Abstract
We report here investigation into the genetic basis of mouse hepatitis virus strain 1 (MHV-1) pneumovirulence. Sequencing of the 3′ one-third of the MHV-1 genome demonstrated that the genetic organization of MHV-1 was similar to that of other strains of MHV. The hemagglutinin esterase (HE) protein was truncated, and reverse transcription-PCR (RT-PCR) studies confirmed previous work that suggested that the MHV-1 HE is a pseudogene. Targeted recombination was used to select chimeric viruses containing either the MHV-1 S gene or genes encoding all of the MHV-1 structural proteins, on an MHV-A59 background. Challenge studies in mice demonstrated that expression of the MHV-1 S gene within the MHV-A59 background (rA59/SMHV-1) increased the pneumovirulence of MHV-A59, and mice infected with this recombinant virus developed pulmonary lesions that were similar to those observed with MHV-1, although rA59/SMHV-1 was significantly less virulent. Chimeras containing all of the MHV-1 structural genes on an MHV-A59 background were able to reproduce the severe acute respiratory syndrome (SARS)-like pathology observed with MHV-1 and reproducibly increased pneumovirulence relative to rA59/SMHV-1, but were still much less virulent than MHV-1. These data suggest that important determinants of pneumopathogenicity are contained within the 3′ one-third of the MHV-1 genome, but additional important virulence factors must be encoded in the genome upstream of the S gene. The severity of the pulmonary lesions observed correlates better with elevated levels of inflammatory cytokines than with viral replication in the lungs, suggesting that pulmonary disease has an important immunological component.
Severe acute respiratory syndrome (SARS) is an infectious disorder that was first diagnosed in China in November 2002 and subsequently spread worldwide (22, 46, 55). SARS was documented in over 8,000 persons, with 778 deaths (55) before the outbreak was extinguished. Electron microscopy, virus isolation, cloning, and sequencing studies demonstrated that a novel coronavirus (SARS-CoV) was the etiologic agent of SARS (10, 19, 26, 35, 41). Shortly thereafter, the coronavirus etiology of SARS was confirmed when Koch's postulates were fulfilled using cynomolgus macaques (Macaca fuscicularis) (12). In 2003 and 2004, laboratory-associated cases in Singapore, Taiwan, and Beijing were reported, as were four non-laboratory-associated cases in Guandong Province, People's Republic of China (PRC) (53, 54), underlining the possibility of reemergence of SARS.
The SARS coronavirus (SARS-CoV) represents a zoonotic outbreak likely transmitted to humans from palm civets and raccoon dogs in wild animal markets, although these animals do not appear to be the natural host for the virus (15). Recently, SARS-like coronaviruses were identified in Chinese horseshoe bats, making these animals the likely animal reservoir of the SARS-CoV (21, 24). Phylogenetic analyses place the SARS-CoV in group 2b, an early branch of the group 2 coronaviruses.
The group 2 coronaviruses also contain the mouse hepatitis viruses (MHVs) (44), a group of viruses that have been studied extensively in rodent models of human diseases (50). A number of diseases are produced by different strains of MHV, and the disease produced is also somewhat dependent upon the route of inoculation and the age and strain of the recipient mouse. Strain MHV-JHM (also called MHV-4) is strongly neurotropic, produces minimal hepatic lesions, and is poorly pneumotropic. MHV-A59 is neurotropic and hepatotropic, and MHV-3 and MHV-2 are strongly hepatotropic. Studies of MHV-JHM, MHV-A59, and MHV-2 utilizing targeted recombination to create chimeric viruses have demonstrated that the viral spike (S) protein has a major effect on pathogenesis and tropism (7, 27, 28, 37, 39). This is not surprising since the S protein recognizes CEACAM-1, the cellular receptor for MHV, on the plasma membrane and mediates virus entry into the cell as well as cell-to-cell spread during infection (51). Other MHV genes influence pathogenesis as well (6, 16, 28, 29, 31, 45).
Recently we identified a strain of MHV, MHV-1, that is highly pneumotropic, produces high mortality, and reproduces most of the pathological features of SARS after intranasal challenge of A/J mice (9). In this work, we began to examine the genetic basis for MHV-1 pneumotropism and pneumovirulence. Sequencing of the 3′ one-third of the MHV-1 genome coding for the structural proteins demonstrated that the genetic organization of MHV-1 was generally similar to that of other strains of MHV. The open reading frame (ORF) encoding the hemagglutinin esterase (HE) protein is truncated, and subgenomic mRNA 2b is not transcribed; thus, the HE gene is a pseudogene. Chimeric viruses containing either the MHV-1 S gene or genes encoding all of the MHV-1 structural proteins, on MHV-A59 and MHV-JHM backgrounds, were generated by targeted recombination. Challenge studies with mice demonstrated that the MHV-1 S gene conferred pneumotropism and a low degree of pneumovirulence in an MHV-A59 background, but not in an MHV-JHM background. Infections with chimeric viruses containing all of the MHV-1 structural genes in an MHV-A59 background reproduced the SARS-like pathology observed with MHV-1 and increased pneumovirulence relative to the MHV-A59/MHV-1-S chimeric virus, but were still considerably less virulent than MHV-1. These data suggest that important determinants of pneumopathogenicity are contained within the 3′ one-third of the MHV-1 genome, but additional virulence factors must lie upstream of the S gene.
MATERIALS AND METHODS
Cells and virus.
DBT and 17Cl-1 were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum (HyClone, Logan, UT). FCWF (Felis catus whole fetus) cells were grown in DMEM supplemented with 10% fetal bovine serum. L2 cells were maintained at 37°C and 3% CO2 in DMEM supplemented with 10% calf serum. The origin and growth of the MHV-1 (9), recombinant MHV-A59 (rA59) (38), recombinant MHV-JHM (rJHM) (29), MHV-3 (23), and MHV-S (9) viruses used in this study have been described previously. The rJHM strain used here is derived from the highly neurovirulent strain JHM.SD (originally called MHV-4) (32). Recombinant feline A549 (fA59) (20) and feline JHM (fJHM) (33) viruses contained the feline infectious peritonitis virus (FIPV) spike protein ectodomain in place of the MHV-A59 and MHV-JHM ectodomains and were propagated in FCWF cells.
Sequencing of MHV-1.
Total RNA was extracted from MHV-1-infected 17Cl-1 cells using an RNeasy minikit (Qiagen), and cDNAs were synthesized with reverse transcriptase (Superscript II; Invitrogen). Overlapping cDNAs were amplified by PCR using primers based on the MHV-1 HE pseudogene (GenBank accession no. M64313), a partial S gene (GenBank accession no. D8333), and the N gene (GenBank accession no. M35253) sequences present in the public databases and a mixture of Vent and recombinant Tth-XL (rTth) DNA polymerases (30). The resulting amplified cDNAs were purified by gel electrophoresis and subjected to automated sequencing without cloning. The complete list of oligonucleotides used for reverse transcription-PCR (RT-PCR) and sequencing can be obtained by contacting the authors.
Generation and amplification of MHV-1 cDNA clones.
DBT cells were infected with MHV-1 at a multiplicity of 0.3, and total RNA was extracted at 7 h postinfection using an RNEasy minikit. The freshly prepared RNA was used as a template for cDNA synthesis (Superscript II; Invitrogen) with primers specific to regions containing the structural genes. For the region encompassing the HE gene from codon 28 extending into the E gene, cDNA was primed with an oligonucleotide designated E60-40rev with the sequence GACTATAAAAATAATCTGCC. The resulting cDNA was PCR amplified with the forward primer HE28-BsmBI-FW-new (sequence GTACGTCTCAACCTCTCAACATCGTTTCAC), containing a BsmBI site at its 5′ end to facilitate later recombinant DNA manipulations not described in this work, and the reverse primer E60-40Rev, using a mixture of Vent and rTth DNA polymerases as described previously (30). The resulting amplicon, designated HE-S, was TA cloned into pGEM-T Easy (Promega). Similarly, a cDNA that would encompass the 3′ end of the genome was synthesized by reverse transcription-PCR (RT-PCR) using primer N-RV-SfiI [GGCCATTTAGGCCTTAATTAA(T)15GTGATTCTTCAATTGGCC] for the cDNA synthesis reaction and amplified with S45-FW5 (GCGATTGGTGCTATACAGG, nucleotides [nt] 4109 to 4128 in GenBank accesssion no. EF682498) and N-RV-SfiI primers for the PCR step. The amplified DNA fragment extending from nucleotide 4109 to the 3′ poly(A) tail in GenBank accession no. EF682498 was subsequently TA cloned into pGEM-T Easy, and two clones, designated SN#4 and SN#13, were obtained. A cDNA extending from positions 1379 to 5477 and containing the S gene from codon 9 was similarly RT-PCR amplified with forward primer CCTAATACCCTCTTGCCTAGGGTATATTGGTGACTT and reverse primer CTGTCCTTCCACCTGCAGGTGTACATCTAGTCAATCCTCG. To allow subsequent recombinant DNA manipulations, the forward primer contained two coding silent mutations in codons 13 and 14 of the spike gene (T1394C and T1395C relative to GenBank accesssion no. EF682498) to introduce an AvrII site (underlined in the primer sequence). The reverse primer introduced mutations G5458C, C5460T, T5461G, T5463A, C5464G, and C5465G (all mutations are in the positive sense) to introduce the SbfI site (underlined in the primer sequence) 3′ to the spike gene termination codon and prior to the gene 4 transcriptional regulatory sequence. The resultant cDNA fragment was TA cloned into PCR 2.1 Topo and designated clone 4B. All clones were sequenced in their entirety.
Plasmid constructions.
All positions for oligonucleotides or mutagenesis are given relative to GenBank accesssion no. EF682498. The MHV-1 spike gene 4B clone contained an AvrII site at positions 4734 to 4739, in addition to the AvrII site engineered into the 5′ end of the clone by RT-PCR. The internal AvrII site at 4734 to 4739 was removed without introducing a coding change by PCR-mediated mutagenesis (mutations T4736A and G4739A), to produce cDNA clone 4B1. This allowed the subsequent use of the AvrII and SbfI sites that were introduced into the 5′ and 3′ ends of this clone during RT-PCR cloning (see above), to transfer the MHV-1 spike gene to the targeted recombination plasmids pMH54 (for MHV-A59) (20) and pJHM (for MHV-JHM) (33), to produce plasmids pMH54/SMHV-1 and pJHM/SMHV-1, which were used in the targeted recombination studies described below. Additionally, site-directed mutagenesis was used to introduce two additional coding silent mutations into clone 4B1 (T2384A and G2552A) to destroy two BsmBI sites that would interfere with subsequent recombinant DNA manipulations not described here. The resultant plasmid was designated 4B1ΔBsmBI.
A cDNA clone extending from codon 28 of the HE gene into the E gene, designated clone HE-S#3, underwent site-directed mutagenesis to insert an AvrII site at nucleotides 1394 to 1399 (T1394C and T1395C). This was accomplished by site-directed mutagenesis of a subcloned AgeI-StuI fragment followed by restriction fragment exchange to introduce the AvrII site into HE-S#3 to produce cDNA clone HE-S-AvrII. Clone SN#13, a cDNA clone extending from position 4109 into the poly(A) tail followed by PacI and SfiI sites, was subjected to site-directed mutagenesis to insert an SbfI site at nucleotides 5459 to 5466, just downstream of the S gene stop codon. Site-directed mutagenesis was performed by PCR amplification of a fragment containing nucleotides 5453 to 6047 using clone SN#13 as template, a mutagenic forward primer (TGTACACCTGCAGGGGAAGGACAGAAAATCTAAAC; the introduced SbfI site is underlined, and a naturally occurring BsrGI site is double underlined), and a reverse primer containing an AvaI restriction site [CTCGGGGTTGGCAAT; the AvaI site is underlined]) followed by TA cloning. The introduced SbfI site was then shuttled back into clone SN#13 by restriction fragment exchange using an AvaI site at positions 6042 to 6047 and a BsrGI site (nt 5453 to 5458) immediately 5′ to the introduced SbfI site. SN#13 then underwent an additional restriction fragment exchange with its sibling clone SN#4 to eliminate two mutations in the N gene to produce a clone, designated SNF2. Clones HE-S-AvrII and SNF2 were ligated together utilizing a shared unique BglII site (nucleotides 4240 to 4205) and a 3′ flanking SalI site in pGEM-T Easy to produce the plasmid pHe-N, a construct that containing the HE gene from codon 28 to the poly(A) tail. The introduced AvrII and SbfI sites were then used to replace the majority of the S coding sequences with those in clone 4B1ΔBsmBI to correct errors that were introduced into the S gene during RT-PCR-based cloning and to facilitate future recombinant DNA manipulations at the 5′ end of the construct that will utilize the BsmBI site at the 5′ end of the construct using “no see'm technology” (58). This clone, designated pMHV-1ΔBsmBI, corresponded to the wild-type sequence from HE codon 28 through the 3′ end, with the exception of the noncoding changes in the S gene described above, a coding silent T6404C change in the E gene, a T6032C mutation in gene 5a that changes a tyrosine to a histidine in the gene 5a protein, and an extra G which was inadvertently inserted after the SbfI site located in a noncoding region between the S and open reading frame 4 (ORF4) genes.
The MHV-1 sequences downstream of S extending to the 3′ end of the genome were excised from pHe-N using SfbI and PacI and were placed into pMH54/SMHV-1 and pJHM/SMHV-1 by restriction fragment exchange to produce the plasmids pMH54/S-3′UTRMHV-1 and pJHM/S-3′UTRMHV-1, respectively. These plasmids were subsequently used in targeted recombination studies described below.
Targeted recombination.
After digestion with PacI to linearize the plasmids, donor RNAs were transcribed with T7 RNA polymerase as previously described from plasmids pMH54/SMHV-1 and pMH54/S-3′UTRMHV-1 for isolation of MHV-A59/MHV-1 recombinant viruses and from pJHM/SMHV-1 and pJHM/S-3′UTRMHV-1 for isolation of MHV-JHM/MHV-1 recombinant viruses (20, 33). Targeted recombination with MHV-A59 and MHV-JHM was performed as previously described (20, 33). Briefly, two viruses, fMHV (20), corresponding to MHV-A59, in which the sequences encoding the spike protein ectodomain had been replaced by the corresponding feline infectious peritonitis virus (FIPV) spike ectodomain coding sequences, and fJHM (20, 33), in which the MHV-JHM spike ectodomain coding sequence had similarly been replaced by the FIPV ectodomain sequences, were used as acceptor viruses. FCWF cells were infected with either fMHV or fJHM and incubated for 4 h. Cells were then electroporated with donor RNAs with two pulses of 300 V and 960 μF and overlaid onto monolayers of 17CL-1 cells. The cultures were incubated for up to 72 h or until cytopathic effect destroyed the 17CL-1 monolayer. Recombinant viruses able to enter and replicate in murine cells, and thus which contained the MHV-1 spike ectodomain, were selected by plaquing on L2 cells. Plaques containing putative recombinant viruses were picked and underwent a second cycle of plaque purification. The viruses twice plaque cloned were expanded in murine cells, and their recombinant nature was confirmed by sequencing.
RNA analyses.
For metabolic labeling studies, replicate cultures of DBT cells in 12-well plates were infected with MHV at a multiplicity of infection (MOI) of 1, or mock infected, and incubated for 6 h. Cultures were then washed twice with phosphate-free DMEM and fed with DMEM containing 2% dialyzed calf serum and 10 μg/ml actinomycin D. After 15 min of incubation at 37°C, cells were labeled by replacing the medium with fresh phosphate-free DMEM containing 10 μg/ml actinomycin D, 2% dialyzed serum, and 200 μCi/ml 32PO4 and incubated for an additional 5.5 h. The labeled cultures were washed twice with cold phosphate-buffered saline (PBS), and RNA was extracted using an RNeasy minikit. Ten micrograms of RNA was electrophoresed in a 1% formaldehyde-agarose gel at 100 V for 5 h. Following electrophoresis, the gel was soaked in 70% methanol for 30 min, dried, and exposed to X-ray film.
RT-PCR analysis was performed on RNA extracted from cells infected with MHV at 7 h postinfection. cDNA synthesis was primed with an antisense oligonucleotide corresponding to positions 420 to 402 of the HE gene (CAAAGGATAGGCTCCTATA). Forty cycles of PCR amplification was carried out with a sense primer corresponding to nucleotides 1 to 23 of the MHV leader sequence (GTATAAGAGTGATTGGCGTCCGT) and the same antisense primer used for cDNA synthesis. Amplified DNA fragments were displayed by agarose gel electrophoresis.
Cytokine and chemokine analyses.
Relative amounts of mRNA encoding macrophage colony-stimulating factor (M-CSF), fibrinogen-like protein 2 (FGL2), tumor necrosis factor alpha (TNF-α), alpha 1 interferon (IFN-α1), IFN-α2, IFN-α4, IFN-α5, IFN-β, IFN-γ, interleukin-6 (IL-6), chemokine (C-C motif) ligand 2 (CCL2), CCL3, CCL5, and chemokine (C-X-C motif) ligand 2 (CXCL2) were determined using the GeXP genetics analysis system (Beckman Coulter, Inc., Fullerton, CA). Total RNA was extracted from lung tissue using the RNeasy minikit following the manufacturer's protocol. Primers for multiplex RT-PCR were designed using the primer design module of the GeXP Express Profiler software system (Beckman Coulter). The 5′ end of each reverse primer was designed to contain the 19-nucleotide universal tag sequence 5′-GTACGACTCACTATAGGGA-3′. Similarly, the 5′ end of each forward primer was designed to contain the 18-nucleotide universal tag sequence 5′-AGGTGACACTATAGAATA-3′. Primer pairs were designed to yield products ranging from 142 bp to 330 bp, with a spacing of at least 5 nucleotides. The reverse primer mix contained 500 nmol/liter of each reverse primer. The forward primer mix contained 200 nmol/liter of each forward primer. cDNAs were synthesized in a reaction mixture containing 1 μl of total RNA, 3 μl of nuclease-free water, 4 μl of 5× reverse transcriptase buffer (Beckman Coulter, Inc., Fullerton, CA), 2 μl of the reverse primer mix, and 1 μl of Moloney murine leukemia virus reverse transcriptase (Beckman Coulter). The reverse transcription reaction mixtures were incubated for 1 min at 48°C, 5 min at 37°C, and 60 min at 42°C and finally for 5 min at 95°C. Multiplex PCRs were assembled as follows: 9.3 μl reverse transcription reaction mixture, 4 μl 5× PCR buffer (Beckman Coulter), 2 μl forward primer mix, 4 μl of 25 mM MgCl2 and 0.7 μl Thermo-Start DNA polymerase (Thermo Scientific). Thirty-five cycles of PCR amplification were performed using the following conditions: 10 min at 95°C, 30 s at 94°C, 30 s at 55°C, and 1 min at 68°C. All PCRs were performed in duplicate. The multiplex PCR products were analyzed by capillary electrophoresis on a GeXP genetic analysis system (Beckman Coulter). Data analysis was conducted using the eXpress Analysis module of the GeXP Express Profiler software system (Beckman Coulter). Relative gene expression in each sample was compared by normalizing it against a housekeeping gene (coding for GAPDH [glyceraldehyde-3-phosphate dehydrogenase]). Data are presented as changes in expression relative to the untreated control samples ± standard error (n = 3 for each cytokine and time point).
Serum cytokine levels (IL-6, IL-10, CCL2 [monocyte chemoattractant protein 1; MCP-1], IFN-γ, TNF-α, and IL-12 p70) were assayed using the Cytometrix bead array kit for inflammation (BD Biosciences Mississauga, ON, Canada) as previously described (9). Serum samples were processed as per the manufacturer's instructions and analyzed using a Becton Dickinson FACScan flow cytometer.
Mice.
Female A/J mice 6 to 8 weeks of age (Charles River Laboratories and Jackson Laboratories) were maintained in microisolator cages and fed standard lab chow diet and water ad libitum. Viral infections were performed in a viral isolation room as previously described (9). Briefly, mice were anesthetized by intraperitoneal injection with pentobarbital diluted in normal saline, 50 μl of virus diluted in ice-cold DMEM was instilled into the nares, and mice were observed until the virus was inhaled. Mice were monitored daily for symptoms of disease, including ruffled fur, tremors, respiratory distress, and lack of activity. Mice having any combination of three of these clinical signs were considered to be in extremis and were euthanized. At various times postinfection, mice were sacrificed by lethal pentobarbital anesthesia. Lungs, livers, brains, and spleens were collected and analyzed for viral titers and histology as previously described (9). Viral titers were determined by plaque assay on monolayers of L2 cells as previously described. All protocols were approved by Animal Welfare Committees and complied with NIH and CIHR guidelines.
Histology.
Tissue samples were formalin fixed, embedded in paraffin, sectioned, stained with hematoxylin and eosin, and read by a board-certified pathologist (M.J.P.) who was blinded to the clinical aspects of the study. Sections of the lung from all groups and controls were examined by light microscopy. Assessment of the degree of lung pathology was made. Since the main pathology was interstitial pneumonitis with reduction in pulmonary open alveolar air spaces, a scoring scheme based on estimated airway space visualized was used as follows: fully expanded airways, normal (N); 0 to 30% reduction in airway space, mild lung disease (+); 31 to 60% reduction in airway space, moderate lung disease (++); 61 to 90% reduction in airways, severe lung disease (+++); and 91 to 100% reduction, severe lung disease with consolidation (++++).
Statistics.
Viral titers were analyzed by analysis of variance (ANOVA) to determine if replication of the recombinant viruses was different from that of parental MHV-1. A P value of 0.05 was considered statistically significant.
RESULTS
Sequence analysis and molecular cloning of the MHV-1 structural gene region.
As a first step in performing reverse genetics studies to investigate the basis of MHV-1 pneumovirulence, we determined the sequence of 8,905 nucleotides of the MHV-1 genome extending from the first nucleotide of the HE pseudogene to the poly(A) tail (GenBank accession no. EF682498). This region includes all of the MHV structural proteins as well as genes 4 and 5a, which encode nonstructural proteins. Analysis of the open reading frames present indicates the genetic structure of MHV-1 is similar to those of other MHV strains (Fig. 1 A). There are several notable features. As reported previously (57), the HE gene was a pseuodgene (see below) containing an open reading frame truncated to 268 amino acids as compared to 440 amino acids in the intact and functional MHV-JHM HE gene. Gene 4 contained a single 139-amino-acid open reading frame like MHV-JHM, rather than the two smaller open reading frames found in MHV-A59. The S gene encoded a protein of 1,363 amino acids generally similar to other MHV S proteins that have been sequenced. The sequence at the MHV-1 S1-S2 cleavage site, TSHRARRS, differed only slightly from the KSRRARRS and KSRRAHRS cleavage sites (cleavage is between the underlined residues) found in MHV-JHM (43) and MHV-A59 S proteins (25), respectively, suggesting that the MHV-1 spike protein will be cleaved as are those of MHV-JHM and MHV-A59. Cell-cell fusion was observed in MHV-1-infected cell cultures, consistent with the presence of a putative cleavage site in MHV-1 S. The MHV-1 S protein has four small deletions of four, four, three, and two amino acids relative to the MHV-JHM S protein, the longest S protein reported. These deletions are all outside of the receptor binding domain, residues 1 to 330. However, the two four-amino-acid deletions in MHV-1 ablate the C57BL/6 immunodominant cytotoxic T-lymphocyte (CTL) epitope (CSLWNGPHL; S-510-518) (3) that is present in the MHV-JHM S protein. The MHV-JHM subdominant epitope (RCQIFANI; S-598-605) (3) is not conserved in MHV-1; the corresponding sequence is rather RCHIFSNL. However, this sequence contains the anchor residues needed for major histocompatibility complex (MHC) binding and is predicted to be a C57BL/6 (H-2b) CTL epitope by the Promiscuous MHC Class-I Binding Peptide Prediction Server (http://www.imtech.res.in/raghava/propred1/). Alignment of the MHV-1, MHV-JHM, MHV-A59, and MHV-2 S proteins by ClustalW and calculation of a distance matrix indicate the MHV-1 S protein sequence is somewhat more similar to the MHV-2 S protein than to either the MHV-JHM or MHV-A59 S proteins.
FIG. 1.
The MHV-1 HE gene is not expressed. (A) The genetic organization of the 3′ 8,905 nucleotides of the MHV-1 genome. The vertical lollipops represent the TRS sequences, including the nonfunctioning TRS (gray lollipop) for the truncated HE open reading frame. (B) RT-PCR analysis of MHV-1-infected cell RNA. Replicate cultures were infected with MHV-S, MHV-1, MHV-3, and MHV-A59, and RNA was extracted at 7 h postinfection. cDNA synthesis was primed with an antisense oligonucleotide corresponding to positions 420 to 402 of the HE gene. PCR was carried out with a sense primer corresponding to nucleotides 1 to 23 of the MHV leader sequence and the same antisense primer used for cDNA synthesis. Amplification was for 25 cycles at 95°C for 15 s, 48°C for 45 s, and 72°C for 50 s followed by a 5-min final elongation step at 72°C. The PCR products were then resolved by agarose gel electrophoresis. The position of the 492-nucleotide HE mRNA amplicon is indicated by the arrow.
The MHV-1 HE gene is a pseudogene.
It has been reported previously that a subgenomic mRNA containing the HE gene cannot be detected in metabolic labeling studies of MHV-1-infected cells (57). In light of the fact that the transcriptional regulatory sequences (TRSs) immediately upstream of the MHV-JHM and MHV-S and MHV-1 HE coding sequences are identical (57), we reexamined this question. Metabolic labeling of MHV-1-infected cells with 32Pi failed to demonstrate the presence of mRNA 2-1 (not shown). Furthermore, the HE gene encoding mRNA 2-1 was not detected in MHV-1-infected cells by a more sensitive RT-PCR assay that employed leader- and HE-specific primers, although the presence of the HE gene encoding mRNA 2-1 was easily demonstrated in MHV-S- and MHV-3-infected cells (Fig. 1B). As expected, cells infected with MHV-A59, a strain reported not to express mRNA 2-1 or the HE protein, did not give rise to a PCR product. Thus, we concluded that an mRNA expressing the predicted truncated version of the HE protein is not detectable in MHV-1-infected cells.
Isolation and in vitro characterization of rA59/MHV-1 recombinant viruses.
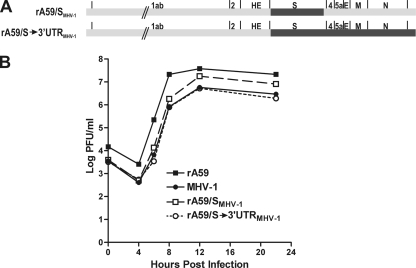
Previous studies have shown that in the context of A59/JHM chimeric viruses, the S gene plays a major role in tropism and virulence (7, 27, 37, 39). Further work with additional A59/JHM chimeras has demonstrated that other structural genes, in addition to the S gene, but not the replicase gene, are major determinants of tropism and level of virulence (14, 28, 29). Thus, we isolated recombinant MHV-A59 viruses in which the A59 S gene was replaced by MHV-1 S (designated rA59/SMHV-1) (Fig. 2 A) by targeted recombination (20), and a second set of recombinants in which the A59 sequences from S through the 3′ end of the genome were replaced by their MHV-1 counterparts (designated rA59/S→3′UTRMHV-1) (Fig. 2A). The genomes of these recombinant viruses were partially sequenced to verify their genetic structure. To ensure that these recombinant viruses replicated comparably to the parental viruses in cell culture, a growth curve experiment comparing the growth rates of rA59, MHV-1, rA59/SMHV-1, and rA59/S→3′UTRMHV-1 in L2 cells was performed (Fig. 2B). All viruses replicated with similar kinetics, and the viruses containing all of the MHV-1 structural genes, MHV-1 and rA59/S→3′UTRMHV-1, reached virtually identical titers. rA59/SMHV-1, a virus that contained A59 structural genes, with the exception of the MHV-1 S gene, reached a higher titer than those of MHV-1 and the rA59/S→3′UTRMHV-1 recombinant, a titer that was only slightly less than that achieved by rA59. We concluded that there were no obvious incompatibilities between the MHV-1 and MHV-A59 structural genes and that the rA59/SMHV-1 and rA59/S→3′UTRMHV-1 viruses were suitable for further in vivo studies.
FIG. 2.
Replication of rA59/MHV-1 viruses in cell culture. (A) Schematic diagrams of the rA59/SMHV-1 and rA59/S→3′UTRMHV-1 viruses. (B) Growth curves of rA59/MHV-1 chimeric recombinants and the parental viruses. Replicate cultures of L2 cells were infected with either rA59/SMHV-1 (plaque isolate C), rA59/S→3′UTRMHV-1 (plaque isolate B), MHV-1, or rA59 at an MOI of 4, and two replicate cultures were harvested for each virus at the indicated times. Viral titers were determined by plaque assay on L2 cells.
MHV-S protein confers increased pneumovirulence on MHV-A59 recombinant viruses.
We examined the ability of the parental MHV-1 and chimeric recombinant viruses for their effect on survival and to produce pulmonary disease in A/J mice. Previously we and others have reported that intranansal infection of A/J mice with MHV-1 produces a lethal pulmonary disease with pathological characteristics similar to that seen in humans with SARS (9, 18). In a preliminary experiment, we infected groups of MHV-1-susceptible A/J mice intranasally with 5,000 PFU of either MHV-1 (10 50% lethal doses [LD50]), the isogenic control parental virus rA59, or recombinant viruses expressing the MHV-1 S gene in an MHV-A59 background (rA59/SMHV-1). Two independent recombinants were tested for rA59/SMHV-1, and identical results were obtained with each recombinant. Mice were observed clinically, and some mice were sacrificed for histopathologic analysis. As expected, all mice infected with MHV-1 developed severe respiratory symptoms (obviously increased respiratory rate), ruffled fur, and decreased activity and were euthanized. Histopathologic examination of the lungs from these mice revealed a severe pneumonitis resembling that observed in SARS patients, as had been reported previously (9). None of the mice infected with rA59/SMHV-1 viruses became clinically ill after infection with this dose of virus, and the mice survived until they were sacrificed on day 7; histopathologic examination of the lungs from these animals revealed a mild interstitial pneumonitis (Fig. 3F and G). Thus, mice infected with recombinant viruses containing the MHV-1 S gene failed to develop significant pulmonary disease at this dose of virus, indicating that at 5,000 PFU, the S gene by itself did not appear to confer pneumovirulence on MHV-A59. Virus titers of lung homogenates of mice infected with rA59/SMHV-1 demonstrated that the chimeric virus replicated in the lung, achieving a mean peak titer of 5.6 × 105 PFU/g at 36 h postinfection. After this time, the titer declined to 3.5 × 103 by day 7.
FIG. 3.
Pulmonary histopathology in mice infected with rA59/SMHV-1, rA59/S→3′UTRMHV-1, rA59, and MHV-1. Mice were infected by intranasal inoculation with 5,000 PFU, sacrificed at 4 and 7 days postinfection, and lungs were removed, fixed with buffered formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin. The sections shown in panels F and G are from a different experiment from the other sections. (A) Lung from normal uninfected animal. This micrograph shows normal lung histology. Note patency of bronchus on the lower right and the well-aerated, open alveolar structure with thin alveolar walls. This is normal murine lung histology (grade 0). (B) MHV-1 at day 4 postinfection. The lung shows diffuse changes in lung parenchyma. There are peribronchitis and thickening of alveolar walls, which is the result of infiltration by mononuclear cells. This is mild-to-moderate interstitial pneumonitis (grade 2). (C) MHV-1 at day 7 postinfection. The lung shows severe changes with near total loss of alveolar spaces by heavy inflammatory mononuclear cell infiltrate of alveolar walls. Note that almost the entire lung appears totally obliterated. This is severe interstitial pneumonitis with consolidation (grade 4). (D) rA59 at day 4 postinfection. This lung shows patent alveoli, but in the upper part of the micrograph, the alveolar lumens are filled with pink-staining fluid which was seen in approximately 25% of the lung tissue. This is normal lung, aside from areas of pulmonary edema (grade 0). (E) rA59 at day 7 postinfection. The lung shows thickening of alveolar walls by mononuclear cell infiltrate. Patency of airways is significantly reduced. There was no pulmonary edema. This is moderate interstitial pneumonitis (grades 2 to 3). (F) rA59/SMHV-1 at day 4 postinfection. The lung shows thickening of alveolar walls by mono- nuclear inflammatory cells, but most airways are still patent, albeit reduced in size, compared to normal. There is also bronchitis. This is mild interstitial pneumonitis (grade 1). (G) rA59/SMHV-1 at day 7 postinfection. The lung shows thickening of alveolar walls by mononuclear cells, but most airways are still patent, albeit reduced in size. It is essentially the same as in day 4. This is interstitial pneumonitis (grade 1). (H) rA59/S→3′UTRMHV-1 at day 4 postinfection. The lung shows peribronchitis and scattered patches where alveolar walls are thickened by mononuclear cells. The changes are mild. This is interstitial pneumonitis (grade 1). (I) rA59/S→3′UTRMHV-1 at day 7 postinfection. The lung shows extensive mononuclear cell infiltration of alveolar walls with great reduction in alveolar lumens. In places, there is early consolidation of the lung parenchyma. This is interstitial pneumonitis (grade 3). All micrographs are ×250. For panels B to I, panels on the left represent day 4, and those on the right represent day 7.
We then infected groups of A/J mice (n = 10 mice/group) intranasally with 5,000 PFU of either MHV-1, with the isogenic control parental virus rA59, or with a recombinant viruses containing all of the MHV-1 genetic information from S to the 3′ end of the genome (rA59/S→3′UTRMHV-1). All of the MHV-1-infected mice developed respiratory distress and died or were euthanized within 8 days of infection. Eight of 10 (80%) of the rA59 mice became severely ill and were euthanized by day 10. All of the mice infected with rA59/S→3′UTRMHV-1 recombinant viruses became severely ill, with increased respiratory rates, ruffled fur, and decreased activity, similar to the clinical findings observed in MHV-1-infected mice. However, only 2 of these 10 mice became sufficiently ill as to require euthanasia (Fig. 4A).
FIG. 4.
Effect of 5,000 PFU of rA59/S→3′UTR MHV-1 viruses on A/J mice. Groups of 14 mice were infected intranasally with 5,000 PFU with two independent rA59/S→3′UTRMHV-1 recombinants, rA59, or MHV-1, and followed clinically as described in Materials and Methods. Mice judged to have severe disease were euthanized. (A) Survival curve. (B) Three mice were selected randomly from each group at 36 h, 4 days, and 7 days postinfection and euthanized. Lung homogenates in DMEM (10% [wt/vol]) were prepared, and virus titers were determined by plaque assay. Results are expressed as PFU/g of tissue. The limit of detection was 170 PFU/g.
MHV-1 and rA59 replicated to high titers. Peak viral titers of 108 and 109 PFU were seen at 36 h in lung tissue from MHV-1 and rA59, respectively, and virus titers greater than 104 PFU were still detected even at 7 days postinfection, at which time most animals had died or had been euthanized (Fig. 4B). Both of the rA59/S→3′UTRMHV-1 recombinant viruses replicated 100-fold less in the lung, with peak titers of 4 × 105 to 8 × 105 PFU at 36 h postinfection, and by day 7, virus was cleared from the lungs of rA59/S→3′UTRMHV-1-infected mice, whereas substantial (104 to 105 PFU/g) amounts of virus were still easily detectable in lungs of mice infected with rA59 and MHV-1 (Fig. 4B). Replication of rA59 in the lung was statistically greater than that of MHV-1 and all recombinant viruses (P < 0.02). However, replication of MHV-1 was not statistically different from that of recombinant viruses (P = 0.996).
Histopathologic examination of the lungs from uninfected A/J mice showed widely patent alveoli with normal architecture (Fig. 3A). Lungs from A/J mice infected with MHV-1 contained a moderate interstitial pneumonitis on day 4 (Fig. 3B), which by day 7 (Fig. 3C) had progressed to severe diffuse interstitial pneumonitis with consolidation and a marked mononuclear cell infiltrate (predominately macrophages, neutrophils, and lymphocytes), as previously described (9). Histologic examination of the lungs of mice infected with rA59 revealed mild-to-moderate interstitial pneumonitis with little or no alveolar changes in spite of vigorous rA59 replication (Fig. 3D and E). Lungs from mice infected with rA59/S→3′UTRMHV-1 recombinant viruses contained pathological changes similar to those observed with MHV-1, although they were less severe (Fig. 3H and I). Histologic examination of livers and brains of MHV-1-infected mice showed only minimal hepatitis with architecture similar to that of uninfected controls (Fig. 5A and B), whereas rA59-infected mice developed severe widespread hepatitis with massive hepatic necrosis (Fig. 5C) and a moderate encephalitis (data not shown) accounting for the death of these animals. Only minimal liver and brain pathology was seen in mice infected with rA59/SMHV-1 (Fig. 5D) and rA59/S→3′UTRMHV-1 (Fig. 5E).
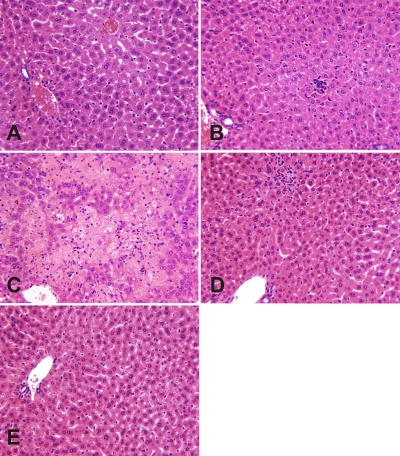
FIG. 5.
Hepatic histopathology in mice infected with rA59/SMHV-1, rA59/S→3′UTRMHV-1, rA59, and MHV-1. Mice were infected by intranasal inoculation with 5,000 PFU and sacrificed at 7 days postinfection, and livers were removed, paraffin embedded, sectioned, and stained with hematoxylin and eosin. (A) Uninfected control liver shows normal histology. (B) MHV-1-infected liver shows minimal focal inflammatory cell collections in lobules and otherwise appeared normal. (C) rA59-infected liver shows severe acute hepatic necrosis. Note large geographic pale areas where liver cells are lost. (D) rA59/SMHV-1-infected liver shows normal liver, aside from small, focal inflammatory cell infiltrates in the lobular parenchyma. Note that this section is from a separate experiment from the other panels. (E) rA59/S→3′UTRMHV-1-infected liver shows normal histology. All micrographs are ×400.
To further examine the pneumovirulence of recombinant viruses containing only the MHV-1 S gene or containing all of the MHV-1 genetic information from S to the 3′ end of the genome, another set of A/J mice (n = 10 mice/group) were infected by intranasal inoculation with 100,000 PFU of rA59, rA59/SMHV-1, or rA59/S→3′UTRMHV-1 recombinant viruses. Mice were observed clinically, and some mice were sacrificed at various times postinfection for viral titers and histopathologic analysis. As shown in Fig. 6, at this higher dose of virus, all of the rA59/S→3′UTRMHV-1-infected mice died, whereas even at 100,000 PFU, only 33% of the rA59/SMHV-1-infected mice died (Fig. 6A). At 1.5 days postinfection, the MHV-1, rA59, rA59/SMHV-1, and rA59/S→3′UTRMHV-1 viruses replicated to high titers in the lung (Fig. 6B), although the titer achieved by the rA59/S→3′UTRMHV-1 recombinant was lower than that achieved by the other viruses. As was the case for the low-challenge experiment, the peak titer achieved by rA59 was significantly (P < 0.05) greater than that achieved by the rA59/SMHV-1 and rA59/S→3′UTRMHV-1 recombinant viruses. By day 4, the lung titers decreased 10- to 100-fold for all viruses except for rA59/SMHV-1. By day 7, all of the rA59-infected mice had died of hepatitis; pneumovirulent MHV-1 and the two chimeric viruses persisted in the lungs. Pulmonary histopathology of mice infected with rA59 isogenic control virus, rA59/SMHV-1, or rA59/S→3′UTRMHV-1, is shown in Fig. 7. Infection with rA59, even with an inoculum of 100,000 PFU, induced only a mild interstitial pneumonitis without alveolar lesions in the lung (Fig. 7A and B) in spite of the presence of high viral titers. Infection with rA59/SMHV-1 resulted in mild interstitial pneumonitis in the peribronchial regions by 4 days postinfection (Fig. 7C), with mild-to-moderate pneumonitis by day 7 (Fig. 7D). Consistent with its somewhat greater lethality, infection with rA59/S→3′UTRMHV-1 produced more severe pulmonary pathology at both day 4 and day 7 (Fig. 7E and F) than rA59/SMHV-1, with severe interstitial pneumonitis with complete consolidation of the lung seen on day 7. The changes observed for rA59/S→3′UTRMHV-1 were very similar to those seen with infection by 5,000 PFU of MHV-1. The histologic changes observed with high doses of the two recombinant viruses were somewhat more severe than those observed in the prior experiments when 5,000 PFU was used as a challenge dose. This parallels the greater lethality achieved at 100,000 PFU. A statistical analysis of pulmonary virus load in mice infected with 100,000 PFU of chimeric and isogenic rA59 recombinant viruses revealed that the peak titer of rA59 in the lung (36 h postinfection) was significantly greater than that of the viral recombinants (P < 0.05), a result similar to that obtained with the low-dose challenge experiment (Fig. 4).
FIG. 6.
Effect of 100,000 PFU of rA59/SMHV-1 and rA59/S→3′UTRMHV-1 viruses on A/J mice. Groups of 14 mice were infected intranasally with 5,000 PFU of MHV-1 or 100,000 PFU of rA59, rA59/SMHV-1 (plaque isolate C), or rA59/S→3′UTRMHV-1(plaque isolate B) and followed clinically as described in Materials and Methods. Mice judged to have severe disease were euthanized. (A) Survival curve. (B to D) Three mice were selected randomly from each group at 36 h, 4 days, and 7 days postinfection and euthanized. Organ homogenates in DMEM (10% [wt/vol]) were prepared, and virus titers were determined by plaque assay. Results are expressed as PFU/g of tissue. The limit of detection was 170 PFU/g.
FIG. 7.
Pulmonary histopathology in mice infected with a high dose of rA59, rA59/S→3′UTRMHV-1, or rA59/SMHV-1. Mice were infected by intranasal inoculation with 100,000 PFU of rA59, rA59/S→3′UTRMHV-1 (plaque isolate B), or rA59/SMHV-1 (plaque isolate C) and sacrificed at 4 and 7 days postinfection. Lungs were removed, fixed with buffered formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin. (A) rA59 at day 4 postinfection. The alveolar walls are diffusely thickened by a mononuclear cell infiltrate, which reduces the size of alveolar lumens. This is mild interstitial pneumonitis (grade 1). (B) rA59 at day 7 postinfection. The alveolar walls are diffusely thickened by mononuclear cells. The appearance is unchanged from day 4. This is mild interstitial pneumonitis (grade 1). (C) r59/SMHV-1 at day 4 postinfection. The alveolar walls are thickened by a mononuclear cell infiltrate which is slightly variable from place to place as shown: the changes are slightly greater on the left side of the micrograph. This is interstitial pneumonitis (grade 2). (D) r59/SMHV-1 at day 7 postinfection. The alveolar walls are thickened throughout. Note that the changes are more severe on the right of the micrograph, where the infiltration is greater and alveolar luminal spaces are smaller. The figure is entirely representative. This is interstitial pneumonitis (grades 2 to 3). (E) rA59/S→3′UTRMHV-1 at day 4 postinfection. The lung is greatly altered by a heavy interstitial infiltrate of alveolar walls, with few patent alveoli, and some of these mononuclear cells can be seen (as in the upper left corner). This is moderate-to-severe interstitial pneumonitis (grades 3 to 4). (F) rA59/S→3′UTRMHV-1 at day 7 postinfection. Shown are severe changes with obliteration of large areas of the lung by thickened alveoalar walls and cellular infiltration. This is severe interstitial pneumonitis with consolidation (grade 4). All micrographs are ×250. The left panels represent day 4, and the right panels represent day 7.
rA59 replicated to high titers in the liver (Fig. 6C), whereas the rA59/SMHV-1 and rA59/S→3′UTRMHV-1 recombinants failed to replicate well in the liver and produced no hepatopathology (not shown), similar to the result obtained with the 5,000-PFU challenge (Fig. 5). Viral replication in the central nervous system of mice infected with MHV-1, rA59/SMHV-1, and rA59/S→3′UTRMHV-1 occurred later than that in mice infected with rA59; similarly the peak titers achieved were more than 100-fold less than those achieved with rA59 (Fig. 6D) and produced only mild histopathologic changes in the brain (not shown).
We also isolated a parallel set of recombinants on a MHV-JHM backbone (designated rJHM/SMHV-1 and rJHM/S→3′UTRMHV-1). Both of these recombinant viruses were completely attenuated, with no illness or death even at a challenge of 100,000 PFU. These viruses grew poorly in lung, not at all in liver, and relatively poorly in brain. Examination of the lungs, liver, and brain, failed to reveal any pathology in lung and liver and found only mild encephalitic changes in brain (not shown).
Cytokine and chemokine profiles.
To examine cytokine expression in the lungs, mice were infected with 5,000 PFU of parental rA59 and rJHM viruses; rA59/SMHV-1, rA59/S→3′UTRMHV-1, rJHM/SMHV-1, or rJHM/S→3′UTRMHV-1 chimeras; or MHV-1. Total RNA was extracted from samples of lung tissue harvested at 36 h, 4 days, and 7 days postinfection. Analysis for selected cytokine and chemokine mRNA levels was performed by multiplex RT-PCR as described in Materials and Methods. The results of the analysis at 36 h postinfection are shown in Table 1. At this early time postinfection, cytokine and chemokine induction correlated well with viral replication in lungs: MHV-1, rA59, rA59/SMHV-1, and rA59/S→3′UTRMHV-1 generally had highly elevated (>4-fold increase relative to uninfected mice) levels of mRNA transcripts for IL-6, IFN-γ, IFN-β, IFN-α2, IFN-α4, and CCL2 and more moderate (2- to 4-fold increase relative to uninfected mice) induction of TNF-α, FGL2, IFN-α1, IFN-α5 CCL3, CCL5, and CXCL2. The induction of relatively high levels of IFN-γ, IL-6, and CCL2 by rA59 was somewhat surprising in light of the paucity of inflammatory changes in the lung. Viruses that replicated poorly in lung, rJHM, rJHM/SMHV-1, and rJHM/S→3′UTRMHV-1, either failed to significantly (>2-fold) induce the synthesis of mRNAs for these proinflammatory molecules or induced a moderate (2- to 4-fold) increase in a small number of these mRNAs. The one exception to this was rJHM/SMHV-1, which induced a 5-fold increase in IL-6.
TABLE 1.
Fold induction of lung cytokine and chemokine mRNAs at 36 h postinfection
| Cytokine or chemokine | Fold induction of mRNAa |
||||||
|---|---|---|---|---|---|---|---|
| MHV-1 | rA59 | rJHM | rA59/SMHV-1 | rA59/S→3′UTRMHV-1 | rJHM/SMHV-1 | rJHM/S→3′UTRMHV-1 | |
| M-CSF | 1.52 ± 0.13 | 2.21 ± 0.65 | 0.72 ± 0.10 | 1.04 ± 0.21 | 1.77 ± 0.07 | 0.95 ± 0.06 | 0.76 ± 0.16 |
| TNF-α | 3.59 ± 0.29 | 3.86 ± 0.11 | 0.95 ± 0.06 | 2.43 ± 0.11 | 1.77 ± 0.13 | 1.38 ± 0.20 | 0.50 ± 0.08 |
| FGL2 | 2.35 ± 0.38 | 1.44 ± 0.11 | 0.82 ± 0.11 | 1.37 ± 0.24 | 1.48 ± 0.12 | 1.10 ± 0.18 | 0.47 ± 0.03 |
| IL-6 | 13.2 ± 1.44 | 18.8 ± 1.2 | 2.33 ± 0.08 | 13.8 ± 1.3 | 13.5 ± 0.76 | 5.05 ± 0.96 | 0 |
| IFN-γ | 26.0 ± 0.92 | 7.20 ± 2.6 | 1.18 ± 0.69 | 16.6 ± 3.5 | 12.8 ± 2.7 | 3.23 ± 1.9 | 0 |
| IFN-β | 42 ± 12 | 4.1 ± 1.7 | 1.3 ± 0.4 | 34 ± 3.6 | 25 ± 4.1 | 1.2 ± 0.8 | 1.6 ± 1.1 |
| IFN-α1 | 3.2 ± 0.5 | 2.6 ± 0.5 | 0.4 ± 0.2 | 4.5 ± 1.1 | 6.2 ± 2.1 | 0.5 ± 0.3 | 0.8 ± 0.5 |
| IFN-α2 | 24 ± 8 | 16 ± 7 | 2.1 ± 0.5 | 38 ± 4.6 | 37 ± 7.3 | 1.4 ± 0.5 | 1.2 ± 0.2 |
| IFN-α4 | 32.2 ± 3.8 | 18.4 ± 3.2 | 2.6 ± 1.2 | 19.4 ± 2.5 | 26.7 ± 5.3 | 0.7 ± 0.3 | 1.2 ± 1.1 |
| IFN-α5 | 1.8 ± 0.5 | 2.2 ± 0.4 | 0.2 ± 0.1 | 3.9 ± 1.2 | 6.2 ± 2.2 | 0 | 0.6 ± 0.3 |
| CCL2 | 30.4 ± 8.6 | 11.4 ± 4.2 | 1.04 ± 0.36 | 7.18 ± 3.6 | 7.13 ± 1.4 | 2.51 ± 0.70 | 0 |
| CCL3 | 4.96 ± 0.76 | 3.34 ± 0.11 | 0.55 ± 0.20 | 1.95 ± 0.41 | 2.09 ± 0.12 | 1.10 ± 0.27 | 0 |
| CCL5 | 3.80 ± 0.48 | 5.06 ± 1.2 | 1.08 ± 0.06 | 2.61 ± 0.38 | 3.44 ± 0.08 | 1.22 ± 0.19 | 0.74 ± 0.02 |
| CXCL2 | 5.17 ± 0.53 | 3.68 ± 0.64 | 1.38 ± 0.43 | 2.10 ± 0.54 | 2.50 ± 0.64 | 1.38 ± 0.12 | 3.39 ± 0.22 |
Results are expressed as fold induction compared to mock-infected control mice. Cytokines induced by more than 4-fold are in bold. Values of 0 are below detectable levels.
The evolution of IFN-γ, TNF-α, IL-6, FGL2, CCL2, and CCL3 mRNA levels over the course of infection is shown in Table 2. These cytokines were studied because they have been shown to be important determinants of disease severity and host outcome in both experimental models of SARS as well as SARS in humans (1, 9, 49, 52). In general, a continually increasing level of cytokines and chemokines after infection was associated with more severe pulmonary pathology. IFN-γ, TNF-α, FGL2, and CCL3 mRNAs in lung increased continually over the course of the infection for the two viruses that produced the most severe pulmonary disease, MHV-1 and rA59/S→3′UTRMHV-1. In mice infected with rA59/SMHV-1, pulmonary CCL3 mRNA levels also increased continuously over the course of infection, although the levels of mRNA observed were lower than those observed in mice infected with MHV-1 and rA59/S→3′UTRMHV-1. In the same mice, CCL2 mRNA levels peaked at 4 days postinfection and declined somewhat on day 7, and IFN-γ levels remained elevated but slowly declined over the course of infection, whereas TNF-α and FGL2 were only modestly elevated and tended to decline over time. Mice infected with rA59 had lower initial levels of IFN-γ, TNF-α, and FGL2 mRNAs than mice infected with rA59/S→3′UTRMHV-1 or rA59/SMHV-1, and these cytokine mRNAs tended to decline to near-normal levels over the course of infection. IL-6 mRNA was present at high levels in mice infected with MHV-1, and all of the viruses with an MHV-A59 background, including rA59, but slowly declined in all but rA59/S→3′UTRMHV-1, where it continued to increase over the course of infection.
TABLE 2.
Kinetics of cytokine/chemokine induction by virus infection
| Cytokine or chemokine and virus | Fold induction at postinfection timea: |
||
|---|---|---|---|
| 36 h | Day 4 | Day 7 | |
| IFN-γ | |||
| MHV-1 | 26.0 ± 0.92 | 48.3 ± 7.2 | 62.7 ± 8.7 |
| rA59 | 7.20 ± 2.6 | 4.1 ± 1.2 | 1.4 ± 2.1 |
| rJHM | 1.18 ± 0.69 | 0 | 0 |
| rA59/SMHV-1 | 16.6 ± 3.5 | 12.2 ± 4.2 | 4.3 ± 1.7 |
| rJHM/SMHV-1 | 3.23 ± 1.9 | 1.1 ± 0.4 | 1.0 ± 1.1 |
| rA59/S→3′UTRMHV-1 | 12.8 ± 2.7 | 28.7 ± 4.2 | 47.6 ± 11.4 |
| rJHM/S→3′UTRMHV-1 | 0 | 0 | 0 |
| TNF-α | |||
| MHV-1 | 3.59 ± 0.29 | 6.7 ± 3.2 | 11.2 ± 2.1 |
| rA59 | 3.86 ± 0.11 | 2.1 ± 0.7 | 1.6 ± 0.8 |
| rJHM | 0.95 ± 0.06 | 1.1 ± 0.4 | 0 |
| rA59/SMHV-1 | 2.43 ± 0.11 | 1.7 ± 0.4 | 1.2 ± 0.1 |
| rJHM/SMHV-1 | 1.38 ± 0.20 | 0 | 1.1 ± 0.15 |
| rA59/S→3′UTRMHV-1 | 1.77 ± 0.13 | 6.3 ± 2.1 | 8.4 ± 2.7 |
| rJHM/S→3′UTRMHV-1 | 0.50 ± 0.08 | 1.61 ± 0.18 | 0.92 ± 0.17 |
| IL-6 | |||
| MHV-1 | 13.2 ± 1.44 | 17.7 ± 2.1 | 8.7 ± 1.6 |
| rA59 | 18.8 ± 1.2 | 14.2 ± 2.1 | 3.7 ± 1.6 |
| rJHM | 2.33 ± 0.08 | 1.3 ± 0.9 | 1.4 ± 0.4 |
| rA59/SMHV-1 | 13.8 ± 1.3 | 11.2 ± 1.7 | 6.1 ± 2.3 |
| rJHM/SMHV-1 | 5.05 ± 0.96 | 2.2 ± 0.9 | 1.9 ± 0.7 |
| rA59/S→3′UTRMHV-1 | 13.5 ± 0.76 | 21.4 ± 3.2 | 32.5 ± 7.2 |
| rJHM/S→3′UTRMHV-1 | 0 | 1.1 ± 0.4 | 0 |
| FGL2 | |||
| MHV-1 | 2.35 ± 0.38 | 6.3 ± 1.7 | 8.1 ± 2.2 |
| rA59 | 1.44 ± 0.11 | 1.9 ± 0.8 | 1.3 ± 0.25 |
| rJHM | 0.82 ± 0.11 | 0.79 ± 0.20 | 0 |
| rA59/SMHV-1 | 1.37 ± 0.24 | 1.32 ± 0.36 | 1.71 ± 0.4 |
| rJHM/SMHV-1 | 1.10 ± 0.18 | 1.03 ± 0.21 | 1.1 ± 0.2 |
| rA59/S→3′UTRMHV-1 | 1.48 ± 0.12 | 6.7 ± 1.1 | 12.2 ± 4.4 |
| rJHM/S→3′UTRMHV-1 | 0.47 ± 0.03 | 2.8 ± 0.4 | 2.9 ± 0.5 |
| CCL2 | |||
| MHV-1 | 30.4 ± 8.6 | 24.8 ± 4.3 | 21.2 ± 7.1 |
| rA59 | 11.4 ± 4.2 | 6.9 ± 1.8 | 5.2 ± 2.1 |
| rJHM | 1.04 ± 0.36 | 1.7 ± 0.45 | 1.5 ± 0.22 |
| rA59/SMHV-1 | 7.18 ± 3.6 | 8.2 ± 4.3 | 5.1 ± 0.7 |
| rJHM/SMHV-1 | 2.51 ± 0.70 | 2.9 ± 1 | 2.4 ± 0.4 |
| rA59/S→3′UTRMHV-1 | 7.13 ± 1.4 | 11.2 ± 1.7 | 4.5 ± 1.3 |
| rJHM/S→3′UTRMHV-1 | 0 | 0 | 0 |
| CCL3 | |||
| MHV-1 | 4.96 ± 0.76 | 7.2 ± 2.3 | 8.4 ± 1.7 |
| rA59 | 3.34 ± 0.11 | 1.8 ± 0.5 | 1.4 ± 0.2 |
| rJHM | 0.55 ± 0.20 | 0.6 ± 0.4 | 1.1 ± 0.2 |
| rA59/SMHV-1 | 1.95 ± 0.41 | 3.2 ± 0.8 | 4.1 ± 0.2 |
| rJHM/SMHV-1 | 1.10 ± 0.27 | 0.7 ± 0.4 | 0.85 ± 0.2 |
| rA59/S→3′UTRMHV-1 | 2.09 ± 0.12 | 6.4 ± 1.7 | 7.7 ± 1.7 |
| rJHM/S→3′UTRMHV-1 | 0 | 0 | 0 |
Results are expressed as fold induction compared to mock-infected control mice. Values of 0 are below detectable levels.
In general, mice infected with JHM or recombinant viruses on a MHV-JHM background (viruses that replicated poorly in lung) all had normal or only moderate induction of cytokine or chemokine mRNAs in lung tissue relative to uninfected mice, and these levels tended to decline over the course of infection. There were two notable exceptions to this: rJHMV/SMHV-1 induced a 5-fold increase in IL-6 mRNA at 36 h postinfection, which declined over time, and rJHM/S→3′UTRMHV-1 induced a modest increase in FGL2 levels starting at day 4.
Serum cytokine levels in mice infected with MHV-1, rA59/SMHV-1, or rA59/S→3′UTRMHV-1 or mock infected were measured at 2 days postinfection as previously described (9). Large increases in all cytokines tested were observed in mice infected with the two most pneumovirulent viruses, MHV-1 and rA59/S→3′UTRMHV-1 (Table 3). A much more modest (∼2-fold) increase was observed in mice infected with the somewhat less pneumovirulent rA59/SMHV-1.
TABLE 3.
Serum cytokine or chemokine levels at 2 days postinfection
| Cytokine or chemokine | Serum cytokine or chemokine level (pg/ml) |
|||
|---|---|---|---|---|
| Uninfected | MHV-1 | rA59/SMHV-1 | rA59/S→3′UTRMHV-1 | |
| IL-10 | 23.5 ± 6 | 204.8 ± 12 | 42.3 ± 12 | 394 ± 18 |
| IL-6 | 48.1 ± 3.6 | 836 ± 9 | 38.6 ± 12 | 1184 ± 64 |
| IL-12p70 | 136 ± 21 | 483 ± 28 | 184 ± 18 | 528 ± 30 |
| IFN-γ | 12 ± 2 | 736 ± 62 | 21 ± 6 | 596 ± 81 |
| TNF-α | 18 ± 4 | 184 ± 22 | 32 ± 5 | 204 ± 21 |
| CCL2 | 104 ± 7 | 6140 ± 280 | 186 ± 9 | 5470 ± 30 |
In summary, these results show that elevated levels of inflammatory cytokine mRNAs in lung and serum cytokine levels are sensitive correlates of disease severity, as assessed by pulmonary pathology. The most highly pneumovirulent chimeric virus, rA59/S→3′UTRMHV-1, elicited cytokine responses that were very similar to those observed with MHV-1. The slightly less pneumovirulent rA59/SMHV-1 elicted cytokine responses that were somewhat more similar to those seen in rA59 than those observed with rA59/S→3′UTRMHV-1, and the MHV-JHM-based viruses, which failed to replicate well in lung, all induced poor cytokine responses in mice.
DISCUSSION
The aim of the present study was to map the MHV determinants of pneumotropism and pneumonvirulence. We report here our initial investigations into the genetic basis of the pneumovirulence of MHV-1 relative to other strains of MHV. Sequencing of the 3′ one-third of the MHV-1 genome containing the genes encoding the structural proteins demonstrates that the genetic organization of MHV-1 is generally similar to that of other strains of MHV. The open reading frame encoding the hemagglutinin esterase (HE) protein is truncated, and as demonstrated by RT-PCR, subgenomic mRNA 2b is not transcribed, confirming earlier studies using metabolic labeling (57) that suggested that the MHV-1 HE was a pseudogene. Thus, it is unlikely that the HE gene contributes to MHV-1 pneumovirulence. Sequencing of the MHV-1 S gene revealed two interesting findings: two 4-amino-acid deletions relative to the MHV-JHM S sequence that result in the loss of the C57BL/6 immunodominant CTL epitope CSLWNGPHL (S-510-518) (3) contained in the MHV-JHM S protein, and three amino acid changes relative to MHV-JHM in the subdominant C57BL/6 CTL epitope in the same gene. The changes in the subdominant epitope relative to MHV-JHM (RCQIFANI to RCHIFSNL) should still permit binding to MHC class I and antigen presentation and a CD8+ T-cell epitope (RCHIFSNLL) at residues 587 to 595 has been identified by John Harty and coworkers (personal communication). It is likely that the region of the spike protein containing this epitope is functionally required; sequencing of multiple MHV spike genes demonstrated that the hypervariable domain of S1 tolerates deletions, but these deletions end at or before this epitope (34). Furthermore, our attempts at recovering recombinant MHV-A59 with mutations within the sequence encoding this epitope were unsuccessful (unpublished data). To our knowledge, there are no published studies that identify T-cell epitopes for A/J mice for any strain of MHV.
Targeted recombination was used to select chimeric viruses containing either the MHV-1 S gene or genes encoding all of the MHV-1 structural proteins, on MHV-A59 and MHV-JHM backgrounds. Growth curves with the rA59/MHV-1 chimeras demonstrated that these recombinant viruses grew efficiently in vitro with growth kinetics similar to those of the parental MHV-A59 and MHV-1 viruses (Fig. 2). The maximal titers achieved by these chimeras were equal to or somewhat greater than those achieved by MHV-1. Taken together, these data suggest that the rA59/MHV-1 chimeric viruses did not have replication defects that would preclude interpreting in vivo findings.
Challenge studies in mice demonstrated that expression of the MHV-1 S gene in the MHV-A59 background (rA59/SMHV-1) increased the pneumovirulence of MHV-A59. Mice infected with this virus developed pulmonary lesions that were less severe than those observed with MHV-1. Compatible with this finding, the rA59/SMHV-1 virus was much less virulent, with an LD50 approximately 200-fold greater than that observed with MHV-1. Chimeras containing all of the MHV-1 structural genes on an MHV-A59 background reproduced the SARS-like pathology observed with MHV-1 and reproducibly increased pneumovirulence relative to the rA59/SMHV-1 virus. However, this chimera was still considerably less virulent than MHV-1, with an estimated LD50 50- to 100-fold greater than that of MHV-1. A summary table depicting the results of scoring for total inflammation along with the histology data of mice infected with the low dose of viruses is shown in Table 4. Collectively, these data suggest that important determinants of pneumopathogenicity are contained within the 3′ one-third of the MHV-1 genome, but additional important virulence factors must be encoded in the genome upstream of the S gene. Similar chimeric viruses containing MHV-1 genes in an MHV-JHM background were completely attenuated. There are several possible interpretations of this result: MHV-JHM could encode upstream genes that somehow impede pneumovirulence; JHM is well adapted to the CNS and does not replicate in other organs. Indeed, the JHM background genes eliminate the ability of a chimeric virus expressing the A59 spike protein to induce hepatitis (28). However, in the context of A59/JHM chimeras, the restriction to the CNS maps to the 3′ third of the genome containing the structural genes (29), perhaps different from the observations with the MHV-1/JHM chimeras. Alternatively, MHV-A59 and MHV-1 genes upstream of HE may make a contribution to pneumovirulence that is absent in the corresponding MHV-JHM genes, or there is the trivial explanation that some incompatibility between MHV-JHM and MHV-1 genes results in “crippled” viruses. We view the latter of these possibilities as unlikely as the replication in cell culture of both types of rJHM/MHV-1 chimeras approximated that of parental MHV-JHM. Currently, we cannot distinguish between the other two possibilities. The lack of neurovirulence of these rJHM/MHV-1 viruses is consistent with previous observations that demonstrate that the JHM spike gene plays a key role in the high neurovirulence of MHV-JHM (14, 38).
TABLE 4.
Summary of histopathological findings in mice infected with low dose of viruses
| Day postinfection and symptom | Histopathology scorea |
||||
|---|---|---|---|---|---|
| Uninfected | MHV-1 | rA59 | rA59/SMHV-1 | rA59/S→3′UTRMHV-1 | |
| Day 4 | |||||
| Edema | − | + | +++ | − | − |
| Bronchitis | − | ++ | − | ++ | +++ |
| Fibrin | − | + | + | − | − |
| Cells in alveolar lumen | − | ++ | + | − | − |
| Inflammatory cells in alveolar wall | − | +++ | + | + | + |
| Airway narrowing | − | +++ | − | + | + |
| Consolidation | − | − | − | − | − |
| Day 7 | |||||
| Edema | − | − | − | − | − |
| Bronchitis | − | − | − | ++ | ++ |
| Fibrin | − | − | − | − | − |
| Cells in alveolar lumen | − | ++++ | − | − | +++ |
| Inflammatory cells in alveolar wall | − | ++++ | − | + | ++ |
| Airway reduction | − | ++++ | − | + | +++ |
| Consolidation | − | ++++ | − | − | ++ |
Shown is a summary of histopathological findings in mice infected with low dose of MHV-1, rA59, rA59/SMHV-1 and rA59/S→3′UTRMHV-1. Scores for disease: −, absent; +, mild; ++, moderate; +++, severe; ++++, very severe.
A number of genes 5′ to the S gene have been shown to contribute to MHV-A59 pathogenesis. These genes might also contribute to the pathogenesis of the pulmonary disease observed in MHV-1-infected mice. These include the ns2 gene encoding a putative viral cyclic phosphodiesterase required for hepatovirulence (42, 45), the ADP-ribose-1′-phosphatase contained within nsp3 (11), nsp14 (45), and the MHV N protein (56). While the papain-like protease (PLP) domains of SARS-CoV and HCoV-NL63 (13) have been demonstrated to be type I interferon antagonists, the role of the analogous MHV PLP-2 is controversial (13) (60). A sequence comparison of MHV-1 with MHV-A59 indicates that MHV-1 and MHV-A59 Orf1a and Orf1b are very similar, with 95.3% and 98.5% sequence identity, respectively. There is a small region within MHV-1 nsp3 that divergences significantly from the MHV-A59 nsp3 sequence. This region does not map to one of the identified functional domains within nsp3, leaving its functional relevance unknown. This sequence difference, as well as smaller differences in Orf1a and Orf1b, could potentially influence the pathogenesis of MHV-1 infection.
Studies with both SARS patients (2, 36, 40, 48, 52, 59) and the MHV-1 model studied here (9, 17) suggest that the pulmonary lesions observed in these two infections correlate with elevated levels of inflammatory cytokines rather than with viral replication in the lungs. Our current work supports these observations; there was a poor correlation between the replication of the different viruses and the severity of the pulmonary pathology. MHV-A59 replicated as well as or better than MHV-1 and all of the rA59/MHV-1 chimeras, but failed to induce significant pulmonary pathology (Fig. 3 and 4). This situation is similar to that observed in the CNS disease induced by MHV-JHM and MHV-A59 infection, both during the acute encephalitic phase of disease and during the largely immunologically mediated demyelinating phase of the disease. In both phases, there is a poor correlation with virus load and pathology (14, 39, 47). Taken together, these data suggest that the chemokine and cytokine inflammatory response of the host to the virus accounts for the severity of the pulmonary pathology.
The mRNAs encoding the cytokines IL-6, IFN-γ, IFN-β, IFN-α2, and IFN-α4 and the chemokine CCL2 were all strongly induced in the lungs of mice infected with MHV-1, rA59, or the rA59/MHV-1 chimeras and either were not induced or only demonstrated moderate increases in mRNA levels in mice infected with rJHM or the rJHM/MHV-1 chimeras. Thus, high levels of mRNAs for these cytokines correlated well with high levels of virus replication in lung. In general, the highest levels of cytokine expression were observed in animals which had the most severe pulmonary pathology, that is mice infected with MHV-1 and rA59/S→3′UTRMHV-1, the mice infected with rA59/SMHV-1 generally having a somewhat less brisk cytokine response and the mice infected with rA59 having a further muted, but still fairly robust, cytokine response. The minimal degree of pulmonary inflammation observed in rA59-infected mice is somewhat surprising in light of their cytokine responses. At this time, we do not have a good explanation for this observation. Significant increases in serum levels of the proinflammatory cytokines IFN-γ, IL-1β, IL-6, and IL-12 correlated with disease severity, as reported previously in an experimental model of SARS (9) and in SARS patients (1, 49).
The pulmonary lesions observed in SARS are thought in part to be mediated by a robust increase in proinflammatory cytokines in response to viral replication in the lungs. A recent study comparing two mouse-adapted strains of the SARS coronavirus (SARS-CoV) and the Urbani strain, which does not produce significant pathology in young mice, reported elevated levels of pulmonary IL-6, CCL3, IFN-γ, CCL2, and CCL5 at 3 days postinfection, with the increases in CCL3 and IL-6 correlating with increasing pneumovirulence in the mouse (8). Although we determined relative cytokine mRNA levels rather than determine the cytokine levels themselves, the cytokine responses we observed with MHV-1 are similar to those reported by Day et al. (8), particularly if we look at our data at 4 days postinfection (Table 2), where the correlation of IL-6 and CCL3 mRNA levels with lung pathology is most apparent. In a study utilizing aged mice, which develop pulmonary disease in response to infection with the Urbani strain of SARS-CoV, Chen et al. (4) demonstrated both an early wave of cytokines (TNF-α and IL-6) and chemokines (CXCL10, CCL2, CCL3, and CCL5) at 2 days postinfection and a late wave of cytokines (TNF-α, IL-6, IFN-γ, IL-2, and IL-5) and chemokines (CXCL9, CXCL10, CCL2, CCL3, and CCL5) and their receptors (CXCR3, CCR2, and CCR5) at 7 days postinfection, when pulmonary pathology had developed. Thus, in both the MHV-1 model of SARS and in several mouse models of SARS-CoV infection, the elaboration of these proinflammatory molecules correlates with pulmonary pathology. The cytokine and chemokine responses of mice to MHV-1 infection (9) presented here and SARS-CoV infections of mice (4, 8) largely mirror those of patients infected during the 2002-2003 SARS outbreak (2, 5, 40, 48, 52, 59), with elevated levels of proinflammatory cytokines and chemokines, particularly IFN-γ, IL-6, and IFN-γ-inducible protein 10 (IP-10; CXCL10).
In a previous report (9), the intrapulmonary cell infiltrates seen in MHV-1-infected A/J mice were predominantly macrophages, with modest numbers of CD3+ T cells and neutrophils, similar to that seen in human SARS. Furthermore, by immunoelectron microscopy we demonstrated that MHV-1 was primarily localized in the intrapulmonary macrophages, similar to what has been reported with SARS patients (9).
Given the similarity of the pathology and the cytokine and chemokine responses of MHV-1-infected mice to human SARS and the advantages of an agent that only requires biosafety level 2 (BSL-2) containment, we believe that this model system has great potential to further the study of SARS pathogenesis, keeping in mind that it is likely that there will also be some differences since MHV-1 lacks the unique accessory genes found in SARS-CoV. Further studies are now under way in our laboratories to examine the role of various coronavirus genes in the pathogenesis of severe pulmonary infections, which hopefully will lead to new therapies to prevent the morbidity and mortality associated with these pathogens.
Acknowledgments
We gratefully acknowledge support from the U.S. National Institutes of Health, grant 1R21 AI078148, and the Canadian Institutes of Health Research, grant CIHR 79561.
We thank Brenna McGruder for a thoughtful reading of the manuscript.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Beijing Group of National for SARS Research Project. 2003. Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Chin. Med. J. 116:1283-1287. [PubMed] [Google Scholar]
- 2.Cameron, M. J., L. Ran, L. Xu, A. Danesh, J. F. Bermejo-Martin, C. M. Cameron, M. P. Muller, W. L. Gold, S. E. Richardson, S. M. Poutanen, B. M. Willey, M. E. DeVries, Y. Fang, C. Seneviratne, S. E. Bosinger, D. Persad, P. Wilkinson, L. D. Greller, R. Somogyi, A. Humar, S. Keshavjee, M. Louie, M. B. Loeb, J. Brunton, A. J. McGeer, and D. J. Kelvin. 2007. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 81:8692-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castro, R. F., and S. Perlman. 1995. CD8+ T-cell epitopes within the surface glycoprotein of a neurotropic coronavirus and correlation with pathogenicity. J. Virol. 69:8127-8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., Y. F. Lau, E. W. Lamirande, C. D. Paddock, J. H. Bartlett, S. R. Zaki, and K. Subbarao. 2010. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 84:1289-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien, J. Y., P. R. Hsueh, W. C. Cheng, C. J. Yu, and P. C. Yang. 2006. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology 11:715-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowley, T. J., S. Y. Long, and S. R. Weiss. 2010. The murine coronavirus nucleocapsid gene is a determinant of virulence. J. Virol. 84:1752-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das Sarma, J., L. Fu, J. C. Tsai, S. R. Weiss, and E. Lavi. 2000. Demyelination determinants map to the spike glycoprotein gene of coronavirus mouse hepatitis virus. J. Virol. 74:9206-9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day, C. W., R. Baric, S. X. Cai, M. Frieman, Y. Kumaki, J. D. Morrey, D. F. Smee, and D. L. Barnard. 2009. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology 395:210-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Albuquerque, N., E. Baig, X. Ma, J. Zhang, W. He, A. Rowe, M. Habal, M. Liu, I. Shalev, G. P. Downey, R. Gorczynski, J. Butany, J. Leibowitz, S. R. Weiss, I. D. McGilvray, M. J. Phillips, E. N. Fish, and G. A. Levy. 2006. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in a/j mice. J. Virol. 80:10382-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drosten, C., S. Gunther, W. Preiser, S. Van Der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 11.Eriksson, K. K., L. Cervantes-Barragan, B. Ludewig, and V. Thiel. 2008. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1"-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 82:12325-12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frieman, M., K. Ratia, R. E. Johnston, A. D. Mesecar, and R. S. Baric. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J. Virol. 83:6689-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iacono, K. T., L. Kazi, and S. R. Weiss. 2006. Both spike and background genes contribute to murine coronavirus neurovirulence. J. Virol. 80:6834-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan, B., M. Wang, H. Jing, H. Xu, X. Jiang, M. Yan, W. Liang, H. Zheng, K. Wan, Q. Liu, B. Cui, Y. Xu, E. Zhang, H. Wang, J. Ye, G. Li, M. Li, Z. Cui, X. Qi, K. Chen, L. Du, K. Gao, Y. T. Zhao, X. Z. Zou, Y. J. Feng, Y. F. Gao, R. Hai, D. Yu, Y. Guan, and J. Xu. 2005. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 79:11892-11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazi, L., A. Lissenberg, R. Watson, R. J. de Groot, and S. R. Weiss. 2005. Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J. Virol. 79:15064-15073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanolkar, A., S. M. Hartwig, B. A. Haag, D. K. Meyerholz, L. L. Epping, J. S. Haring, S. M. Varga, and J. T. Harty. 2009. Protective and pathologic roles of the immune response to mouse hepatitis virus type 1: implications for severe acute respiratory syndrome. J. Virol. 83:9258-9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanolkar, A., S. M. Hartwig, B. A. Haag, D. K. Meyerholz, J. T. Harty, and S. M. Varga. 2009. Toll-like receptor 4 deficiency increases disease and mortality after mouse hepatitis virus type 1 infection of susceptible C3H mice. J. Virol. 83:8946-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 20.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, N., D. Hui, A. Wu, P. Chan, P. Cameron, G. M. Joynt, A. Ahuja, M. Y. Yung, C. B. Leung, K. F. To, S. F. Lui, C. C. Szeto, S. Chung, and J. J. Sung. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986-1994. [DOI] [PubMed] [Google Scholar]
- 23.Levy, G. A., J. L. Leibowitz, and T. S. Edgington. 1982. Lymphocyte-instructed monocyte induction of the coagulation pathways parallels the induction of hepatitis by the murine hepatitis virus, p. 399-410. In F. Schaffner (ed.), Progress in liver diseases. Grune & Stratton, New York, NY. [PubMed]
- 24.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 25.Luytjes, W. C., L. S. Sturman, P. J. Bredenbeek, J. Charite, B. A. M. van der Zeijst, M. C. Horzinek, and W. J. M. Spaan. 1987. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161:479-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 27.Navas, S., S. H. Seo, M. M. Chua, J. D. Sarma, E. Lavi, S. T. Hingley, and S. R. Weiss. 2001. Murine coronavirus spike protein determines the ability of the virus to replicate in the liver and cause hepatitis. J. Virol. 75:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navas, S., and S. R. Weiss. 2003. Murine coronavirus-induced hepatitis: JHM genetic background eliminates A59 spike-determined hepatotropism. J. Virol. 77:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navas-Martin, S., M. Brom, M. M. Chua, R. Watson, Z. Qiu, and S. R. Weiss. 2007. Replicase genes of murine coronavirus strains A59 and JHM are interchangeable: differences in pathogenesis map to the 3′ one-third of the genome. J. Virol. 81:1022-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navas-Martin, S., S. T. Hingley, and S. R. Weiss. 2005. Murine coronavirus evolution in vivo: functional compensation of a detrimental amino acid substitution in the receptor binding domain of the spike glycoprotein. J. Virol. 79:7629-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning, Q., S. Lakatoo, M. Liu, W. Yang, Z. Wang, M. J. Phillips, and G. A. Levy. 2003. Induction of prothrombinase fgl2 by the nucleocapsid protein of virulent mouse hepatitis virus is dependent on host hepatic nuclear factor-4 alpha. J. Biol. Chem. 278:15541-15549. [DOI] [PubMed] [Google Scholar]
- 32.Ontiveros, E., T. S. Kim, T. M. Gallagher, and S. Perlman. 2003. Enhanced virulence mediated by the murine coronavirus, mouse hepatitis virus strain JHM, is associated with a glycine at residue 310 of the spike glycoprotein. J. Virol. 77:10260-10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 34.Parker, S. E., T. M. Gallagher, and M. J. Buchmeier. 1989. Sequence analysis reveals extensive polymorphism and evidence of deletions within the E2 glycoprotein gene of several strains of murine hepatitis virus. Virology 173:664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. V. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perlman, S., and A. A. Dandekar. 2005. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5:917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips, J. J., M. Chua, S. H. Seo, and S. R. Weiss. 2001. Multiple regions of the murine coronavirus spike glycoprotein influence neurovirulence. J. Neurovirol. 7:421-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips, J. J., M. M. Chua, G. F. Rall, and S. R. Weiss. 2002. Murine coronavirus spike glycoprotein mediates degree of viral spread, inflammation, and virus-induced immunopathology in the central nervous system. Virology 301:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reghunathan, R., M. Jayapal, L. Y. Hsu, H. H. Chng, D. Tai, B. P. Leung, and A. J. Melendez. 2005. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 42.Roth-Cross, J. K., H. Stokes, G. Chang, M. M. Chua, V. Thiel, S. R. Weiss, A. E. Gorbalenya, and S. G. Siddell. 2009. Organ-specific attenuation of murine hepatitis virus strain A59 by replacement of catalytic residues in the putative viral cyclic phosphodiesterase ns2. J. Virol. 83:3743-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt, I., M. Skinner, and S. Siddell. 1987. Nucleotide sequence of the gene encoding the surface projection glycoprotein of coronavirus MHV-JHM. J. Gen. Virol. 68:47-56. [DOI] [PubMed] [Google Scholar]
- 44.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sperry, S. M., L. Kazi, R. L. Graham, R. S. Baric, S. R. Weiss, and M. R. Denison. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 79:3391-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsang, K. W., P. L. Ho, G. C. Ooi, W. K. Yee, T. Wang, M. Chan-Yeung, W. K. Lam, W. H. Seto, L. Y. Yam, T. M. Cheung, P. C. Wong, B. Lam, M. S. Ip, J. Chan, K. Y. Yuen, and K. N. Lai. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1977-1985. [DOI] [PubMed] [Google Scholar]
- 47.Wang, F. I., S. A. Stohlman, and J. O. Fleming. 1990. Demyelination induced by murine hepatitis virus JHM strain (MHV-4) is immunologically mediated. J. Neuroimmunol. 30:31-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, W. K., S. Y. Chen, I. J. Liu, C. L. Kao, H. L. Chen, B. L. Chiang, J. T. Wang, W. H. Sheng, P. R. Hsueh, C. F. Yang, P. C. Yang, and S. C. Chang. 2004. Temporal relationship of viral load, ribavirin, interleukin (IL)-6, IL-8, and clinical progression in patients with severe acute respiratory syndrome. Clin. Infect. Dis. 39:1071-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward, S. E., M. R. Loutfy, L. M. Blatt, K. A. Siminovitch, J. Chen, A. Hinek, B. Wolff, D. H. Pham, H. Deif, E. A. LaMere, K. C. Kain, G. A. Farcas, P. Ferguson, M. Latchford, G. Levy, L. Fung, J. W. Dennis, E. K. Lai, and E. N. Fish. 2005. Dynamic changes in clinical features and cytokine/chemokine responses in SARS patients treated with interferon alfacon-1 plus corticosteroids. Antiviral Ther. 10:263-275. [PubMed] [Google Scholar]
- 50.Weiss, S. R., and J. L. Leibowitz. 2007. Pathogenesis of murine coronavirus infections p. 259-278. In S. Perlman, T. Gallagher, and E. J. Snijder (ed.), Nidoviruses. ASM Press, Washington, DC.
- 51.Williams, R. K., G. S. Jiang, and K. V. Holmes. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 88:5533-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, C. K., C. W. Lam, A. K. Wu, W. K. Ip, N. L. Lee, I. H. Chan, L. C. Lit, D. S. Hui, M. H. Chan, S. S. Chung, and J. J. Sung. 2004. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 136:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.World Health Organization. 30 April 2004, posting date. China confirms SARS infection in another previously reported case; summary of cases to date—update 5. World Health Organization, Geneva, Switzerland.
- 54.World Health Organization. 31 January 2004, posting date. New case of laboratory-confirmed SARS in Guangdong, China—update 5. World Health Organization, Geneva, Switzerland.
- 55.World Health Organization. 2003. The World Health Report 2003—shaping the future. World Health Organization, Geneva, Switzerland.
- 56.Ye, Y., K. Hauns, J. O. Langland, B. L. Jacobs, and B. G. Hogue. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 81:2554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yokomori, K., L. R. Banner, and M. M. Lai. 1991. Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology 183:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y., J. Li, Y. Zhan, L. Wu, X. Yu, W. Zhang, L. Ye, S. Xu, R. Sun, Y. Wang, and J. Lou. 2004. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect. Immun. 72:4410-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng, D., G. Chen, B. Guo, G. Cheng, and H. Tang. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 18:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]