Abstract
The 9-aminoacridine (9AA) derivative quinacrine (QC) has a long history of safe human use as an antiprotozoal and antirheumatic agent. QC intercalates into DNA and RNA and can inhibit DNA replication, RNA transcription, and protein synthesis. The extent of QC intercalation into RNA depends on the complexity of its secondary and tertiary structure. Internal ribosome entry sites (IRESs) that are required for initiation of translation of some viral and cellular mRNAs typically have complex structures. Recent work has shown that some intercalating drugs, including QC, are capable of inhibiting hepatitis C virus IRES-mediated translation in a cell-free system. Here, we show that QC suppresses translation directed by the encephalomyocarditis virus (EMCV) and poliovirus IRESs in a cell-free system and in virus-infected HeLa cells. In contrast, IRESs present in the mammalian p53 transcript that are predicted to have less-complex structures were not sensitive to QC. Inhibition of IRES-mediated translation by QC correlated with the affinity of binding between QC and the particular IRES. Expression of viral capsid proteins, replication of viral RNAs, and production of virus were all strongly inhibited by QC (and 9AA). These results suggest that QC and similar intercalating drugs could potentially be used for treatment of viral infections.
Identification of novel properties and new potential therapeutic uses for existing well-known drugs is an enticing approach to “drug discovery” (5, 6). The pharmacokinetics and safety profiles of these drugs are usually well established, and since they have typically been approved by the Food and Drug Administration (FDA), they can enter clinical trials for new indications via a streamlined process.
Quinacrine (QC) (also known as atebrine or mepacrine) is a well-known compound with favorable pharmacological and toxicological properties that has been clinically available since the 1930s and is widely used for a number of indications. QC is a derivative of 9-aminoacridine (9AA); the two compounds are closely related structurally (see Fig. 1A) and have some characteristics and activities in common. To date, however, 9AA has not been used for therapeutic applications (2, 12, 15). Historically, the main use of QC has been in treatment of malaria and giardiasis (13, 18). In addition, QC has been used to treat discoid and subcutaneous lupus erythematosus (47, 50) and prescribed as a general anti-inflammatory drug (46, 50, 51). More recently, QC has been tested for efficacy in the treatment of sporadic Creutzfeldt-Jakob disease (CJD) (17, 24) and cancer (16). Despite this widespread use of QC in humans for almost a century, its mechanism of action remains poorly understood (13, 16, 33, 51). Some properties of QC and 9AA that likely contribute to their biological activities have been identified. It has been well documented that both QC and 9AA can intercalate into double-stranded DNA and structured RNA molecules (31, 34, 44) and are capable of inhibiting DNA replication, RNA transcription, and protein synthesis (7, 10, 26, 29). The extent of intercalation of QC into RNA molecules depends on the complexity of RNA secondary and tertiary structures (such as hairpin loops that produce regions of double-stranded RNA) and pseudoknots (2, 52). QC has also been shown to inhibit protein synthesis in a cell-free translation system either by inhibiting the peptidyl transferase activity of the ribosome or by preventing binding of mRNAs to the ribosome (28). However, the effect of QC on translation of various mRNA species that differ in their secondary structure and/or utilize different modes of translation initiation (in eukaryotes) has been only partially documented (28, 29).
FIG. 1.
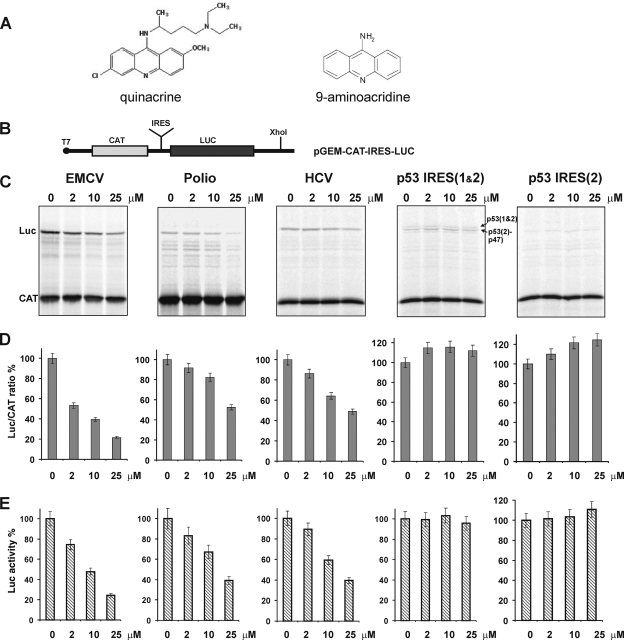
Effect of quinacrine (QC) on IRES-mediated translation in a cell-free system. (A) Chemical structures of QC and the structurally related compound 9-aminoacridine (9AA). (B) Schematic diagram of the bicistronic pGEM-CAT-IRES-LUC plasmid constructs. Five different constructs containing different IRES insertions were generated (see Materials and Methods). To generate mRNA for use in cell-free translation assays, the plasmids were linearized with XhoI and transcribed in vitro with T7 polymerase. (C) Relative translation efficiencies of chloramphenicol acetyltransferase (CAT) (cap-dependent initiation) and firefly luciferase (Luc) (IRES-dependent internal initiation) produced from CAT-IRES-LUC mRNAs in the presence of the indicated concentrations of QC. CAT-IRES-LUC mRNAs contained the EMCV, poliovirus (Polio), HCV, p53 IRES(1&2) or p53 IRES(2) IRES elements (described in the text) as indicated above each gel. 35[S]Met-labeled products were resolved using denaturing gel electrophoresis and visualized by autoradiography. (D) Quantitation of the translation products shown in panel C was done using a Typhoon 9410 imaging scanner. For each concentration of QC, the ratio of luciferase protein production to CAT protein production is shown relative to the ratio in the absence of QC (set at 100%). The mean value obtained from three independent sets of measurements is shown. Values are shown as means ± standard deviations (error bars). (E) Relative firefly luciferase activity values. The mean values ± standard deviations (error bars) obtained from four independent sets of measurements are shown.
Translation initiation is a complex process in eukaryotes. For the majority of eukaryotic mRNAs, initiation involves binding of the mRNA 5′ m7G cap structure by a group of proteins referred to as the cap-binding complex, or eukaryotic initiation factor 4F (eIF4F) (21, 45). This is followed by recruitment of the 40S ribosomal subunit and a number of initiation factors and movement of the resultant 43S initiation complex along the 5′ untranslated region (5′UTR) of the mRNA in search of the initiation codon (21, 45). This cap-dependent mode of translation initiation is also known as “ribosomal scanning” (21, 45). The 5′UTRs of mRNAs that utilize this mechanism of initiation typically do not have complex secondary structures (21, 45). However, some viral and eukaryotic cellular mRNAs can be translated via internal initiation, a process that involves direct binding of the ribosome to specific mRNA regions termed internal ribosome entry sites (IRESs) (19, 20, 22, 25). Most IRES elements possess very complex secondary and tertiary structures (involving stem-loops and pseudoknots), which are believed to allow for multiple interactions with components of the translational machinery (i.e., canonical initiation factors, IRES-transacting factors [ITAFs], and 40S ribosomal subunits) and, hence, for direct recruitment of 40S ribosomal subunits to IRES-containing mRNAs without involvement of the 5′ end (19, 20, 22, 25). It should be mentioned that the vast majority of IRES elements are located within 5′UTRs in close proximity to initiation codons (19, 20, 22, 25). It has recently been found that a number of nucleic acid intercalating agents are capable of inhibiting IRES-mediated initiation of translation to a much greater extent than cap-dependent initiation (28, 29). These studies have, for example, shown that QC is capable of inhibiting IRES-mediated translation of hepatitis C virus (HCV) in an in vitro cell-free system by impairing binding of the HCV IRES to the 40S/80S ribosome (28). Nevertheless, many questions regarding this activity of QC and its efficacy in cases of different IRES elements remain unanswered (28, 29). Furthermore, despite identification of IRES elements in an increasing number of viral and cellular mRNAs, the mechanism of internal initiation is not well understood (19, 20, 22, 25). Notably, different IRES elements have significantly different levels of structural complexity, with some cellular IRESs being much less structured than other eukaryotic and viral IRESs (3, 4, 11, 25). This suggests that translation driven by IRES elements that differ in the complexity of their secondary structures or utilize different subsets of initiation factors and ITAFs might be affected by intercalating agents to different extents (28).
Here we test this hypothesis by comparing the effects of QC on translation driven by three different viral IRES elements (HCV, encephalomyocarditis virus [EMCV], and poliovirus) and one human cellular IRES element (p53). Our results show that initiation of translation from these four different IRES elements was suppressed by QC to substantially different extents, with translation from the EMCV IRES being most affected and that from the p53 IRES being unaffected. Competition dialysis experiments demonstrated that the degree of inhibition of IRES-mediated cell-free translation correlated strongly with the ability of QC to bind the corresponding IRES elements. Furthermore, experiments performed in EMCV- and poliovirus-infected HeLa cells showed that synthesis of viral capsid proteins, production of viral RNAs, and virus replication were all strongly inhibited by QC (as well as 9AA) in a dose-dependent manner.
These results indicate that QC and other similar intercalating compounds might have clinical potential as drugs to treat certain viral infections.
MATERIALS AND METHODS
Plasmids.
The bicistronic constructs used in this study were based on the pGEM-CAT-LUC vector backbone (CAT stands for chloramphenicol acetyltransferase and LUC stands for luciferase) (37). The pGEM-CAT-EMCV-LUC (49) and pGEM-CAT-polio-LUC plasmids were kindly provided by Graham Belsham (National Veterinary Institute, Lindholm, Denmark) and Yuri Svitkin (McGill University, Montreal, Canada), respectively. The pGEM-CAT-HCV-LUC, pGEM-CAT-p53IRES(1&2)-LUC, and pGEM-CAT-p53IRES(2)-LUC plasmids were constructed as follows. The HCV IRES was amplified by PCR from the HCV IRES-containing plasmid pHCV(40-372).NS′ (32) kindly provided by Tatyana Pestova (State University of New York [SUNY] Downstate Medical Center, Brooklyn, NY) using primers 5′-TTTTTGGATCCCTCCCCTGTGAGGAACTACTGTCTTCA-3′ and 5′-TTTTTGGATCCTTTTTCTTTGAGGTTTAGGATTCGTGCT-3′. The p53 IRES(1&2) was amplified from the p53 cDNA-containing plasmid (39) kindly provided by Peter Chumakov (Cleveland Clinic, Cleveland, OH) using primers 5′-TTTTTGGATCCTCTAGAGCCACCGTCCAGGGAGC-3′ and 5′-TTTTTGGATCCACATTGCTTGGGACGGCAAGGGG-3′. The p53 IRES(2) was amplified from the same plasmid using primers 5′-TAATACGACTCACTATAGGGAGGAGGAGCCGCAGTCAGATCCTAGC-3′ and 5′-TTTTTGGATCCACATTGCTTGGGACGGCAAGGGG-3′. The PCR fragments were digested with BamHI and inserted into BamHI-digested pGEM-CAT-HCV-LUC plasmid. It should be noted that p53 IRES(1&2) triggers synthesis of two p53 isoforms (p53 and p47) using two alternative start sites, while p53 IRES(2) triggers synthesis of just the p47 isoform (35). The nucleotide sequences of the plasmids and the orientation of the inserted fragments were verified by DNA sequencing.
In vitro transcription and translation.
In vitro transcription reactions were performed using T7 polymerase and Ambion's mMessage mMachine T7 ultrakit (Ambion, Austin, TX) following the manufacturer's instructions (1a). Bicistronic 5′-capped mRNAs were transcribed from the corresponding plasmids after digestion of the plasmids with XhoI. Transcription reactions were carried out at 37°C for 2.5 h, and the resultant mRNA was purified by LiCl precipitation and washing with 70% ethanol. Aqueous mRNA solutions (∼1 mg/ml) were used for in vitro cell-free translation. Cell-free translation was performed using Promega's rabbit reticulocyte lysate nuclease-treated extract in vitro translation system (Promega, Madison, WI) in the presence of [35S]methionine (15 mCi/ml; GE Healthcare, Piscataway, NJ) and in the presence or absence of quinacrine (QC) (Sigma-Aldrich, St. Louis, MO) as indicated in each figure. Typically, ∼0.5 μl of the reaction mixture was resolved on 10% SDS-polyacrylamide gels, and the radiolabeled translation products were visualized by autoradiography using a Typhoon 9410 imaging scanner (GE Healthcare, Piscataway, NJ).
Competition dialysis.
The competition dialysis assay, which is based on the fundamental thermodynamic principle of equilibrium dialysis, was done essentially as described by Chaires (9, 36). RNA fragments corresponding to various IRES elements were generated by in vitro transcription from corresponding PCR fragments carrying the T7 promoter. The IRES-containing plasmids described above were used as templates. The following primers were used to obtain the relevant PCR fragments: for the EMCV IRES, 5′-TAATACGACTCACTATAGGGAGCCCCCCTAACGTTACTGGCCG-3′ and 5′-GATCGTGTTTTTCAAAGGAAAACCACG-3′; for the poliovirus IRES, 5′-TAATACGACTCACTATAGGGAGTTAAAACAGCTCTGGGGTTGTACCC-3′ and 5′-TTTTTAGATCTTCATTATGATACAATTGTCTGATTGAAATAACTGT-3′; for the HCV IRES, 5′-TAATACGACTCACTATAGGGAGACTCCCCTGTGAGGAACTACTGTCTTCA-3′ and 5′-TTTTTGGATCCTTTTTCTTTGAGGTTTAGGATTCGTGCT-3′; for the p53 IRES(1&2), 5′-TAATACGACTCACTATAGGGAGTCTAGAGCCACCGTCCAGGGAGC-3′ and 5′-TTTTTGGATCCACATTGCTTGGGACGGCAAGGGG-3′; and for the p53 IRES(2), 5′-TAATACGACTCACTATAGGGAGGAGGAGCCGCAGTCAGATCCTAGC-3′ and 5′-TTTTTGGATCCACATTGCTTGGGACGGCAAGGGG-3′. In vitro transcription was carried out using Ambion's MEGAscript T7 kit (Ambion, Austin, TX), and the resulting RNA was purified using the MEGAclear kit (Ambion, Austin, TX). In addition, we have also used poly(rU) (average length of 250 to 300 nucleotides [nt]) obtained from Midland Certified Reagent Company (Midland, TX). The RNA samples were dissolved in a buffer containing 6 mM Na2HPO4, 2 mM H2PO4, 1 mM EDTA, 185 mM NaCl, 5 mM MgCl2 (pH 7.0) (buffer A) at a final concentration of 50 μM (per base). All RNA samples were heated to 90°C for 2 min and then slowly (∼15 to 20 min) cooled to room temperature before use in dialysis experiments to allow proper folding of secondary structures. For each competition dialysis assay, 800 ml of dialysate solution (1 μM quinacrine [Sigma-Aldrich, St. Louis, MO] in buffer A) was placed in a beaker, and the RNA sample to be tested (180 μl of 50 μM solution) was placed in a Slide-A-Lyzer mini-dialysis unit with a 7,000-molecular-weight-cutoff (MWCO) membrane (Pierce Biotechnology, Inc., Rockford, IL) floating in a mini-dialysis flotation device (Pierce Biotechnology, Inc., Rockford, IL). Dialysis was performed with continuous stirring for 24 h at room temperature (22 to 24°C). The concentrations of free and bound ligand (QC) were determined by using a FluoroMax-2 spectrofluorimeter to measure the specific fluorescence of QC at 495 nm after excitation at 389 nm. The amount of RNA-bound QC was determined by calculating the difference between the concentration of QC in the dialysis unit and the concentration of free QC as described previously (2, 9, 36). In each experiment, every RNA sample was tested at least in duplicate.
Cell-based assays of EMCV, poliovirus, and VSV replication.
The effects of QC and 9AA on EMCV, poliovirus, and vesicular stomatitis virus (VSV) protein expression and replication in cultured cells were determined by the following: (i) assessing production of viral proteins using Western blotting with anti-EMCV/poliovirus capsid protein antibodies or anti-p-VSV protein antibodies (p-VSV is VSV P-protein mRNA), (ii) measuring accumulation of viral RNA by Northern blotting, and (iii) assessing virus titer. HeLa cells were infected at a multiplicity of infection (MOI) of 2 50% cell culture infectious doses (CCID50)/cell of EMCV (gift of Robert Silverman, Cleveland Clinic, Cleveland, OH), at a MOI of 2 CCID50/cell of poliovirus (strain Mahoney; gift of Konstantin Chumakov, FDA, Bethesda, MD), or at a MOI of 2 CCID50/cell of VSV (strain New Jersey; gift of Amiya Banerjee, Cleveland Clinic, Cleveland, OH). The indicated concentrations of QC and 9AA (Sigma-Aldrich, St. Louis, MO) were added to cell cultures at the time of infection. All three types of assays were performed on samples collected 3, 4, or 5 h postinfection for poliovirus, VSV, and EMCV, respectively. Western and Northern blotting was performed as described below. To determine the virus titer, we used a modified version of the CCID50 assay in which 10-fold dilutions (1:10 to 1:109) of conditioned culture medium were added to duplicate wells of HeLa cells in 48-well plates. Virus titer was determined by cytopathic effects visible after 3 days. All MOI data are given in CCID50 units/cell.
Western blotting.
Western blotting was performed following standard procedures (48). Total protein extracts from HeLa cells were prepared in 1 ml of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% SDS, 10 mM Tris-HCl [pH 8.0], 1% sodium deoxycholate, 1% NP-40) containing a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). The protein extracts were separated by electrophoresis in denaturing 4 to 20% polyacrylamide Novex gels (Invitrogen, Carlsbad, CA) and transferred to nylon polyvinylidene difluoride (PVDF) membranes (Hybond P; GE Healthcare, Piscataway, NJ). The following antibodies were used: antipoliovirus capsid protein and anti-EMCV capsid protein antibodies (gift of Ann Palmenberg, University of Wisconsin—Madison, Madison, WI) and anti-p-VSV protein antibodies (Abcam, Inc., Cambridge, MA). To control for protein loading, levels of housekeeping proteins were assessed using rabbit anti-keratin 18 antibodies (gift of Robert Oshima, Burnham Institute for Medical Research, La Jolla, CA), and rabbit anti-Hsp90 antibodies (Abcam, Inc., Cambridge, MA). Horseradish peroxidase (HRP)-conjugated secondary anti-rat, anti-rabbit, and anti-mouse antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Northern blotting.
Ten micrograms of total RNA isolated from poliovirus-, EMCV-, or VSV-infected HeLa cells was resolved by denaturing gel electrophoresis and analyzed by Northern blotting (1) with 32P-end-labeled probes specific to either poliovirus RNA (3C-coding PCR fragment), EMCV IRES RNA, p-VSV mRNA, or human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA. Probes (PCR fragments) were generated using poliovirus genomic cDNA (with primers specific for the poliovirus 3C coding sequence 5′-GGGCCTGGGTTTGACTAT-3′ and 5′-TTGGCTCTGAGTGAAGTATGA-3′ downstream), pGEM-CAT-EMCV-LUC plasmid (with primers specific to the EMCV IRES sequence [see above]) or GAPDH full-length cDNA. The probe for VSV P-protein mRNA (p-VSV) was generated by PCR from cloned VSV genome using p-VSV-specific primers: 5′-AATTATGAGTTGTTCCAAGA-3′ and 5′-TTCAAGGATATGGTGAGAGG-3′. The GAPDH cDNA clone was obtained from OriGene Technologies, Inc. (Rockville, MD).
Miscellaneous.
Molecular cloning was performed following the general procedures described by Sambrook et al. (41). DNA sequencing was performed by the Molecular Biology Core Facility at Cleveland Clinic, Cleveland, OH. Measurements of luciferase activity were done using the Promega luciferase assay system (Promega, Madison, WI) and the Turner BioSystems 20/20n luminometer.
RESULTS
QC inhibits initiation of translation from EMCV, HCV, and poliovirus IRESs, but not mammalian p53 IRESs in a cell-free system.
High concentrations of intercalating agents (>50 μM) have been shown to inhibit ribosomal peptidyl transferase activity and, hence, overall protein synthesis, including (in eukaryotes) both cap-dependent and IRES-mediated expression (28). However, at lower concentrations (∼10 to 20 μM), intercalating drugs only minimally affected this activity (28). At these lower concentrations, intercalators preferentially inhibited the initiation step of protein synthesis, specifically initiation driven by highly structured IRES elements (28). Given these results, it was postulated that the extent to which IRES-mediated translation will be affected by intercalating drugs might differ depending on characteristics of the particular IRES, including its sequence, secondary structure, and use of different canonical initiation factors and ITAFs (28). To test this possibility, we used a bicistronic translation assay to assess the effect of the intercalating small-molecule QC on translation initiation driven by four different IRES elements. In this assay (Fig. 1B), translation of the first cistron (encoding chloramphenicol acetyltransferase [CAT]) occurs through the cap-dependent mechanism, while translation of the second cistron (encoding firefly luciferase [LUC]) occurs by a cap-independent, IRES-dependent mechanism. Five different bicistronic constructs were generated to allow testing of initiation from highly structured viral IRES elements (EMCV, HCV, and poliovirus [for a review, see references 4 and 11]) and presumably less-structured mammalian cellular IRES elements [p53(1&2) and p53(2)] (14, 35).
We performed in vitro translation of the bicistronic constructs in a cell-free rabbit reticulocyte lysate-based system in the presence or absence of different physiologically relevant concentrations of QC. The relative efficiency of translation of the two proteins encoded by each construct (LUC relative to “control” CAT) was determined by detection of labeled proteins by SDS-PAGE and autoradiography, followed by quantification of the bands using a Typhoon 9410 imaging scanner. We found that QC inhibited translation from the EMCV, HCV, and poliovirus IRES elements in a dose-dependent manner yet had no effect on translation from the two p53 IRES elements (Fig. 1C and D). When used at a concentration of 25 μM, QC inhibited translation from the EMCV IRES by more than 80% and inhibited translation from the HCV and poliovirus IRESs by about 50 to 60% (Fig. 1C and D). The extent of HCV IRES inhibition was found to be similar to that reported previously (28). The p53 IRES(1&2)-containing construct directs production of two p53 isoforms, p53 and p47 (14, 35), using two different start sites; thus, two translation products are observed in assays with this construct (Fig. 1C). In contrast, p53 IRES(2) triggers production of only the p47 isoform (14, 35). However, neither p53 IRES(1&2) nor p53 IRES(2) was sensitive to QC-mediated inhibition (Fig. 1C). Cap-dependent translation initiation (production of CAT in the bicistronic system) was unaffected by QC in all cases (Fig. 1C). Since cap-dependent initiation (first cistron) was unaffected at the indicated concentrations of QC, we have also assessed the extent of inhibition of IRES-mediated translation by direct measurements of the luciferase activity (second cistron). Luciferase assays (Fig. 1E) corroborated the findings above (Fig. 1D). Taken together, these results confirm that QC can specifically suppress IRES-mediated translation initiation and that the effect of QC can vary greatly between different IRESs.
QC displays enhanced binding to the EMCV, HCV, and poliovirus IRESs, but not to the p53 IRESs.
The ability of intercalating drugs to bind nucleic acids (particularly “single-stranded” RNA molecules) depends on a number of parameters, including the complexity of the secondary structure, the flexibility of the molecule, the interplay of nonspecific electrostatic forces, and other factors (2, 9, 36, 51). Competition dialysis has been proven to be a powerful tool for probing the structural selectivity of ligand-nucleic acid binding and for assessing the affinity of such interactions (2, 9, 36). Here we set up a dialysis system in which QC (the “ligand”) was present in the dialysis solution, and different IRES-containing RNAs were placed within dialysis chambers floating in the solution. At equilibrium, the concentration of free ligand will be identical throughout the dialysis system; however, if the ligand selectively binds to the RNA being tested, the ligand will accumulate in the RNA-containing dialysis chamber to a higher concentration than that in the surrounding solution (2, 9, 36). In our case, the concentration of ligand accumulated in the dialysis chamber after 24 h of dialysis was determined by using a FluoroMax-2 spectrofluorimeter to measure the specific fluorescence of QC at 495 nm after excitation at 389 nm.
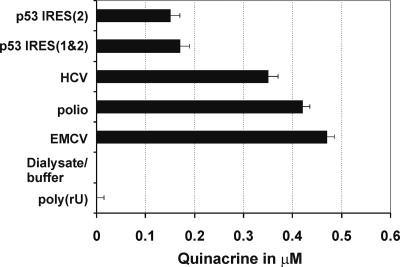
These experiments demonstrated that QC binds to the tested viral IRES elements with 2- to 3-fold greater efficiency than to the cellular p53 IRESs and displays the highest binding affinity to the EMCV IRES element (∼0.47 μM) (Fig. 2). The ability of IRES elements to bind QC correlated strongly with the extent to which QC inhibited cell-free translation driven by the respective IRES elements (Fig. 1C, D, and E and Fig. 2). This correlation indicates that inhibition of IRES-mediated translation initiation by QC involves binding of the drug to the IRES-containing transcript and that the affinity of this interaction is not the same for all IRESs. This finding is consistent with the proposal that QC intercalation into the RNA molecule depends upon its complexity (9, 36). No binding to poly(rU) (∼250 to 300 nt) was observed, in accordance with the previously reported data (9, 36). Poly(rU) is well-known to be unstructured at ambient temperature (40).
FIG. 2.
Binding of QC to IRES elements in competition dialysis experiments. The graph shows the concentration of QC present in the dialysis chamber containing RNAs with the indicated IRES elements after 24 h of dialysis against buffer containing 1 μM QC. Poly(rU) (unstructured RNA [40]) was used as a negative control of QC binding (9, 36). Values are means plus standard deviations (error bars).
QC and 9AA efficiently suppress production of the EMCV and poliovirus capsid proteins, but not the p-VSV protein, in infected HeLa cells.
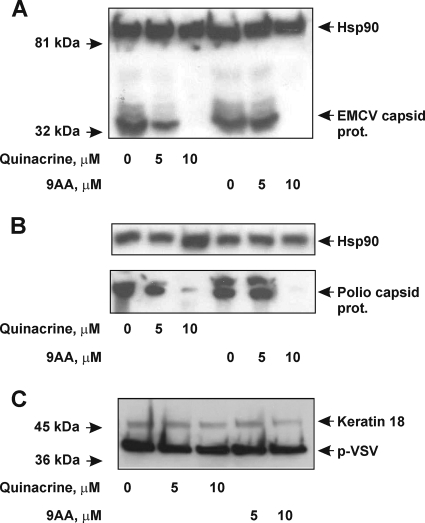
Our cell-free translation assays showed that QC inhibits translation from the EMCV, HCV, and poliovirus IRESs, but not from the p53 IRESs (Fig. 1). To confirm these findings in a more natural scenario, we tested the ability of QC, as well as the structurally related compound 9-aminoacridine (9AA) (Fig. 1A), to inhibit production of viral proteins in EMCV- and poliovirus-infected cultured HeLa cells. Both EMCV and poliovirus have a single-stranded positive-sense RNA genome, which can act as a mRNA (encoding a polyprotein), as a template for genomic RNA replication, or as a nascent genome to be packaged into virus particles (reviewed in reference 27). For both EMCV and poliovirus, expression of the viral genome and replication of the virus are critically dependent on the function of IRES elements located in the 5′-UTRs of their genomes. The IRESs recruit the small ribosomal subunit to the vicinity of the initiation codon and initiate synthesis of a viral polyprotein (27). Upon cleavage, the N-terminal part of the polyprotein gives rise to the structural (capsid) protein, while the C-terminal part gives rise to a number of nonstructural proteins, including an RNA-dependent RNA polymerase and a number of virus-specific proteases (27). Therefore, expression of viral capsid proteins can be used as a measure of viral IRES activity (27). Here, we assessed capsid protein expression by Western blotting of total cell lysates obtained from virus-infected HeLa cells (Fig. 3). We found that production of EMCV and poliovirus capsid proteins was suppressed by both QC and 9AA in a dose-dependent manner in HeLa cells infected with the corresponding virus (Fig. 3A and B). When used at a final concentration of 10 μM, both drugs inhibited production of the EMCV and poliovirus capsid proteins almost completely (Fig. 3A and B). It should be noted that we recently found that QC actively accumulates inside cultured cells to a concentration ∼5- to 8-fold higher than that in the culture medium (data not shown). This observation might explain why the effective QC concentration required for inhibition of initiation from EMCV and poliovirus IRESs in cell-free translation assays was higher than the concentration found to block viral protein production in infected cells. We also tested the effects of QC and 9AA on production of the p-VSV protein in HeLa cells infected with VSV. The p-VSV protein plays a pivotal role in transcription and replication of the VSV genome (30). In contrast to EMCV and poliovirus, VSV is a negative-sense single-stranded RNA virus, which means that the plus-strand (messenger) RNA is produced from the negative-strand RNA, is capped, does not possess any complex secondary structures in the 5′UTR, and likely supports the cap-dependent mode of initiation (30). Interestingly, production of p-VSV protein was largely unaffected by the concentrations of QC and 9AA that had a significant effect on EMCV and poliovirus protein synthesis (Fig. 3C). These findings provide confirmation of our cell-free data in living cells and provide further support for structure-based binding of QC to IRESs leading to inhibition of IRES-dependent translation initiation.
FIG. 3.
Effects of QC and 9-aminoacridine (9AA) on production of EMCV, poliovirus, and VSV proteins in HeLa cells infected with the corresponding viruses. HeLa cells were infected with EMCV (A), poliovirus (B), or VSV (C) at an MOI of 2 CCID50/cell and treated with various amounts of QC or 9AA at the time of infection. Control cells were infected with virus, but not treated with either drug. Cells were incubated with virus and/or drugs for 5 h (EMCV), 3 h (poliovirus), or 4 h (VSV). Five micrograms of total protein extract was analyzed by Western blotting with anti-EMCV capsid protein antibody and anti-Hsp90 antibody (A), anti-poliovirus capsid protein antibody and anti-Hsp90 antibody (B), and anti-p-VSV protein antibody and anti-keratin 18 (K18) antibody (C). Hsp90 and keratin 18 served as protein loading controls. prot., protein.
QC and 9AA suppress replication of the EMCV and poliovirus RNAs in infected HeLa cells and reduce virus production.
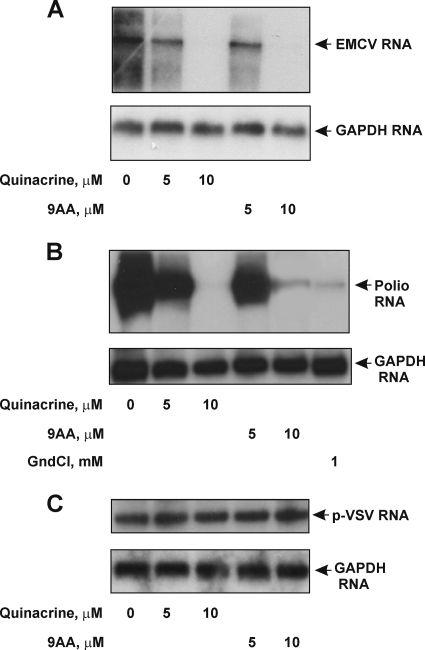
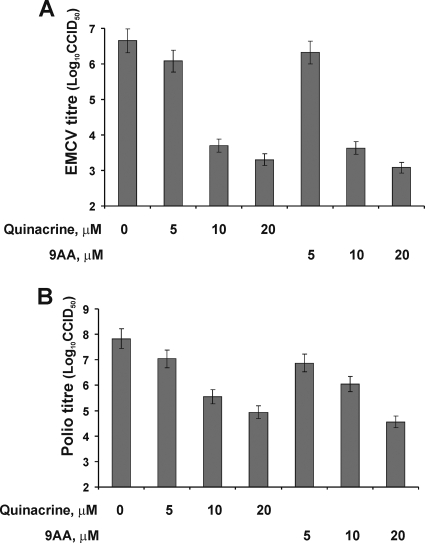
The ability of QC and 9AA to suppress synthesis of the EMCV and poliovirus capsid proteins strongly suggested that overall replication of the viral RNAs and virus production would also be inhibited. To test this, we assessed the amount of viral RNA that accumulated in infected HeLa cells in the absence or presence of various concentrations of QC and 9AA and also quantified virus production by determining the titer of virus in the medium in cell cultures. Northern hybridization showed that synthesis of viral RNA in EMCV- and poliovirus-infected cells was inhibited by both QC and 9AA in a dose-dependent manner (Fig. 4A and B). The effect was specific to viral RNAs bearing IRES elements, as the p-VSV mRNA (Fig. 4C) and cellular GAPDH mRNA were not affected by either QC or 9AA. Guanidine hydrochloride (GndCl), a known inhibitor of the replication of many picornaviruses (without any apparent effect on cellular metabolism) (8), was used as a positive control of drug action and showed an effect similar to that of QC and 9AA (Fig. 4B, bottom gel). The primary mechanism of guanidine inhibition of picornavirus replication is the inhibition of initiation of viral RNA synthesis (8). We also found that QC and 9AA reduce accumulation of virus in the medium in infected cell cultures (Fig. 5). At a concentration of 10 μM, both compounds suppressed the virus titer by at least 100-fold. The inhibitory effect of the drugs depended on their concentration and was substantial at concentrations higher than 5 μM. These results demonstrate that impairment of viral IRES activity by QC has substantial biological effects on the virus that can limit the effectiveness of its infection of cells.
FIG. 4.
Effects of QC and 9AA on EMCV, poliovirus RNA, and p-VSV mRNA accumulation. Ten micrograms of total RNA prepared from HeLa cells 5 h postinfection (p.i.) with EMCV (MOI of 2 CCID50/cell) (A), 3 h p.i. with poliovirus (MOI of 2 CCID50/cell) (B), and 5 h p.i. VSV (C) was analyzed by Northern blotting with an EMCV IRES-specific probe, a poliovirus protein 3C-specific probe, and a p-VSV mRNA-specific probe, respectively. A human GAPDH probe was used as a control. Guanidine hydrochloride (GndCl) (final concentration of 1 mM) was used as a positive control of antiviral drug action.
FIG. 5.
Effects of QC and 9AA on EMCV and poliovirus virus replication. Titration of EMCV (A) and poliovirus (B) growth in infected HeLa cells (MOI of 2 CCID50/cell) treated with the indicated amounts of QC or 9AA. HeLa cells were grown in the presence of virus and drugs for 5 h (EMCV) or 3 h (poliovirus). Control cells were infected, but not treated with drugs. The growth of virus was determined by titration of virus particles in the medium (see Materials and Methods) and is presented on a log scale. Cell culture infectious doses (CCID50)/cell served as viral inocula. Mean values ± standard deviations (error bars) (from three independent sets of experiments) are presented.
DISCUSSION
Compared to double-stranded DNA, RNA displays a greater variety of folds and structures due to a variety of base pairing and other interactions between different regions of the single-stranded molecule (43). The complex and dynamic architecture of RNA makes this nucleic acid especially difficult to target using small molecules (2, 9, 36). On the other hand, it is precisely this complex and distinct architecture that makes RNA attractive for targeting, since small molecules that selectively bind such distinct structures could potentially interfere with many distinct cellular processes depending on the biological functions associated with these structures (2, 9, 28, 29, 36). To date, pharmacological targeting of RNA has not been as successful as targeting of DNA; however, recent experiments have clearly shown the feasibility of targeting specific RNA folds with small molecules (28, 29). This opens up a new avenue in research aimed at modulating gene expression by utilizing ligands that recognize specific RNA folds, such as those present in viral IRES elements.
IRES-mediated initiation is critical for translation of many viral mRNAs as well as some host/cellular mRNAs. At certain physiological concentrations (∼10 to 20 μM), some intercalating agents were found to preferentially inhibit translation initiation (by impairing RNA-ribosome binding) from highly structured mRNAs such as those harboring IRES elements (e.g., HCV and cricket paralysis virus [CrPV] [28]). However, the extent of inhibition varied with different intercalating drugs, showing dependence on the structural complexity of both the IRES and the drug (28, 29). Unfortunately, at the present time, there is still limited understanding of why certain intercalating drugs preferentially inhibit IRES-mediated translation, while others do not or do so with reduced efficiency (28). Nevertheless, the accumulated data suggested that mRNA structural complexity is likely a critical determinant of this process (28).
Undoubtedly, the design of antagonists able to target specific RNA elements is of great interest and importance. Since expression of many pathogenic viruses relies on the presence of IRES elements in their genomes, it might be possible to treat such viral infections with small intercalating compounds that specifically target the IRESs. As a first step in this line of reasoning, we have tested the effect of quinacrine (QC), a well-known intercalating drug with a long history of human clinical use, on translation initiation directed by different IRES elements. Our results demonstrate that QC inhibits cell-free translation from three highly structured viral IRES elements (EMCV, HCV, and poliovirus) (4, 11), but not from presumably less structured cellular p53 IRESs (14, 35) (Fig. 1). Competition dialysis experiments revealed that QC has higher binding affinity (per nucleotide) toward the viral IRES elements possessing complex secondary structure than toward the cellular p53 IRESs (Fig. 2). Thus, a positive correlation between QC-IRES binding and QC-mediated inhibition of IRES-directed translation was established. Our data from cell-free assays were corroborated by experiments using HeLa cells infected with EMCV and poliovirus. Here QC (and the structurally related molecule, 9AA) were shown to suppress production of viral capsid proteins, replication of viral RNAs, and virus titer (Fig. 3 to 5).
To the best of our knowledge, this is the first report showing that QC is able to inhibit EMCV and poliovirus replication in infected cultured cells. The inhibitory effect of QC on EMCV and poliovirus replication was specific, since expression of cellular protein(s) (HSP90, keratin 18) and RNA(s) (GAPDH), as well as expression of the p-VSV protein and mRNA (VSV is a single-stranded negative-sense RNA virus) (Fig. 3 and 4), was not affected or only minimally affected in comparison with expression of EMCV and poliovirus proteins and RNAs.
Interestingly, it was recently shown that replication of a plant virus (Tomato bushy stunt virus [TBSV]), a model positive-sense RNA virus can also be inhibited by QC (42). TBSV RNA accumulation was inhibited by QC both in plants and in protoplasts (42). In addition, QC and some other structurally similar intercalating antimalarial drugs, such as chloroquine and hydroxychloroquine, were suggested to have some anti-human immunodeficiency virus (HIV) effects (23, 38). These studies, together with the results reported here, indicate that QC or QC-like molecules might be effective tools to inhibit replication of various positive-sense RNA viruses that rely on translation initiation from highly structured IRES elements in their genomes. Additional work, including experiments in animal models, will be required to fully demonstrate the potential of this strategy for treatment of viral infections.
Acknowledgments
We thank Amiya Banerjee, Graham Belsham, Konstantin Chumakov, Peter Chumakov, Robert Oshima, Ann Palmenberg, Tatyana Pestova, Robert Silverman, and Yuri Svitkin for their generous gifts of strains, antibodies, and plasmids used in this study. Patricia Baker is gratefully acknowledged for help in manuscript preparation.
This work was supported by grant CA75179 from NIH and funds from Cleveland BioLabs, Inc., to A.V.G. and a grant from the National American Heart Association (0730120N) to A.A.K.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Alwine, J. C., D. J. Kemp, and G. R. Stark. 1977. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl. Acad. Sci. U. S. A. 74:5350-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Ambion. 2008. mMessage mMachine T7 Ultra kit technical manual. Ambion, Austin, TX.
- 2.Arya, D. P., L. Xue, and B. Willis. 2003. Aminoglycoside (neomycin) preference is for A-form nucleic acids, not just RNA: results from a competition dialysis study. J. Am. Chem. Soc. 125:10148-10149. [DOI] [PubMed] [Google Scholar]
- 3.Baird, S. D., M. Turcotte, R. G. Korneluk, and M. Holcik. 2006. Searching for IRES. RNA 12:1755-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balvay, L., R. S. Rifo, E. P. Ricci, D. Decimo, and T. Ohlmann. 2009. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta 1789:542-557. [DOI] [PubMed] [Google Scholar]
- 5.Barry, U., and Z. Zuo. 2005. Opioids: old drugs for potential new applications. Curr. Pharm. Des. 11:1343-1350. [DOI] [PubMed] [Google Scholar]
- 6.Blankenship, J. R., W. J. Steinbach, J. R. Perfect, and J. Heitman. 2003. Teaching old drugs new tricks: reincarnating immunosuppressants as antifungal drugs. Curr. Opin. Invest. Drugs 4:192-199. [PubMed] [Google Scholar]
- 7.Bogdanovsky, D., and M. Pierre. 1979. Effect of tilorone and quinacrine on translation in reticulocyte lysates. Biochem. Pharmacol. 28:3683-3685. [DOI] [PubMed] [Google Scholar]
- 8.Caliguira, L. G., and I. Tamm. 1973. Guanidine and 2-(α-hydroxybenzyl)-benzimidazole (HBB): selective inhibitors of picornavirus multiplication, p. 257-294. In W. A. Carter (ed.), Selective inhibitors of viral function. CRC Press, Cleveland, OH.
- 9.Chaires, J. B. 2005. Competition dialysis: an assay to measure the structural selectivity of drug-nucleic acid interactions. Curr. Med. Chem. Anticancer Agents 5:339-352. [DOI] [PubMed] [Google Scholar]
- 10.Ciak, J., and F. E. Hahn. 1967. Quinacrine (atebrin): mode of action. Science 56:655-656. [DOI] [PubMed] [Google Scholar]
- 11.Filbin, M. E., and J. S. Kieft. 2009. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 19:267-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay, G. J., W. R. Wilson, and B. C. Baguley. 1989. Chemoprotection by 9-aminoacridine derivatives against the cytotoxicity of topoisomerase II-directed drugs. Eur. J. Cancer Clin. Oncol. 25:1695-1701. [DOI] [PubMed] [Google Scholar]
- 13.Gardner, T. B., and D. R. Hill. 2001. Treatment of giardiasis. Clin. Microbiol. Rev. 14:114-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grover, R., M. M. Candeias, R. Fåhraeus, and S. Das. 2009. p53 and little brother p53/47: linking IRES activities with protein functions. Oncogene. 28:2766-2772. [DOI] [PubMed] [Google Scholar]
- 15.Guo, C., A. V. Gasparian, Z. Zhuang, D. A. Bosykh, A. A. Komar, A. V. Gudkov, and K. V. Gurova. 2009. 9-Aminoacridine-based anticancer drugs target the PI3K/AKT/mTOR, NF-kappaB and p53 pathways. Oncogene 28:1151-1161. [DOI] [PubMed] [Google Scholar]
- 16.Gurova, K. V., J. E. Hill, C. Guo, A. Prokvolit, L. G. Burdelya, E. Samoylova, A. V. Khodyakova, R. Ganapathi, M. Ganapathi, N. D. Tararova, D. Bosykh, D. Lvovskiy, T. R. Webb, G. R. Stark, and A. V. Gudkov. 2005. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kappaB-dependent mechanism of p53 suppression in tumors. Proc. Natl. Acad. Sci. U. S. A. 102:17448-17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haïk, S., J. Brandel, D. Salomon, V. Sazdovitch, N. Delasnerie-Lauprêtre, J. Laplanche, B. Faucheux, C. Soubrié, E. Boher, C. Belorgey, J. Hauw, and A. Alpérovitch. 2004. Compassionate use of quinacrine in Creutzfeldt-Jakob disease fails to show significant effects. Neurology 63:2413-2415. [DOI] [PubMed] [Google Scholar]
- 18.Hays, C. W. 2000. The United States Army and malaria control in World War II. Parassitologia 42:47-52. [PubMed] [Google Scholar]
- 19.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 20.Hellen, C. U. 2009. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta 1789:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershey, J. W. B., and W. C. Merrick. 2000. The pathway and mechanism of eukaryotic protein synthesis, p. 33-38. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Jackson, R. J. 2000. Comparative view of initiation site selection mechanisms, p. 127-183. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews, (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Jiang, M. C., J. K. Lin, and S. S. Chen. 1996. Inhibition of HIV-1 Tat-mediated transactivation by quinacrine and chloroquine. Biochem. Biophys. Res. Commun. 226:1-7. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, Y., K. Hirata, H. Tanaka, and T. Yamada. 2003. Quinacrine administration to a patient with Creutzfeldt-Jakob disease who received a cadaveric duramater graft-an EEG evaluation. Rinsho Shinkeigaku 43:403-408. [PubMed] [Google Scholar]
- 25.Komar, A. A., and M. Hatzoglou. 2005. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 280:23425-23428. [DOI] [PubMed] [Google Scholar]
- 26.Leduc, E. H., W. Bernhard, A. Viron, J. Fain, and E. Puvion. 1981. Effect of quinacrine on nuclear structure and RNA synthesis in cultured rat hepatocytes. Cancer Res. 41:2832-2841. [PubMed] [Google Scholar]
- 27.Liu, Y., E. Wimmer, and A. V. Paul. 2009. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim. Biophys. Acta 1789:495-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malina, A., S. Khan, C. B. Carlson, Y. Svitkin, I. Harvey, N. Sonenberg, P. A. Beal, and J. Pelletier. 2005. Inhibitory properties of nucleic acid-binding ligands on protein synthesis. FEBS Lett. 579:79-89. [DOI] [PubMed] [Google Scholar]
- 29.Novac, O., A.-S. Guenier, and J. Pelletier. 2004. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 32:902-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novella, I. S. 2003. Contributions of vesicular stomatitis virus to the understanding of RNA virus evolution. Curr. Opin. Microbiol. 6:399-405. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien, R. L., J. G. Olenick, and F. E. Hahn. 1966. Reactions of quinine, chloroquine and quinacrine with DNA and their effects on the DNA and RNA polymerase reactions. Proc. Natl. Acad. Sci. U. S. A. 55:1511-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phuan, P. W., J. A. Zorn, J. Safar, K. Giles, S. B. Prusiner, F. E. Cohen, and B. C. May. 2007. Discriminating between cellular and misfolded prion protein by using affinity to 9-aminoacridine compounds. J. Gen. Virol. 88:1392-1401. [DOI] [PubMed] [Google Scholar]
- 34.Pritchard, N. J., A. Blake, and A. R. Peacocke. 1966. Modified intercalation model for the interaction of amino acridines and DNA. Nature 212:1360-1361. [DOI] [PubMed] [Google Scholar]
- 35.Ray, P. S., R. Grover, and S. Das. 2006. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 7:404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren, J., and J. B. Chaires. 2001. Rapid screening of structurally selective ligand binding to nucleic acids. Methods Enzymol. 340:99-108. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, L. O., R. A. Seamons, and G. J. Belsham. 1998. Recognition of picornavirus internal ribosome entry sites within cells: influence of cellular and viral proteins. RNA 4:520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romanelli, F., K. M. Smith, and A. D. Hoven. 2004. Chloroquine and hydroxychloroquine as inhibitors of human immunodeficiency virus (HIV-1) activity. Curr. Pharm. Des. 10:2643-2648. [DOI] [PubMed] [Google Scholar]
- 39.Sablina, A. A., A. V. Budanov, G. V. Ilyinskaya, L. S. Agapova, J. E. Kravchenko, and P. M. Chumakov. 2005. The antioxidant function of the p53 tumor suppressor. Nat. Med. 11:1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saenger, W. 1984. Forces stabilizing associations between bases: hydrogen bonding and base stacking, p. 117-158. In C. R. Cantor (ed.), Principles of nucleic acid structure. Springer advanced texts in chemistry. Springer-Verlag, New York, NY.
- 41.Sambrook, J., F. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Sasvari, Z., S. Bach, M. Blondel, and P. D. Nagy. 2009. Inhibition of RNA recruitment and replication of an RNA virus by acridine derivatives with known anti-prion activities. PLoS One 4:e7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shcherbakova, I., S. Mitra, A. Laederach, and M. Brenowitz. 2008. Energy barriers, pathways, and dynamics during folding of large, multidomain RNAs. Curr. Opin. Chem. Biol. 12:655-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha, R., M. Hossain, and G. S. Kumar. 2007. RNA targeting by DNA binding drugs: structural, conformational and energetic aspects of the binding of quinacrine and DAPI to A-form and H(L)-form of poly(rC).poly(rG). Biochim. Biophys. Acta 1770:1636-1650. [DOI] [PubMed] [Google Scholar]
- 45.Sonenberg, N., and A. G. Hinnebusch. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun, S., N. L. Rao, J. Venable, R. Thurmond, and L. Karlsson. 2007. TLR7/9 antagonists as therapeutics for immune-mediated inflammatory disorders. Inflamm. Allergy Drug Targets 6:223-235. [DOI] [PubMed] [Google Scholar]
- 47.Toubi, E., A. Kessel, I. Rosner, M. Rozenbaum, D. Paran, and Y. Shoenfeld. 2006. The reduction of serum B-lymphocyte activating factor levels following quinacrine add-on therapy in systemic lupus erythematosus. Scand. J. Immunol. 63:299-303. [DOI] [PubMed] [Google Scholar]
- 48.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Velden, A., A. Kaminski, R. J. Jackson, and G. J. Belsham. 1995. Defective point mutants of the encephalomyocarditis virus internal ribosome entry site can be complemented in trans. Virology 214:82-90. [DOI] [PubMed] [Google Scholar]
- 50.Wallace, D. J. 1989. The use of quinacrine (Atabrine) in rheumatic diseases: a reexamination. Semin. Arthritis Rheum. 18:282-296. [DOI] [PubMed] [Google Scholar]
- 51.Whitehouse, M. W., and H. Boström. 1965. Biochemical properties of anti-inflammatory drugs. VI. The effects of chloroquine (resochin), mepacrine (quinacrine) and some of their potential metabolites on cartilage metabolism and oxidative phosphorylation. Biochem. Pharmacol. 14:1173-1184. [DOI] [PubMed] [Google Scholar]
- 52.Yan, Z., and A. M. Baranger. 2004. Binding of an aminoacridine derivative to a GAAA RNA tetraloop. Bioorg. Med. Chem. Lett. 14:5889-5893. [DOI] [PubMed] [Google Scholar]