the interaction of NS5A with the 3′ UTR RNA is not modulated in vitro by the presence of the 3′-terminal stem-loop elements in NS5B.
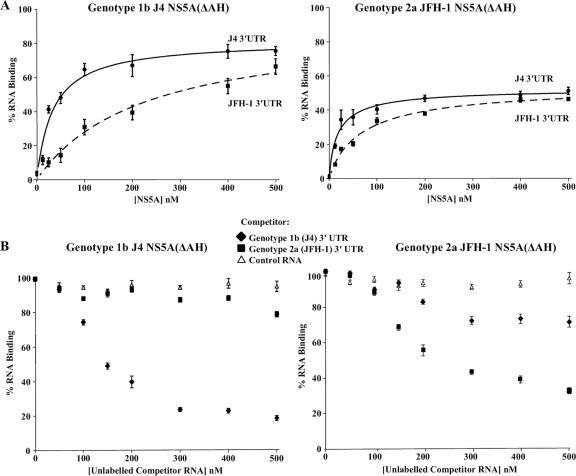
The genotype specificity of the interaction of NS5A(ΔAH) constructs for their cognate 3′ UTR sequences was also investigated using the filter binding method (Fig. 4A). Between these two genotypes the 3′ UTRs possess 80% sequence similarity, with differences in the length of the poly(U/UC) region and in the sequence of the upstream variable region. Interestingly, NS5A(ΔAH) of genotype 1b showed a lower affinity for the genotype 2a 3′ UTR RNA than for its cognate 3′ UTR RNA (Kd values of 85 ± 3.1 nM and 54 ± 5.4 nM, respectively) (Table 1). In contrast, the genotype 2a NS5A(ΔAH) interaction with the corresponding genotype 2a 3′ UTR RNA was of lower affinity than its interaction with the genotype 1b 3′ UTR RNA (Kd values of 59 ± 5.8 nM and 37 ± 6.7 nM, respectively) (Table 1). To address the possibility of a genotype-dependent interaction between NS5A and the 3′ UTR, competition experiments were undertaken. Filter binding assays were performed in which genotype 1b or 2a NS5A(ΔAH) was incubated with the corresponding radiolabeled 3′ UTR RNA (1 nM) in the presence of increasing concentrations (0 to 500 nM) of unlabeled competitor RNA (genotype 1b or 2a 3′ UTR or control aptamer RNA) (Fig. 4B). For either genotype the interaction of NS5A(ΔAH) with the 3′ UTR was efficiently competed with unlabeled 3′UTR RNA of the corresponding genotype but not with an unlabeled negative-control aptamer RNA. For genotype 1b NS5A(ΔAH), it was observed that unlabeled genotype 2a 3′ UTR RNA did not efficiently compete for the binding to radiolabeled genotype 1b 3′ UTR RNA. Surprisingly, in comparison, unlabeled genotype 1b 3′ UTR RNA was able to compete (albeit inefficiently) for the binding of genotype 2a NS5A(ΔAH) to radiolabeled genotype 2a 3′ UTR RNA. These data correlate with the relative affinities of the NS5A proteins to the noncognate 3′ UTR RNA molecules but do not clearly support a reciprocal genotype-dependent interaction between NS5A and the 3′ UTR. Rather, the competition experiment suggests that J4 NS5A binds more efficiently to its cognate 3′ UTR, whereas JFH-1 NS5A is a less selective and less efficient RNA binding protein.
FIG. 4.
Filter binding analysis of the genotype specificity of the NS5A-3′ UTR interaction. (A) Purified NS5A(ΔAH) of genotype 1b (J4, left) or genotype 2a (JFH-1, right) was incubated with radiolabeled 3′ UTR RNA (1 nM) or either genotype 1b (J4, solid lines) or genotype 2a (JFH-1, dashed lines) and analyzed as described for Fig. 3. The percentage of RNA bound to the nitrocellulose membrane was quantified and plotted as a function of the NS5A concentration. The data were fitted to a hyperbolic equation. (B) Purified NS5A(ΔAH) of genotype 1b (J4, left) or genotype 2a (JFH-1, right) was incubated with 1 nM radiolabeled 3′ UTR RNA in the presence of increasing concentrations (0 to 500 nM) of unlabeled 3′ UTR of either genotype 1b or 2a or control FMDV 3C aptamer RNA. The percentage of RNA bound to the nitrocellulose membrane was quantified and plotted as a function of the competitor RNA concentration.
Mapping the RNA binding domains of NS5A.
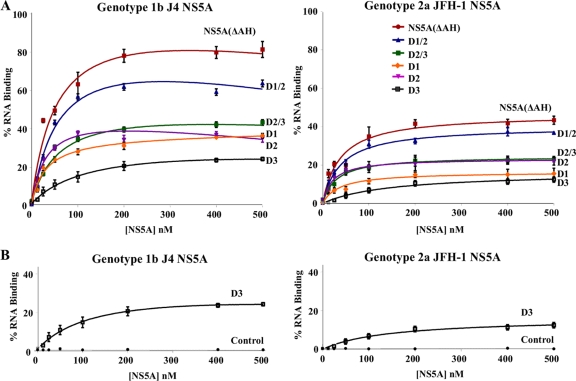
To define the domains of NS5A responsible for binding the 3′ UTR RNA, a series of individual domains (D1, D2, and D3) or pairs of domains (D1/2 and D2/3) for both genotype 1b and 2a NS5A proteins were expressed and purified as for the NS5A(ΔAH) protein (Fig. 2), and their RNA binding abilities were assessed by filter binding assay (Fig. 5). The results of these assays, comparing the domain affinities and efficiencies of binding to the 3′ UTR, are displayed in Table 2. Interestingly, we found that domains I and II of NS5A efficiently bound to the 3′ UTR RNA, whereas domain III bound weakly. Both the efficiency and affinity of the interaction with domain III were lower than for the interaction with either domain I or II alone (Table 2). An additive effect in the efficiency of interaction was observed when both domains I and II were present in a single construct (D1/2). This observation suggests that the conformation of domains I and II when binding the RNA separately is not dissimilar to that when the two domains are expressed as a single protein. The two-domain construct does, however, bind a higher percentage of the available RNA. The results described were observed for the binding interactions for both genotypes 1b and 2a (Fig. 5A and Table 2). None of the proteins bound the control aptamer RNA (data not shown).
FIG. 5.
The three domains of NS5A exhibit different RNA binding properties in vitro. (A) Purified NS5A(ΔAH) and either combinations of domains or individual domains of genotype 1b (J4, left) or genotype 2a (JFH-1, right) were incubated with radiolabeled 3′ UTR RNA (1 nM) of the corresponding genotype and analyzed as described for Fig. 3. The percentage of RNA bound to the nitrocellulose membrane was quantified and plotted as a function of the NS5A concentration. The data were fitted to a hyperbolic equation. (B) Purified D3 (domain III alone) of genotype 1b (J4, left) or genotype 2a (JFH-1, right) was incubated with radiolabeled 3′ UTR RNA (1 nM) of the corresponding genotype or a control FMDV 3C aptamer RNA and analyzed as described for Fig. 3. In all cases experiments were performed in triplicate, and the means and standard errors are plotted.
TABLE 2.
Interactions between 3′ UTR RNA and NS5A protein domain constructs as assessed by filter binding assaya
| Genotype (clone) and NS5A domain construct | Kd, nM (mean ± SE) | Endpoint, % (mean ± SE) |
|---|---|---|
| 1b (J4) | ||
| NS5A(ΔAH) | 55 ± 10 | 82 ± 4.0 |
| D1/2 | 58 ± 7 | 66 ± 1.9 |
| D2/3 | 63 ± 13 | 42 ± 3.7 |
| D1 | 41 ± 6 | 36 ± 1.8 |
| D2 | 43 ± 5 | 39 ± 1.6 |
| D3 | 145 ± 9 | 17 ± 2.4 |
| D3 + control RNA | NDb | 0.67 ± 0.07 |
| 2a (JFH-1) | ||
| NS5A(ΔAH) | 57 ± 7 | 42 ± 4.1 |
| D1/2 | 48 ± 8 | 37 ± 1.5 |
| D2/3 | 51 ± 5 | 22 ± 1.8 |
| D1 | 56 ± 9 | 18 ± 2.4 |
| D2 | 52 ± 4 | 21 ± 2.7 |
| D3 | 166 ± 11 | 11 ± 2.4 |
| D3 + control RNA | ND | 1.3 ± 0.15 |
The binding affinity and percentage of the total RNA bound to the NS5A proteins are represented for all domain constructs of genotypes 1b and 2a with 3′ UTR RNA and control FMDV 3C aptamer RNA.
ND, not determinable. The low affinity of D3 for the control aptamer RNA meant that it was not possible to calculate the Kd from the binding curve.
The capacity of domain III NS5A to interact with the 3′ UTR was substantially lower than those of domains I and II. However, neither genotype 1b nor 2a domain III interacted with the negative-control FMDV 3D polymerase aptamer RNA, indicating that the low-affinity and -efficiency interaction seen between domain III and the 3′ UTR RNA is specific (Fig. 5B). As a clear additive effect on the affinity or efficiency of the interaction is not seen for domains II and III combined (D2/3) compared to domain II alone, this implies that domain III of NS5A does not contribute greatly to RNA binding in vitro.
NS5A interacts preferentially with the poly(U/UC) region of the 3′ UTR RNA.
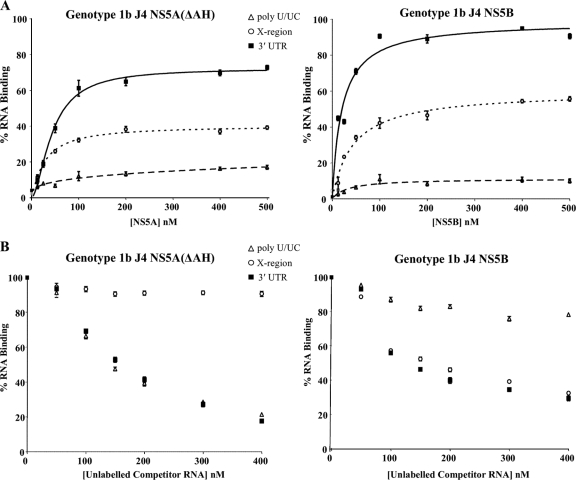
The 3′ UTR contains three distinct regions, i.e., a variable region not required for RNA replication followed by a polypyrimidine tract [poly(U/UC)] and a conserved 98-nucleotide X region which contains three distinct stem-loops. The poly(U/UC) and X region are both essential for RNA replication (Fig. 1). Previous studies have reported that NS5A binds preferentially to uridylate- and guanylate-rich RNA sequences. In order to define more precisely the region of the 3′ UTR to which NS5A binds, a series of filter binding assays and competition experiments were performed, focusing on genotype 1b interactions. 3′ UTR binding by NS5A was also compared to the interaction of the NS5B polymerase (Fig. 6) with the 3′ UTR. NS5A and NS5B interact both in vivo and in vitro (26, 29). In vitro, this interaction has a stimulatory affect on NS5B polymerase activity, suggesting that NS5A enhances RNA replication (29). In contrast, the ability of NS5A to directly bind RNA may reflect a role in the regulation or control of the switch between translation and replication. We investigated whether there was any preference for RNA sequence or region for NS5A or NS5B binding to provide insight into this. Recombinant genotype 1b (J4) NS5B protein was purified to high homogeneity (Fig. 2), and RNA transcripts of the poly(U/UC) and X region of the 3′ UTR were synthesized in vitro in the presence of [α-32P]UTP and used as substrates in filter binding analyses (Fig. 6A). The results clearly demonstrate that for NS5A(ΔAH), a preference for binding of the poly(U/UC) RNA over the X region RNA exists. The affinity of the NS5A interaction with poly(U/UC) RNA was approximately 3-fold higher than that for the X region RNA [Kd values of 44 ± 4.0 nM for poly(U/UC) and 123 ± 5.3 nM for the X region] (Table 3). In contrast, NS5B protein showed a high-affinity interaction with 3′ UTR RNA (Kd value of 33 ± 1.2 nM) and with X region RNA (Kd value of 75 ± 2.1 nM) but lower affinity for the poly(U/UC) region (Table 3; Fig. 6A).
FIG. 6.
Binding specificity of the in vitro 3′ UTR-NS5A interaction. (A) Purified NS5A(ΔAH) (left) or NS5B (right) of genotype 1b was incubated with radiolabeled 3′ UTR RNA (solid lines), poly(U/UC) (dashed lines), or X region RNA (dotted lines) (all at 1 nM) and analyzed as described for Fig. 3. The percentage of RNA bound to the nitrocellulose membrane was quantified and plotted as a function of the NS5A concentration. The data were fitted to a hyperbolic equation. Experiments were performed in triplicate, and the means and standard errors are plotted. (B) NS5A(ΔAH) (left) and NS5B (right) proteins were bound to 1 nM radiolabeled 3′ UTR in the presence of increasing concentrations (0 to 400 nM) unlabeled 3′ UTR, poly(U/UC), or X region RNA.
TABLE 3.
Comparison of the binding interaction between specific regions of the 3′ UTR RNA and NS5A(ΔAH) and NS5B proteins constructs as evaluated by filter binding assaya
| Genotype 1b (J4) protein and RNA region | Kd, nM (mean ± SE) | Endpoint, % (mean ± SE) |
|---|---|---|
| NS5A(ΔAH) | ||
| 3′ UTR | 56 ± 1.7 | 72 ± 3.8 |
| Poly(U/UC) | 44 ± 4.0 | 39 ± 1.9 |
| X region | 123 ± 5.3 | 16 ± 2.2 |
| NS5B | ||
| 3′ UTR | 25 ± 1.6 | 89 ± 1.8 |
| Poly(U/UC) | 75 ± 2.1 | 9.0 ± 2.9 |
| X region | 33 ± 1.2 | 54 ± 1.7 |
The binding affinity and percentage of the total RNA bound to the NS5A and NS5B proteins are shown for experiments conducted with proteins and RNA of genotype 1b.
To further demonstrate the preference and possible specificity of the NS5A and NS5B-3′ UTR interaction, competition experiments were performed with unlabeled RNA transcripts derived from the 3′ UTR. We investigated the ability of unlabeled 3′ UTR RNA transcripts to compete for binding of NS5A(ΔAH) or NS5B to radiolabeled 3′ UTR RNA. NS5A(ΔAH) or NS5B (100 nM) was incubated with 1 nM radiolabeled 3′ UTR RNA in the presence of increasing concentrations (0 to 400 nM) of unlabeled competitor RNA [3′ UTR, the poly(U/UC) region and the X region], and binding was measured by the filter binding method (Fig. 6B). The interaction of NS5A(ΔAH) with the 3′ UTR was effectively competed by both unlabeled 3′ UTR and poly(U/UC) RNA but not by the X region RNA. Contrastingly, it was observed that the unlabeled X region and 3′ UTR RNAs effectively competed for the binding of NS5B to radiolabeled 3′ UTR RNA, whereas poly(U/UC) RNA was not significantly able to compete for the binding of NS5B to the radiolabeled 3′ UTR (Fig. 6B). Combined with the binding analyses, these data suggest that the poly(U/UC) region of the 3′ UTR RNA provides the specificity of the interaction with NS5A but that the X region of the 3′ UTR is a better substrate for the NS5B binding than the poly(U/UC) region. These distinctive preferences may have implications for the interplay of NS5A and NS5B in RNA replication.
EMSA analysis of NS5A RNA binding.
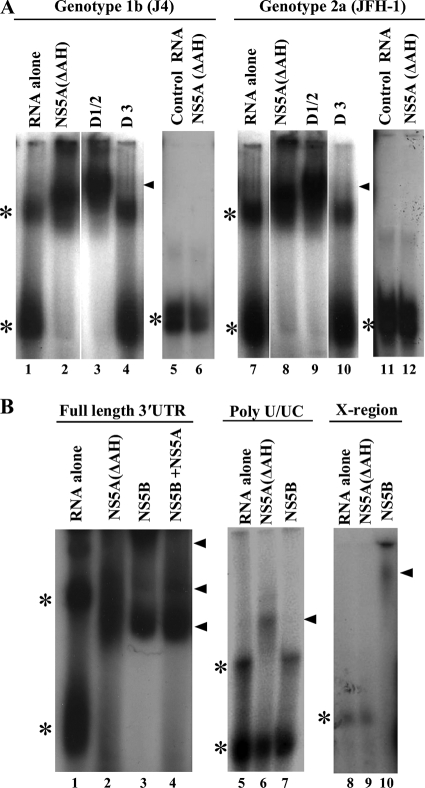
The interaction between the panel of NS5A proteins and 3′ UTR RNA was also assessed by gel mobility shift assays (Fig. 7). NS5A proteins at a concentration of 100 nM were incubated with in vitro-transcribed radiolabeled 3′ UTR RNA, and complexes formed were resolved on native acrylamide gels. As shown in Fig. 7A, when analyzed by native gel electrophoresis, the 3′ UTR RNA exists in two distinct conformations with different mobilities (lanes 1 and 7). For both genotype 1b and genotype 2a, NS5A(ΔAH) and D1/2 were able to stably interact with the 3′ UTR RNA, causing its retardation in the gel (lanes 2, 3, 8, and 9). Distinct protein-RNA complexes were observed for the latter binding reactions, along with the loss of free 3′ UTR RNA in their presence. However, no stable, retarded protein-RNA complexes were observed in the presence of D3 and 3′ UTR RNA (either genotype 1b or 2a) (lanes 4 and 10). These data are broadly consistent with the results of the filter binding assays and, combined, define the major RNA binding region of NS5A to be within domains I and II. Gel mobility shift assays were also performed using the negative-control FMDV 3C aptamer RNA, and no distinct protein-RNA complexes were observed in the presence of NS5A(ΔAH) of either genotype 1b or 2a (lanes 5, 6, 11, and 12), again demonstrating the specificity of the 3′ UTR-NS5A interaction.
FIG. 7.
Electrophoretic mobility shift analysis of NS5A-3′ UTR RNA binding. (A) Radiolabeled 3′ UTR RNA or control FMDV 3C aptamer was incubated with 200 nM NS5A(ΔAH), D1/2, or D3 proteins as indicated, prior to separation by native acrylamide gel electrophoresis and visualization by autoradiography. The positions of the major retarded NS5A-RNA complexes are indicated by black triangles, and the asterisks mark the positions of the unbound 3′ UTR RNA (the two bands most likely represent alternative conformations of the RNA). (B) Radiolabeled RNA corresponding to the 3′ UTR, poly(U/UC), or X region was incubated with 100 nM NS5A(ΔAH) or NS5B or a combination. The positions of the major retarded NS5A-RNA and NS5B-RNA complexes are indicated by black triangles, and the asterisks mark the positions of the unbound 3′ UTR RNA.
We also examined the interaction of either NS5A(ΔAH) or NS5B with the full-length 3′ UTR, the poly(U/UC), or the X region. As shown in Fig. 7B, consistent with the filter binding data, complexes were formed between NS5A(ΔAH) and poly(U/UC) RNA but not between NS5A(ΔAH) and the X region of the 3′ UTR (compare lanes 5 and 6 to lanes 8 and 9). The opposite result was seen for NS5B, where complexes with the X region were detectable but not so with the poly(U/UC) RNA (lanes 7 and 10). Lastly, we examined whether both NS5A(ΔAH) and NS5B could interact simultaneously with full-length 3′ UTR RNA. Although there were subtle changes in the amounts of each of the retarded complexes compared to either NS5A(ΔAH) or NS5B alone, no new complexes were observed (lane 4). Indeed, the pattern of retarded complexes most closely resembled that seen in the presence of NS5B alone (compare lanes 2 and 4). These data suggest that binding of NS5A(ΔAH) or NS5B may be mutually exclusive, perhaps indicative of different roles of the two proteins in the utilization of genomic RNA.
DISCUSSION
HCV genome replication is a complex event in the viral life cycle, requiring both viral and host proteins. NS5A plays an essential role in these interactions, not only by interacting with other viral and cellular proteins but also via binding to viral RNA sequences. In this in vitro study, we sought to investigate in detail the ability of NS5A to bind the initiation site of negative-strand RNA synthesis, namely, the HCV genomic (positive-strand) 3′ UTR.
The first major conclusion from this study is that all three domains of NS5A were able to bind to the 3′ UTR independently, albeit with differing affinities. From the analysis of both individual domains and two-domain combinations (D1/2 and D2/3), it is clear that the major contributions to RNA binding are provided by domains I and II. Although the Kd of RNA binding for NS5A(ΔAH) was similar to that for D1/2 (Fig. 5 and Table 2), the efficiency of binding (defined by the percentage of RNA bound) was higher, indicating that whereas domain III showed weak binding alone, in the context of the complete NS5A protein it did make a contribution to overall binding. The observation that all three domains bind RNA independently provides functional support for the proposed domain structure of NS5A, which has been derived by a combination of in silico predictions, tryptic digests of bacterially expressed NS5A(ΔAH), and X-ray crystallographic data in the case of domain I. As NS5A lacks any canonical RNA binding motifs, it follows that the protein must possess multiple, novel RNA binding surfaces. Notably, for domain I one of the published crystal structures predicts that dimerization creates a groove with flat basic surfaces that might function in RNA binding (33). However, a more recent study predicted an alternative orientation of the dimer, lacking the basic groove but still presenting two flat basic surfaces, in this case spatially distant from each other (17).
Our data are consistent with the results of the only other published study of NS5A RNA binding (8), which showed binding of NS5A(ΔAH) to both 5′ and 3′ UTRs of the positive strand, as well as binding to the 3′ UTR of the negative strand, with Kd values in a range (80 to 130 nM) similar to those determined in our study. Interestingly, that study showed reduced binding of NS5A(ΔAH) to a 3′ UTR RNA lacking the polypyrimidine tract; our data extend this observation, showing that NS5A(ΔAH) binds to the polypyrimidine tract but not to the X region. In contrast, NS5B binds preferentially to the X region. In the context of genome replication, it is intuitive that the polymerase would specifically interact with the extreme 3′ region of the template RNA in order to synthesize a full-length copy. The differential binding of NS5A and NS5B to regions of the 3′ UTR suggests that they could interact simultaneously with a single RNA. The role of NS5A might therefore be to promote polymerase activity or processivity, and in this context, the two proteins have been shown to directly interact, with effects on NS5B activity (28, 32). However, the EMSA analysis presented here (Fig. 7B) is consistent with the suggestion that in the presence of NS5B, NS5A does not interact significantly with the 3′ UTR. Perhaps cellular proteins known to interact with NS5A and/or NS5B (e.g., cyclophilins [7]), which play as-yet-undefined roles in genome replication, may regulate the interactions of these proteins with the 3′ UTR.
One further implication of the independent RNA binding activity of the three domains, coupled with the previously identified ability of NS5A to bind the 5′ UTR (8), is the potential to mediate genome circularization. This is likely to be important for translation, as circularization of cellular mRNA is mediated by protein-bridging factors such as polypyrimidine tract binding protein (PTB), but would potentially inhibit replication. This raises the possibility that binding of NS5A to the 3′ UTR might drive the necessary switch from the utilization of positive-stranded genomic RNA for replication to translation. A somewhat overlooked requirement of this switch would be the transport of genomic RNA from the replication factories (membranous web) to ribosomes within the cytosol. It has been proposed that the recently identified involvement of NS5A in the process of virus assembly may be explained by a role in the transport of genomic RNA to sites of assembly. This would require a further regulation which could conceivably be driven by differential phosphorylation of NS5A. Alternatively, it has recently been shown that the interaction between domain III of NS5A and the core protein is important for assembly (22), suggesting that core might also regulate the binding of NS5A to the 3′ UTR.
In conclusion, the specific binding of NS5A to viral genome sequences directly implicates NS5A in the multiple processes within the viral life cycle that involve the genome (translation, RNA replication, and assembly). In this regard, the interaction of NS5A with the 3′ UTR could be a valid target for antiviral intervention. However, progress toward this goal will require not only more detailed molecular information about the interaction but also a robust, high-throughput assay for selection of compounds able to block NS5A RNA binding. Future work in our laboratory will be focused toward addressing these challenging issues.
Acknowledgments
We are grateful to Sophie Forrest for providing the FMDV 3D polymerase aptamer, to Matthew Bentham and Stephen Griffin for advice and critical comments on the manuscript, and to Jens Bukh and Takaji Wakita for molecular clones of HCV genotypes 1b (J4) and 2a (JFH-1), respectively.
This work was supported by the Wellcome Trust. T.L.F. is a postgraduate student within the Wellcome Trust 4-year Ph.D. program, entitled “The Molecular Basis of Biological Mechanisms,” awarded to the Astbury Centre for Structural Molecular Biology. A.R.P. is a Research Councils UK Academic Research Fellow. This work was additionally supported by Wellcome Trust grant 082812. Work in the N.J.S. laboratory is supported by the Biotechnology and Biological Sciences Research Council and Yorkshire Cancer Research.
Footnotes
Published ahead of print on 30 June 2010.
REFERENCES
- 1.Appel, N., M. Zayas, S. Miller, J. Krijnse-Locker, T. Schaller, P. Friebe, S. Kallis, U. Engel, and R. Bartenschlager. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng, J., C. Ming-Fu, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73:7044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, and P. J. Barr. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster, G., and P. Mathurin. 2008. Hepatitis C virus therapy to date. Antivir. Ther. 13:1-8. [PubMed] [Google Scholar]
- 5.Friebe, P., J. Boudet, J.-P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanoulle, X., D. Verdegem, A. Badillo, J. M. Wieruszeski, F. Penin, and G. Lippens. 2009. Domain III of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem. Biophys. Res. Commun. 381:634-638. [DOI] [PubMed] [Google Scholar]
- 7.Hanoulle, X., A. Badillo, J. M. Wieruszeski, D. Verdegem, I. Landrieu, R. Bartenschlager, F. Penin, and G. Lippens. 2009. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 284:13589-13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, L., J. S. D. Hwang, M. R. S. Hargittai, Y. Chen, J. J. Arnold, K. D. Raney, and C. E. Cameron. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417-36428. [DOI] [PubMed] [Google Scholar]
- 9.Huang, Y., K. Staschke, R. De Francesco, and S. L. Tan. 2007. Phosphorylation of hepatitis C virus NS5A non-structural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Hughes, M., S. Griffin, and M. Harris. 2009. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J. Gen. Virol. 90:1329-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khabar, K. S. A., and S. J. Polyak. 2002. Hepatitis C virus-host interactions: the NS5A protein and the interferon/chemokine systems. J. Interferon Cytokine Res. 22:1005-1012. [DOI] [PubMed] [Google Scholar]
- 12.Kim, Y. K., C. S. Kim, S. H. Lee, and S. K. Jang. 2002. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 290:105-112. [DOI] [PubMed] [Google Scholar]
- 13.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang, Y., C. B. Kang, and H. S. Yoon. 2006. Molecular and structural characterization of the domain II of hepatitis C virus non-structural protein 5A. Mol. Cells 22:13-20. [PubMed] [Google Scholar]
- 15.Liang, Y., H. Ye, C. B. Kang, and H. S. Yoon. 2007. Domain II of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 46:11550-11558. [DOI] [PubMed] [Google Scholar]
- 16.Lindenbach, B. D., and C. M. Rice. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933-938. [DOI] [PubMed] [Google Scholar]
- 17.Love, R. A., O. Brodsky, M. J. Hickey, P. A. Wells, and C. Cronin. 2009. Crystal structure of a novel dimeric form of NS5A domain I from hepatitis C virus. J. Virol. 83:4395-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macdonald, A., K. Crowder, A. Street, C. McCormick, and M. Harris. 2004. The hepatitis C virus NS5A protein binds to members of the Src family of tyrosine kinases and regulates kinase activity. J. Gen. Virol. 85:721-729. [DOI] [PubMed] [Google Scholar]
- 19.Macdonald, A., K. Crowder, A. Street, C. McCormick, K. Saksela, and M. Harris. 2003. The hepatitis C virus non-structural NS5A protein inhibits activating protein-1 function by perturbing ras-ERK pathway signalling. J. Biol. Chem. 278:17775-17784. [DOI] [PubMed] [Google Scholar]
- 20.Macdonald, A., and M. Harris. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485-2502. [DOI] [PubMed] [Google Scholar]
- 21.Macdonald, A., S. Mazaleyrat, C. McCormick, A. Street, N. J. Burgoyne, R. M. Jackson, V. Cazeaux, H. Shelton, K. Saksela, and M. Harris. 2005. Further studies on hepatitis C virus NS5A-SH3 domain interactions: identification of residues critical for binding and implications for viral RNA replication and modulation of cell signalling. J. Gen. Virol. 86:1035-1044. [DOI] [PubMed] [Google Scholar]
- 22.Masaki, T., R. Suzuki, K. Murakami, H. Aizaki, K. Ishii, A. Murayama, T. Date, Y. Matsuura, T. Miyamura, T. Wakita, and T. Suzuki. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 24.Moradpour, D., V. Brass, and F. Penin. 2005. Function follows form: the structure of the N-terminal domain of HCV NS5A. Hepatology 42:732-735. [DOI] [PubMed] [Google Scholar]
- 25.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453-463. [DOI] [PubMed] [Google Scholar]
- 26.O'Farrell, D., R. Trowbridge, D. Rowlands, and J. Jäger. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025-1035. [DOI] [PubMed] [Google Scholar]
- 27.Penin, F., V. Brass, N. Appel, S. Ramboarina, R. Montserret, D. Ficheux, H. E. Blum, R. Bartenschlager, and D. Moradpour. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A J. Biol. Chem. 279:40835-40843. [DOI] [PubMed] [Google Scholar]
- 28.Quezada, E. M., and C. M. Kane. 2009. The hepatitis C virus NS5A stimulates NS5B during in vitro RNA synthesis in a template specific manner. Open Biochem. J. 3:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riley, L. W., V. Rehak, and L. Krekulova. 2006. Structure and functions of hepatitis C virus proteins: 15 years after. Folia Microbiol. 51:665-680. [DOI] [PubMed] [Google Scholar]
- 31.Shelton, H., and M. Harris. 2008. Hepatitis C virus NS5A protein binds the SH3 domain of the Fyn tyrosine kinase with high affinity: mutagenic analysis of residues within the SH3 domain that contribute to the interaction Virol. J. 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 33.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tellinghuisen, T. L., K. L. Foss, J. C. Treadaway, and C. M. Rice. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weigand, K., W. Stremmel, and J. Encke. 2007. Treatment of hepatitis C virus infection. World J. Gastroenterol. 13:1897-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zoulim, F., M. Chevallier, M. Maynard, and C. Trepo. 2003. Clinical consequences of hepatitis C virus infection. Rev. Med. Virol. 13:57-68. [DOI] [PubMed] [Google Scholar]
- 38.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]