Abstract
The human cytomegalovirus (HCMV) tegument protein UL69 is important for efficient viral replication at low multiplicities of infection. Several molecular mechanisms by which UL69 contributes to HCMV replication have been proposed, including UL69's ability to interact with the mRNA export factors UAP56 and URH49 to facilitate the shuttling of viral mRNAs from the nuclei of infected cells. Using a UL69 viral mutant that is unable to bind UAP56 and URH49, we demonstrated that UL69's interaction with UAP56 or URH49 does not contribute to the growth phenotype associated with the UL69 deletion mutant.
Human cytomegalovirus (HCMV) encodes roughly 25 proteins that compose the tegument layer, which resides between the nucleocapsid and the viral envelope. These proteins are packaged within the mature virion, are delivered to the host cell immediately upon infection, and play important roles in entry, gene regulation, immune evasion, DNA replication, virion assembly, and viral egress (8, 9). The UL69 tegument protein has previously been shown to play an important role in regulating efficient replication of HCMV (5). Infection with a UL69 deletion mutant results in a severely multiplicity-dependent growth phenotype (5). However, the mechanism whereby UL69 contributes to viral replication has remained elusive. Previous studies have provided clues as to how UL69 may participate in regulating HCMV replication. Activities associated with UL69 include its ability to regulate cell cycle progression (5, 13), regulate viral gene expression (5), shuttle between the nucleus and cytoplasm (10, 12), bind RNA (19), and interact with itself (11) and several cellular proteins (1, 12, 16, 18, 21).
UL69 and its herpesvirus homologues are thought to function in part by regulating the export of intronless viral mRNAs from the nucleus to the cytoplasm within infected cells (10, 12, 20). In support of this, UL69 has been shown to interact with the cellular factors U2AF65-associated protein 56 (UAP56) and the 90% identical UAP56-related helicase 49 (URH49) (12). UAP56 and URH49 are DEAD-box helicases, which are RNA-dependent ATPases that play important roles in connecting pre-mRNA splicing with mature mRNA export (14, 15). The ability of UL69 to bind UAP56 and/or URH49 has been hypothesized to be critical for its ability to promote the export of viral transcripts during infection and to play a critical role in controlling viral replication (12, 20). Even though previous studies have clearly demonstrated that UL69 can bind to UAP56 and URH49 and that these interactions are required for the efficient nuclear export of an unspliced reporter gene, the significance of UL69's interaction with UAP56 and/or URH49 has not been determined in the context of a productive viral infection where these proteins are expressed at physiological levels and in the presence of the full complement of viral proteins. Therefore, this study utilizes a UL69 UAP56/URH49 viral binding mutant to determine if UL69's interaction with UAP56 or URH49 is required for efficient HCMV replication and to determine if these interactions contribute to the growth phenotype observed with a UL69 deletion mutant.
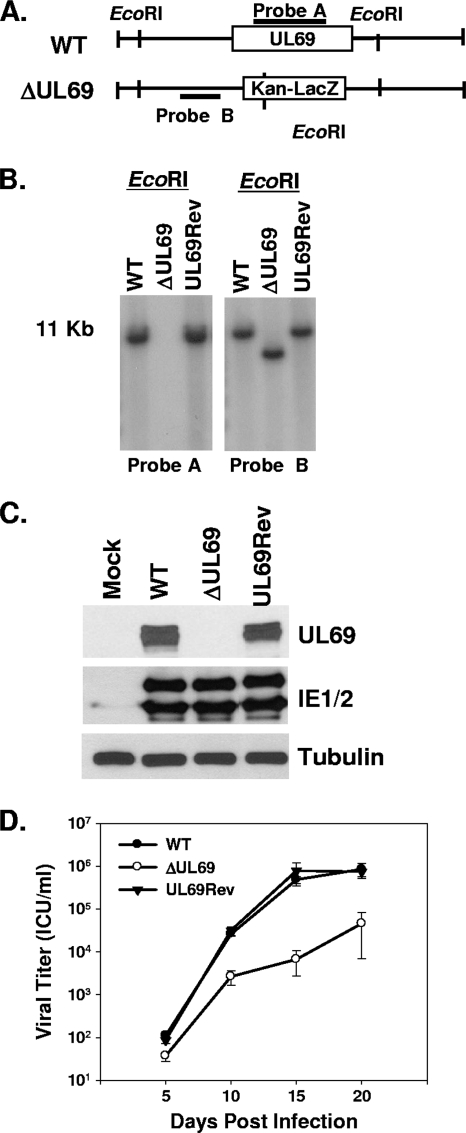
The generation and characterization of a UL69 deletion mutant has previously been described (5). This mutant, termed TNsubUL69, was created by homologous recombination within human fibroblasts. The generation of large deletion mutants using this method is well established; however, it is not conducive to the generation of point mutants or small deletion mutants. Therefore, in order to create a UL69 mutant that is unable to bind UAP56 and URH49, we utilized a bacterial artificial chromosome (BAC) that contains the HCMV genome to generate our UL69 viral mutants. First, we generated a UL69 deletion mutant that could subsequently be used to reinsert a variety of mutated UL69 gene sequences. The UL69 deletion mutant was generated by replacing the UL69 open reading frame (ORF) within the ADCREGFP BAC (3) with a kanamycin (Kan) resistance and LacZ cassette, using standard allelic exchange protocols that have been described previously (17, 22). This UL69 deletion mutant BAC has the same UL69 sequence deleted as does TNsubUL69 (corresponding to nucleotides 98,289 to 100,433 of the AD169 genome) and is termed pADΔUL69 (Fig. 1 A). Southern blot analysis of the pADΔUL69 DNA digested with EcoRI confirmed that pADΔUL69 lacks the UL69 coding region and that the marker cassette recombined properly within the viral genome (Fig. 1B). A revertant BAC, termed pADUL69Rev, was also generated, using the pADΔUL69 BAC and the wild-type (WT) UL69 sequence to demonstrate our ability to reincorporate either the WT sequence or a mutated UL69 sequence back into the viral genome. Southern blot analysis of pADUL69Rev DNA digested with EcoRI confirmed that pADUL69Rev had undergone proper recombination and now contained the UL69 coding sequence (Fig. 1B). Stocks of WT ADCREGFP, ADΔUL69, and ADUL69Rev viruses were then generated by transfecting purified BAC DNA into human foreskin fibroblast (HFF) cells via electroporation as previously described (3). To confirm that ADΔUL69 was unable to express UL69, HFF cells were either mock infected or infected with WT, ADΔUL69, or ADUL69Rev virus at a multiplicity of infection (MOI) of 3 PFU/cell, and cell lysates were harvested for Western blot analysis 72 h postinfection. As shown in Fig. 1C, UL69 was expressed in cells infected with either the WT virus or the UL69 revertant virus but was not expressed in cells infected with the UL69 deletion mutant. The HCMV immediate-early proteins IE1 and IE2 were expressed at similar levels in cells infected with any of the viruses, indicating equivalent levels of infection.
FIG. 1.
Generation and characterization of the UL69 deletion and UL69 revertant viruses. (A) Schematic representation of WT HCMV BAC and ADΔUL69 BAC depicting insertion of the Kan-LacZ cassette and the EcoRI restriction sites. (B) Southern blot analysis of WT pADCREGFP, pADΔUL69 (ΔUL69), and pADUL69Rev (UL69Rev) BACs digested with EcoRI restriction enzyme and probed for either UL69 (probe A) or the UL69 left flanking region (probe B). (C) Western blot analysis of UL69 expression at 72 h postinfection in HFF cells that were either mock infected or infected with WT, ADΔUL69 (ΔUL69), or ADUL69Rev (UL69Rev) virus. Lysates were also assayed for the abundances of IE1 and IE2. Anti-tubulin (α-tubulin) abundance was included as a loading control. (D) HFF cells were infected (0.01 PFU/cell) with either WT ADCREGFP, ADΔUL69, or ADUL69Rev virus. Cultures were harvested at the indicated times postinfection, and infectious virus was quantified by infectious-center assay. Results represent three independent experiments. ICU, infectious-center units.
Viral growth curves were conducted to compare the replication of WT, ADΔUL69, and ADUL69Rev viruses at an MOI of 0.01 PFU/cell. As previously shown for TNsubUL69 (5), ADΔUL69 replicated with delayed kinetics and to lower levels compared to those of the WT virus. We observed a 1.5- to 2-log reduction in viral titers for the UL69 deletion mutant compared to those for the WT virus (Fig. 1D). However, no growth defect was observed when cells were infected with the UL69 revertant virus, demonstrating that the defect associated with the ADΔUL69 virus is due to deletion of the UL69 ORF. The ADΔUL69 BAC was subsequently used to generate UL69 binding mutants.
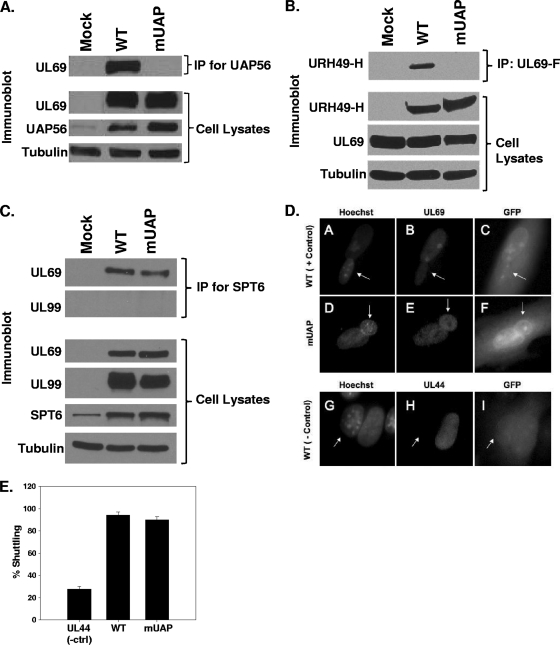
Lischka et al. previously described a UL69 mutant construct that was defective for binding to UAP56 and URH49 (12). Mutation of arginines at positions 22, 23, 25, and 26 within UL69 to alanines abolished UL69's ability to interact with UAP56 and URH49 (12). Using allelic exchange and the pADΔUL69 BAC, we replaced the arginines at positions 22, 23, 25, and 26 within the UL69 ORF with alanines. The resulting recombinant BAC, termed pADmUAPUL69, was screened by restriction enzyme analysis, PCR, and direct sequencing of BAC DNA to confirm incorporation of the desired mutations (data not shown). pADmUAPUL69 BAC DNA was then transfected into HFF cells to generate viral stocks. Upon generation of the ADmUAPUL69 virus stock, the entire UL69 ORF was sequenced from the viral genome to confirm that the desired mutations were present and that no other mutations were incorporated into the UL69 ORF (data not shown). To confirm that the ADmUAPUL69 mutant was unable to bind UAP56, HFF cells were either mock infected or infected with ADmUAPUL69 or WT virus at an MOI of 3 PFU/cell. Cell lysates were prepared 72 h postinfection and subjected to immunoprecipitation with antibody against UAP56 (a gift from Michael Greene). Immunoprecipitates and cell lysates were then separated by SDS-PAGE, transferred to a membrane, and probed with antibody. As shown in Fig. 2 A, the levels of UL69 and UAP56 are similar in cells infected with either the WT virus or the ADmUAPUL69 binding mutant. Interestingly, we observed an increase in UAP56 expression following infection with the WT virus or the ADmUAPUL69 mutant virus compared to that in mock-infected cells. UL69 coprecipitated with UAP56 in lysates infected with WT HCMV. In contrast, UL69 did not coprecipitate with UAP56 following infection with the ADmUAPUL69 virus (Fig. 2A). To confirm that the mUAPUL69 mutant protein could no longer interact with URH49, HFF cells were cotransfected with a hemagglutinin (HA)-tagged URH49 and FLAG-tagged UL69 expression construct. Cotransfections were done, due to the lack of antibodies specific for URH49. Cell lysates were prepared 48 h posttransfection and subjected to immunoprecipitation with anti-FLAG antibody. Immunoprecipitates and cell lysates were then separated by SDS-PAGE, transferred to a membrane, and probed with HA antibody. As shown in Fig. 2B, URH49 coprecipitated with WT UL69. However, we were unable to detect an interaction between URH49 and the mUAPUL69 protein. These results confirm the previous results of Lischka et al. with regard to the alanine substitutions within UL69 that inhibit binding to UAP56 and URH49 (12) and also demonstrate the successful generation of a HCMV UL69 mutant defective for UAP56 and URH49 binding.
FIG. 2.
Interaction of WT and mutant UL69 with UAP56 and Spt6. (A through C) HFF cells were mock infected or infected with WT virus or ADmUAPUL69 (mUAP) virus at an MOI of 3 PFU/cell (A) or cotransfected with expression constructs expressing HA-tagged URH49 (URH49-H) and either FLAG-tagged WT UL69 or mUAPUL69 (mUAP) (B). Lysates were prepared 72 h postinfection or transfection and incubated with antibody against UAP56 (A), the FLAG epitope (B), or Spt6 (C). Immune complexes were collected via protein-A/G-agarose beads, separated by SDS-PAGE, transferred to membranes, and probed for UL69 or URH49 via Western blotting. Cell lysates were also examined for UL69, UAP56, URH49, and Spt6 expression by Western blot analysis. α-Tubulin was included as an internal loading control. (D) HFF cells were infected with WT virus or ADmUAPUL69 virus at an MOI of 0.5 PFU/cell. Forty-eight hours postinfection (hpi), cells were split onto coverslips. At 72 hpi, murine 3T3 cells were added to the coverslips and incubated for 4 h. Cycloheximide (50 μg/ml) was added to the cultures 30 min prior to and during the fusion process. One and a half hours after the fusion process, the cells were fixed with paraformaldehyde and stained with UL69 antibody and Hoechst stain. Virus-encoded green fluorescent protein (GFP) was used to aid in the identification of heterokaryons. Hoechst stain was used to distinguish HFF from 3T3 nuclei (3T3 nuclei display a punctate staining pattern, indicated with arrows). (E) Quantitative data for UL69 shuttling from three independent heterokaryon experiments. UL44 staining was included as a negative control for shuttling.
To confirm the specificity of the mutations and to demonstrate that the alanine substitutions did not abolish UL69's ability to bind other proteins, we assayed for the mutant UL69's ability to bind the cellular protein Spt6. Spt6 has previously been shown to interact with UL69 during infection (21). HFF cells were infected as described above, and the cell lysates were harvested at 72 h postinfection and subjected to immunoprecipitation with antibody directed against Spt6 (Bethyl Laboratories). As shown in Fig. 2C, we detected UL69 bound to Spt6 following infection with either WT or ADmUAPUL69 virus. However, we were unable to detect an interaction between Spt6 and the viral protein UL99, confirming the specificity of the Spt6/UL69 interaction. These results demonstrate that the mutations in UL69 that abolish binding to UAP56 and URH49 do not disrupt the global structure of the UL69 protein and that the UL69 mutant protein still retains its ability to bind Spt6 during infection.
Since UL69's nucleocytoplasmic shuttling activity is required for UL69's ability to shuttle unspliced transcripts from the nucleus in a reporter assay (12), we also examined UL69's ability to shuttle following infection with ADmUAPUL69 using a heterokaryon assay as previously described (10). HFF cells were infected with WT or ADmUAPUL69 virus at an MOI of 0.5 PFU/cell. At 72 hpi, virus-infected cells were fused with murine 3T3 cells in the presence of cycloheximide and then fixed and stained for UL69 expression. The presence of UL69 in the fused murine 3T3 nucleus indicates that nucleocytoplasmic shuttling has occurred. As shown in the representative images (Fig. 2D) or the quantitative heterokaryon data (Fig. 2E), infection with either WT or ADmUAPUL69 virus resulted in efficient UL69 nucleocytoplasmic shuttling. Staining for the viral UL44 protein served as a negative control for shuttling. The data in Fig. 2 demonstrate that UL69 expressed following infection with the ADmUAPUL69 virus is unable to bind UAP56/URH49 but still retains its ability to bind Spt6 and shuttle between the nucleus and the cytoplasm.
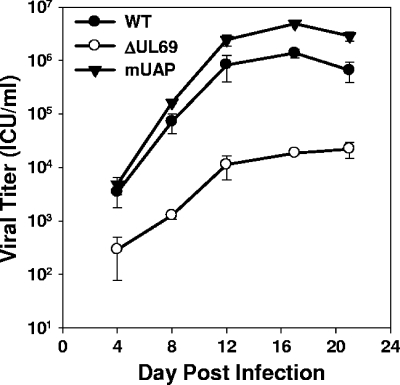
We next determined if UL69's ability to bind UAP56 and/or URH49 was required for efficient virus replication. HFF cells were infected with either WT, ADΔUL69, or ADmUAPUL69 virus at an MOI of 0.01 PFU/cell. Viral harvests were done at various time points postinfection, and titers were determined by infectious center assay as described previously (6). Despite being unable to bind UAP56 or URH49, the ADmUAPUL69 virus replicated with kinetics and to levels equal to or greater than those of the WT virus (Fig. 3), whereas the UL69 deletion mutant replicated more slowly and demonstrated a 1.5- to 2-log reduction in viral titers.
FIG. 3.
Inhibition of UL69's ability to interact with UAP56/URH49 does not affect virus replication. HFF cells were infected at an MOI of 0.01 PFU/cell with WT, ADΔUL69 (ΔUL69), or ADmUAPUL69 (mUAP) virus. Cultures were harvested at the indicated times postinfection, and infectious virus was quantified by infectious-center assay. Results represent the averages of three independent experiments.
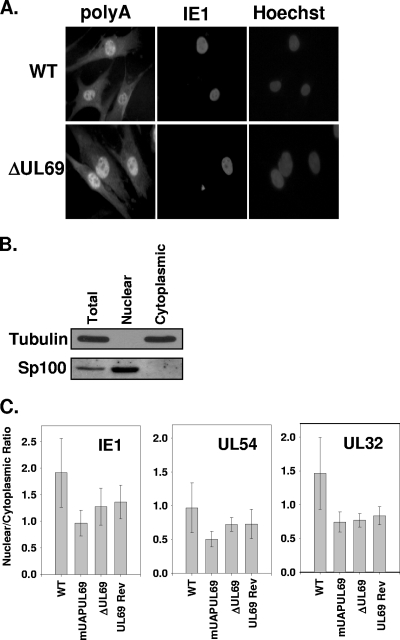
Given the lack of a growth phenotype associated with the UAP56/URH49 binding mutant, we asked if UL69 was playing a role in the export of RNAs from the nucleus to the cytoplasm during infection. It has been hypothesized that UL69 functions, at least in part, to transport viral mRNAs from the nucleus to the cytoplasm during infection. However, this hypothesis has not been tested in the context of a viral infection. To test the hypothesis, we infected HFF cells grown on coverslips with either WT virus or the UL69 deletion mutant at an MOI of 0.5 PFU/cell. Cells were then fixed at 72 h postinfection, and total mRNA was detected by in situ hybridization using a streptavidin-labeled oligo(dT) probe followed by fluorescence detection using a streptavidin-Alexa Fluor 546-conjugated antibody as previously described (7). In addition, the cells were stained for IE1 expression using a monoclonal antibody, and nuclei were stained with Hoechst stain. As shown in Fig. 4 A, we did not observe a difference in the mRNA staining pattern when comparing WT- and ADΔUL69-infected cells. Similar results were obtained at 24 and 48 h postinfection (data not shown). Since the data in Fig. 4A represent staining of total mRNA, we cannot rule out the possibility that UL69 selectively transported viral RNAs during infection. To begin to address this possibility, we analyzed the nuclear and cytoplasmic levels of three HCMV transcripts following infection. Cells were infected with WT, ADmUAPUL69, ADΔUL69, or ADUL69Rev virus at an MOI of 1 PFU/cell. At 72 h postinfection, cells were harvested, and nuclear and cytoplasmic fractions were isolated, using a PARIS kit (Ambion) according to the manufacturer's instructions. To confirm our fractionation protocol, total, nuclear, and cytoplasmic fractions were isolated and examined for expression of either the nuclear protein SP100 or the cytoplasmic protein tubulin. As shown in Fig. 4B, tubulin is present only in the total and cytoplasmic fractions, and SP100 is present only in the total and nuclear fractions. We then isolated RNA from the nuclear and cytoplasmic fractions and assayed each fraction for the abundance of an HCMV immediate-early (IE1), an early (UL54), and a late (UL32) transcript by quantitative PCR as described previously (7). Briefly, 200 ng of RNA was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen), and cDNA was amplified using validated TaqMan primers/probes on an ABI 7900HT real-time thermal cycler (Applied Biosystems) running SDS version 2.3 software. The resulting threshold cycle (CT) values were then compared to the CT values from a standard curve generated by 10-fold serial dilutions of HCMV genomic DNA for each transcript to determine the absolute starting quantity of RNA in each fraction. The average nuclear-to-cytoplasmic ratio of the transcripts from four independent experiments was calculated and plotted. If UL69 were involved in the nuclear export of viral transcripts during infection, we would expect to observe an increase in the nuclear accumulation of viral transcripts following infection with the UL69 deletion mutant compared to that for the WT virus. As shown in Fig. 4C, we did not observe a significant difference (all P values greater than 0.2) in the nuclear-to-cytoplasmic ratio of the three transcripts tested following infection with the WT, ADmUAPUL69, ADΔUL69, or ADUL69Rev virus. These results suggest (at least for the transcripts tested) that UL69 does not play a significant role in the transport of at least a subset of viral transcripts from the nucleus to the cytoplasm during infection. Taken together, our data indicate that UAP56 or URH49 binding to UL69 is not necessary for efficient viral replication or the transport of viral transcripts from the nucleus to the cytoplasm. In addition, it demonstrates that another function associated with UL69 may be responsible for the growth defect associated with the UL69 deletion mutant.
FIG. 4.
Expression of UL69 does not affect shuttling of RNA during HCMV infection. (A) HFF cells were infected at an MOI of 0.5 PFU/cell with either WT or ADΔUL69 (ΔUL69) virus. Seventy-two hours postinfection, cells were fixed and assayed for mRNA localization within the cells by in situ hybridization, using a biotinylated oligo(dT) probe, and were visualized by fluorescence staining with streptavidin-Alexa Fluor 546-conjugated antibody, followed by immunofluorescence staining for the HCMV IE1 protein. (B) HFF cells were infected with either WT, ADmUAPUL69 (mUAPUL69), ADΔUL69 (ΔUL69), or ADUL69Rev (UL69Rev) virus at an MOI of 1 PFU/cell. Seventy-two hours postinfection, cells were harvested, and nuclear and cytoplasmic fractions were isolated using a PARIS kit (Ambion) according to the manufacturer's instructions. Total, nuclear, and cytoplasmic proteins were separated via SDS-PAGE, transferred to nitrocellulose membranes, and subjected to Western blot analysis using antibodies directed against either the cytoplasmic protein α-tubulin or the nuclear protein Sp100. (C) Quantitative PCR was performed on cDNA generated from both the nuclear and cytoplasmic fractions of cells infected with the indicated viruses. Primers and probes specific to an immediate early (IE1), an early (UL54), and a late (UL32) gene were used to amplify and quantify the amount of each transcript present in each fraction. No statistical difference (all P values were >0.2) in the ratios was observed for any of the viral infections.
One mechanism by which UL69 has been proposed to regulate HCMV replication is by functioning as an mRNA export factor that facilitates the shuttling of unspliced viral messages from the nucleus to the cytoplasm. UL69's interaction with the cellular ATP-dependent RNA helicases UAP56 and URH49 has been proposed to be necessary for the export of viral transcripts from the nucleus and to facilitate efficient viral replication (10, 12, 20). In this study, we confirmed that UL69 is required for efficient HCMV replication at low MOIs and demonstrated through the use of a viral mutant that binding of UL69 to UAP56 or URH49 is not required for efficient HCMV replication and does not contribute to the growth phenotype observed with a UL69 deletion mutant. We cannot, however, conclude from our data that UL69 does not function as a viral mRNA export factor during HCMV infection, but rather that UL69 binding of UAP56/URH49 is not necessary for this proposed function. As suggested by Toth and Stamminger, UL69 may function as an export factor by binding to the cellular protein Spt6. Spt6 binds to Ser2-phosphorylated RNA polymerase II and recruits the mRNA export factor REF to target genes via Spt6's direct interaction with the Iws1 protein, thereby providing a potential mechanism for mRNA export that does not involve UL69 binding to UAP56 or URH49 (20). However, that we did not observe a difference in the overall nuclear retention of mRNA or the nuclear-to-cytoplasmic ratio of three HCMV transcripts following infection with the UL69 deletion mutant suggests that UL69 may not function as an export factor but rather regulates viral replication or gene expression through another mechanism. One possible mechanism may involve UL69's interaction with Spt6, a known chromatin-remodeling factor that binds histone H3 (2) and functions as a transcriptional elongation factor (4). UL69 has also recently been shown to interact with the mRNA cap binding complex, and it has been proposed that UL69 functions to support translation in infected cells by excluding the inhibitory 4EBP1 protein from the cap binding complex (1). These hypotheses are currently being examined through the generation of UL69 mutants that are unable to bind Spt6 and eIF4A1. Future experiments with other UL69 mutants should help identify the mechanism by which UL69 functions during HCMV infection and contributes to the growth phenotype observed with the UL69 deletion mutant.
Acknowledgments
We are grateful to Michael Greene for providing the UAP56 antibody, Tom Shenk for the UL69 antibody, and Steve Rice and Peter Southern for critical readings of the manuscript.
This work was supported by NIH grant AI059340 (to W.A.B).
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Aoyagi, M., M. Gaspar, and T. E. Shenk. 2010. Human cytomegalovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4EBP1. Proc. Natl. Acad. Sci. U. S. A. 107:2640-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 3.Cantrell, S. R., and W. A. Bresnahan. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80:6188-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endoh, M., W. Zhu, J. Hasegawa, H. Watanabe, D. K. Kim, M. Aida, N. Inukai, T. Narita, T. Yamada, A. Furuya, H. Sato, Y. Yamaguchi, S. S. Mandal, D. Reinberg, T. Wada, and H. Handa. 2004. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 24:3324-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. U. S. A. 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huh, Y. H., Y. E. Kim, E. T. Kim, J. J. Park, M. J. Song, H. Zhu, G. S. Hayward, and J. H. Ahn. 2008. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J. Virol. 82:10444-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson, L. A., and R. M. Sandri-Goldin. 2009. Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. J. Virol. 83:1184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalejta, R. F. 2008. Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression. Curr. Top. Microbiol. Immunol. 325:101-115. [DOI] [PubMed] [Google Scholar]
- 9.Kalejta, R. F. 2008. Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 72:249-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lischka, P., M. Thomas, Z. Toth, R. Mueller, and T. Stamminger. 2007. Multimerization of human cytomegalovirus regulatory protein UL69 via a domain that is conserved within its herpesvirus homologues. J. Gen. Virol. 88:405-410. [DOI] [PubMed] [Google Scholar]
- 12.Lischka, P., Z. Toth, M. Thomas, R. Mueller, and T. Stamminger. 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 26:1631-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo, M. L., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644-647. [DOI] [PubMed] [Google Scholar]
- 15.Pryor, A., L. Tung, Z. Yang, F. Kapadia, T. H. Chang, and L. F. Johnson. 2004. Growth-regulated expression and G0-specific turnover of the mRNA that encodes URH49, a mammalian DExH/D box protein that is highly related to the mRNA export protein UAP56. Nucleic Acids Res. 32:1857-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rechter, S., G. M. Scott, J. Eickhoff, K. Zielke, S. Auerochs, R. Muller, T. Stamminger, W. D. Rawlinson, and M. Marschall. 2009. Cyclin-dependent kinases phosphorylate the cytomegalovirus RNA export protein pUL69 and modulate its nuclear localization and activity. J. Biol. Chem. 284:8605-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas, M., S. Rechter, J. Milbradt, S. Auerochs, R. Muller, T. Stamminger, and M. Marschall. 2009. Cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. J. Gen. Virol. 90:567-578. [DOI] [PubMed] [Google Scholar]
- 19.Toth, Z., P. Lischka, and T. Stamminger. 2006. RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 34:1237-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toth, Z., and T. Stamminger. 2008. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 13:2939-2949. [DOI] [PubMed] [Google Scholar]
- 21.Winkler, M., T. aus Dem Siepen, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]