Abstract
The predominance and continual emergence of new variants in GII-4 noroviruses (NVs) in recent years have raised questions about the role of host immunity and histo-blood group antigens (HBGAs) in NV evolution. To address these questions, we performed a genetic and phenotypic characterization of GII-4 variants circulating in the past decade (1998 to 2008). Ninety-three GII-4 sequences were analyzed, and of them, 16 strains representing 6 genetic clusters were selected for further characterization. The HBGA binding properties were determined by both saliva- and oligosaccharide-binding assays using P particles as a model of NV capsid. The antigenic properties were also examined by enzyme immunoassay (EIA), Western blot analysis, and receptor blocking assay, using P-particle-specific antibodies from immunized mice and GII-4 virus-infected patients. Our results showed that 15 of the 16 GII-4 viruses bound to saliva of all A, B, and O secretors. Oligosaccharide binding assays yielded largely consistent results, although the binding affinities to some oligosaccharides varied among some strains. The only nonbinder had a mutation in the binding site. While antigenic variations were detected among the 16 strains, significant cross-blocking on the HBGA binding was also noted. Sequence alignment revealed high conservation of HBGA binding interfaces with some variations in adjacent regions. Taken together, our data suggested that the ability of GII-4 to recognize different secretor HBGAs persisted over the past decade, which may explain the predominance of GII-4 over other genotypes. Our data also indicated that both the host immunity and HBGAs play a role in NV evolution. While host immunity may continue driving NV for antigenic change, the functional selection by the HBGAs tends to lock the architecture of the capsid/HBGA interfaces and allows only limited variations outside the HBGA binding sites. A potential outcome of such counterselection between theses two factors in NV evolution is discussed.
Noroviruses (NVs) have been recognized as the most important cause of nonbacterial acute gastroenteritis in both developed and developing countries, affecting people of all ages (13, 35, 39, 44, 48, 56). They are single-stranded positive-sense RNA viruses belonging to the family Caliciviridae. NVs are highly contagious, spreading by a fecal/oral pathway through person-to-person contact and by contaminated food and/or water and usually causing large outbreaks within closed communities in a variety of settings, such as hospitals, nursing homes, schools, childcare centers, restaurants, cruise ships, and the military (11, 63). Human NVs have been difficult to study due to diverse members and the lack of an efficient cell culture and animal model for human NVs. The cloning of the NV genomes (33, 36, 73) and subsequent expression of the viral capsid proteins in baculovirus and other expression systems (3, 31, 32) have greatly advanced the research of NVs, including host-virus interaction, immunology, diagnosis, molecular virology, and epidemiology (16, 17, 19, 20, 25, 28-30, 46, 51, 59, 73).
Several lines of evidence indicate that NVs recognize human histo-blood group antigens (HBGAs) as a ligand or receptor in a strain-specific manner (63, 64). HBGAs are complex carbohydrates presenting on red blood cells and on the epithelia of digestive, respiratory, and genitourinary tracts. They also exist in biologic fluid, such as milk and saliva. NVs are highly diverse in recognizing the human HBGAs, and a number of HBGA-binding patterns involving the ABO, secretor, and Lewis families of human HBGAs have been described (19, 20, 23, 24, 26, 28, 43, 45, 55). The association of HBGA binding with clinical infection and illness has been demonstrated by volunteer challenge studies and outbreak investigations (25, 27, 42, 62, 66), although exceptions also have been reported (41, 50, 53). Further study has mapped the HBGA binding site in the protruding (P) domain of the viral capsid protein (60). Using the P domain as a model, the atomic structures of the HBGA binding interfaces have been resolved by X-ray crystallography (5, 7, 9). The interfaces are comprised of several amino acids located on the top of the P dimer, within the outermost surface of the viral capsid. Extensive hydrogen bond networks between the P dimer and the HBGAs were elucidated and further confirmed by mutagenesis analyses (61, 68, 69). Despite significant differences in genetics and HBGA binding patterns, the sequences of the HBGA-binding interfaces are highly conserved within, but not between, the two major human-related genogroups (GI and GII) of NVs, suggesting that HBGAs are important factors in NV evolution (9, 69).
The NV capsid is composed of a single major structural protein, the capsid protein (VP1), which can be divided into two major domains: the shell (S) and the protruding (P) domains (52). Expression of the full-length VP1 by a eukaryotic system forms empty virus-like particles (VLPs) that have been used as a surrogate for NVs for many years, e.g., in diagnostic tests. Recent studies showed that expression of the P domain alone results in the formation of a subviral particle, the P particle (54, 60). Owing to its easy production in an Escherichia coli system and the same HBGA-binding properties and antigenicity as its parental VLP, the P particle has been used as a research tool of NV-HBGA interaction in a number of studies (54, 59, 60, 67, 68, 69). This report took advantage of the convenient P particle model to study the phenotypic HBGA-binding properties and antigenicity of GII-4 NVs that have circulated in the past decade.
NVs are grouped into five genogroups (GI to GV), of which GI and GII are involved in the majority of acute viral gastroenteritis cases in humans. Strains within each genogroup can be further divided into genotypes, and up to 30 genotypes of GI and GII NVs have been described (75). NVs can be detected throughout the year, with peaks during the fall and winter seasons. Strains representing multiple genotypes can be found cocirculating in the same geographical area during a season. However, a single genotype of NVs, GII-4 (genogroup II genotype 4), has been the predominant cause of major acute gastroenteritis epidemics in many countries since the mid-1990s, and the number of GII-4 epidemics has increased in recent years (49). Overall, the GII-4 genotype is estimated to be responsible for 60 to 80% of all NV-associated outbreaks worldwide (43).
Molecular surveillance has found that the GII-4 viruses are continuously changing, with new variants emerging every 2 or 3 years (1, 2, 57, 71, 72). One hypothesis suggests that the GII-4 viruses might be under selection pressure of the herd immunity, similar to the epochal evolution model used to describe the evolution of influenza (flu) viruses (56). New antigenic variants of GII-4 derived by genetic shift (replacement) accompanied by changes of HBGA binding specificities have been reported (43). However, the HBGA-binding interfaces of NVs have been found to be highly conserved among NVs within each of the two major genogroups, supporting HBGAs as an important factor in NV evolution (69). In fact, it has been shown that the major HBGA-binding pattern of GII-4 viruses to the H3, Leb, and Ley antigens has remained unchanged from 1974 to 1997 (4, 23, 24).
The objective of this study was to elucidate the roles of HBGAs and host immunity in NV evolution using GII-4 viruses as a model. Since most of the studies on the epochal evolution of GII-4 were based on genetic analysis and focused on GII-4 variants identified in the past decade, we performed a study on the GII-4 variants in the same period by both genetic and phenotypic characterizations. Phylogenetic analysis revealed 6 genetic clusters of GII-4 viruses similar to those reported before. Characterization of HBGA-binding patterns of the GII-4 viruses revealed a consensus phenotype of binding to all A, B, and O secretor HBGAs, with some variations in affinity to these antigens. We also discussed the role of both host immunity and HBGAs in NV evolution. While the host immunity may drive NVs for change, as a functional selection factor, the HBGAs may restrict variation. This counterselection mechanism may help in understanding the epochal evolution hypothesis. The principles found through the study of GII-4 NVs can also be applied to other genotypes, which may eventually lead to a refined functional classification of all NVs.
MATERIALS AND METHODS
Clinical samples.
Stool samples from patients infected with GII-4 viruses in outbreak investigations in the United States and Canada were used as the source of viruses for cloning and sequencing of the viral capsid genes, for genetic analysis, and for expression of viral P particles for characterization of antigenic and HBGA binding variations. The U.S. studies involved mainly outbreaks that occurred in the states of Virginia and Ohio and in the U.S. military from 1998 to 2008. The Canada study included a statewide surveillance of NV gastroenteritis outbreaks in the province of Alberta from 2000 to 2007. In addition, the capsid gene cDNAs of the 2004-Hunter and 2005-Sakai (43) strains were kindly provided by Ralph Baric (University of North Carolina—Chapel Hill, Chapel Hill, NC) as the templates for expression of the viral P particles for the antigenic and HBGA characterization. Finally, the capsid gene cDNA from one GII-4 strain (06Y06bC1) in Japan was included in our study (obtained from T. Tanaka's laboratory in the Sakai City Institute of Public Health, Sakai, Osaka, Japan).
Expression and purification of P particles in E. coli.
The P proteins of different GII-4 strains in this study were made according to the procedures described previously (54, 67, 68, 69). The capsid sequences of individual strains were determined by DNA sequencing on the PCR-amplified products following cloning of the cDNA in a pGEM vector. A cysteine-containing peptide was linked to the N (CNGRC-P) or C (P-CDCRGDCFC) terminus of the P domains to enhance P-particle formation (65). The cDNAs encoding the capsid P domain without the hinge were cloned into the expression vector pGEX-4T-1 (GST-gene fusion system; Amersham Biosciences, Piscataway, NJ) at the SalI and NotI sites. After sequence confirmation of each expression vector, the P proteins were expressed in E. coli as described previously (59, 60, 70). Briefly, strain BL21 of E. coli was used as the host, and the bacterial cultures were induced by IPTG (isopropyl-β-d-thiogalactopyranoside) (0.4 mM) at room temperature (∼22°C) overnight. Purification of P protein-glutathione S-transferase (GST) fusion proteins from bacteria was performed using glutathione Sepharose 4 Fast Flow (Amersham Bioscience) according to the manufacturer's protocol. The P proteins were released from GST by thrombin (GE Healthcare Life-Sciences, Piscataway, NJ) digestion at room temperature overnight (59, 65). The determination of P-particle formation was performed by gel filtration using a Superdex 200 (GE Healthcare Life-Sciences, Piscataway, NJ) size exclusion column, in which the P particles form a peak at ∼830 kDa (60, 67, 70). The formation of P particles was also confirmed by SDS-PAGE of the P-particle preps under a partially denaturing condition (no boiling, no reducing reagent, and a low concentration [0.2%] of SDS) which usually resulted in a band with a large molecular size at the top of the gel (data not shown). Over 80 to 90% of the P proteins usually were found in the high-molecular-weight band.
HBGA binding assay.
The binding of P particles to HBGAs was measured by saliva- and synthetic oligosaccharide-based binding assays as described previously (23, 24). The saliva samples were selected from a panel of saliva with well-defined A, B, O, secretor, and Lewis types described previously (23, 24). Boiled saliva samples were diluted 1:1,000 and coated on 96-well microtiter plates (Dynex Immulon; Dynatech, Franklin, MA) at 4°C overnight. After blocking with 5% nonfat milk, 100 μl of P particles (16 μg/ml) were added and incubated at 37°C for 1 h. The bound P particles were detected using a guinea pig serum anti-NV (1:3,330) followed by horseradish peroxidase (HRP)-conjugated goat anti-guinea pig IgG (ICN, Aurora, OH). The signal intensities were displayed using a TMB kit (Kierkegaard and Perry Laboratory, Gaithersburg, MD).
For the oligosaccharide-based binding assays, after coating the plates with pooled rabbit anti-NV (1:2,000) at 4°C overnight, P particles were captured during a 1-h incubation at 37°C. The conditions for their binding to synthetic oligosaccharide-based HBGAs, as reported previously (23, 24), were within a comparable range of protein concentrations (24 ± 9 μg/ml). Oligosaccharides conjugated with two types of carriers, polyacrylamide (PAA)-biotin or bovine serum albumin (BSA), were used in this study and included the following: conjugates of H type 3, type A, and type B disaccharides; Lea, Lex, H type 1, H type 2, type A, and type B trisaccharides; and Leb, Ley, salic-Lex, and salic-Lex tetrasaccharides (GlycoTech Corporation, Rockville, MD). Bound HBGAs were detected using HRP-conjugated streptavidin (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) and displayed using the TMB kit described above.
Mice immunization and Western blot analysis.
Two BALB/c mice per group for each GII-4 strain were immunized with P particles by intramuscular (i.m.) route four times at 2-week intervals. Each immunization consisted of 100 μg P protein per mouse, with Freund's complete adjuvant in the first injection and Freund's incomplete adjuvant in the following injections. Sera were collected 1 week following the last immunization. The immunogenicities of P proteins were determined by Western blot analysis as follows: 1.5 μg of P proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with 5% nonfat milk and incubated with antisera collected from immunized mice at a 1:1,000 dilution. After incubation with a secondary antibody-HRP conjugate at 1:2,000, the HRP was detected by using ECL detection reagent (Amersham Bioscience, Buckinghamshire, England). The ECL signals were captured by HyBlot CL autoradiography film (Denville Scientific, Inc., Metuchen, NJ).
Blocking of HBGA binding by hyperimmune antisera from mice.
The saliva binding assays described above were modified in order to measure the ability of the hyperimmune antisera from mice to block P particles binding to HBGAs. Briefly, the same format of saliva binding assay was used, except the murine antisera were preincubated with P proteins in a separate 96-well microtiter plate at 37°C for 1 h before they were transferred into the saliva-coated plate. Murine antisera were first diluted to 1:50 and then subjected to a 3-fold serial dilution to 1:150 and 1:450. The serum dilutions at which 50% (BT50) or 90% (BT90) of binding was blocked were calculated from the optical density at 450 nm (OD450) values between wells with and without incubation with the murine antisera as described previously (23, 24, 59).
Serology and HBGA blocking assays using serum samples from patients.
Paired serum samples at the acute phase and 2 weeks after the onset of the illness from patients involved in a GII-4 outbreak in a nursing home for the elderly in 1998 were used in this study. After coating 96-well microtiter plates (Dynex Immulon; Dynatech, Franklin, MA) with pooled rabbit antinorovirus antibody (1:2,000) (34) at 4°C overnight, the P-particle proteins were captured by incubation for 1 h at 37°C. Acute- and convalescent-phase sera were added to wells with 2-fold serial dilutions, and then the HRP-conjugated goat anti-human IgG (ICN, Aurora, OH) was added. The signal intensities were displayed by a TMB kit (Kierkegaard and Perry Laboratory, Gaithersburg, MD). The blocking assays were performed using the same conditions of synthetic oligosaccharide-based HBGA binding assays described above, except an extra step of incubation with human antisera was included before the oligosaccharides were added.
Sequencing and phylogenetic analysis.
The sequences of previously uncharacterized GII-4 strains considered in this study were determined by sequencing the reverse transcription (RT)-PCR products after cloning into the pGEX-4T-1 vector (Amersham Biosciences, Piscataway, NJ). The remaining 74 GII-4 capsid sequences were downloaded from GenBank. Multiple alignments (MAs) were generated using ClustalW2 (37) and MUSCLE (12), available at http://www.ebi.ac.uk and http://www.phylogeny.fr, respectively. Due to high sequence identity and the nearly consistent length of the input sequences (ranging from 317 to 319 amino acids), unambiguous multiple alignments were obtained that are identical except for the placement of the insertion at positions 174/175. MA generated by MUSCLE is included in Fig. S3 in the supplemental material and has been used for further phylogenetic analysis after processing with Gblocks (8) to exclude weakly aligned and gapped positions.
Reconstruction and analysis of phylogenetic relationships between the GII-4 P2 domain sequences (without the hinge region) was performed using primarily the widely used PhyML (18) maximum likelihood (ML)-based program, which is available through the MABL (10) and Mobile (47) phylogenetic analysis portals used here. The assessment of the resulting phylogenetic trees has been performed using resampling and statistical models. The support for PhyML trees has been computed using the approximate likelihood ratio test (aLRT) and standard bootstrapping, with values obtained using the nonparametric version of aLRT (which were very similar to those obtained in 100 runs of bootstrapping) reported in Fig. S1 in the supplemental material. The effects of using different substitution matrices, including Blosum62, PAM, and modified WAG matrices (38), as well as different search strategies available in PhyML (nearest neighbor interchanges [NNI] versus subtree pruning and regrafting [SPR]) were also assessed, yielding similar results, with 6 main clusters of sequences identified with high support independent of the parameters used. For additional analysis, we used another maximum likelihood-based approach, as implemented in Puzzle (58), as well as standard parsimony and distance-based approaches, as implemented in the Phylip ProtPars (14) and MegAlign (DNAStar; DNASTAR Inc., Madison, WI) programs. The MegAlign tree obtained using the Hein method (21) is reported in Fig. S2 in the supplemental material for comparison with PhyML trees.
Statistical analysis.
To test differences among serum samples in the HBGA blocking assays, we used the Kruskal-Wallis test and the Ansari-Bradley one-way analysis as provided by SAS 9.1.3 (SAS Institute Inc., Cary, NC).
RESULTS
Phylogenetic analysis and strain selection.
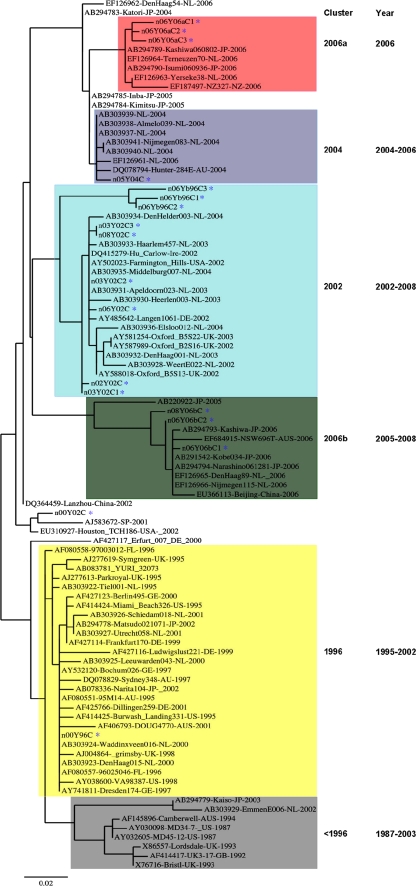
A phylogenetic analysis of the P domains (∼320 amino acids) of 93 GII-4 strains circulating between 1987 and 2008 was performed. Seventy-four sequences were selected from GenBank as referenced in previous reports (43, 56), and 18 were from our sample collection (11 from the United States, 6 from Canada, and 1 from Japan). The minimum (pairwise) amino acid identity among the 93 sequences was 86.8%, and due to nearly identical lengths and a limited number of insertions and deletions, essentially identical multiple alignments (MAs) were obtained, irrespective of the alignment algorithm used. Starting from these MAs, 6 major genetic clusters (named <1996, 1996, 2002, 2004, 2006a, and 2006b) similar to those described by previous studies have been identified using maximum likelihood (ML) phylogenetic analysis (Fig. 1) (56). It should be noted that two main branches, comprising clusters denoted as 1996 and <1996 in one branch and the remaining clusters in the other branch, were consistently identified by PhyML (with both bootstrap and aLRT support of more than 95%) as well as other programs, providing support for these two groups evolving independently, rather than in a linear type of progression from an earliest to extant clusters.
FIG. 1.
Phylogenic tree of GII-4 norovirus P domain sequences (without the hinge region) generated by using the PhyML maximum likelihood analysis. GII-4 sequences from 1987 to 2008 are shown in different colored clusters. Strains isolated in this study are as follows: 00Y96C (GU937455), 06Yb96C1 (GU937451), 06Yb96C2 (GU937451), 06Yb96C3 (GU937453), 00Y02C (GU937448), 03Y02C1 (GU937449), 03Y02C2 (GU937450), 06Y02C (GU937454), 02Y02C (GU937456), 08Y02C (GU937463), 03Y02C3 (GU937465), 05Y04C (GU937457), 06Y06aC1 (GU937458), 06Y06aC2 (GU937459), 06Y06aC3 (GU937460), 08Y06bC (GU937464), 08Y06bC1 (GU937462), and 08Y06bC2 (GU937461). These strains are highlighted by blue stars.
Sixteen strains representing the 5 major clusters (Fig. 1) were selected for further investigation. These included two strains of the 1996 cluster that were isolated in 1998, and 2000; six strains of the 2002 cluster that were isolated in 2002, 2003, 2006, and 2008; two strains of the 2004 cluster that were isolated in 2004 and 2005; two 2006a strains that were isolated in 2006; and three 2006b strains that were isolated in 2005 and 2006; one strain isolated in 2000 is an outlier (Fig. 2). For a direct comparison, two other strains (2004-Hunter and 2005-Sakai) that were reported to be non-HBGA binders (43) were also studied.
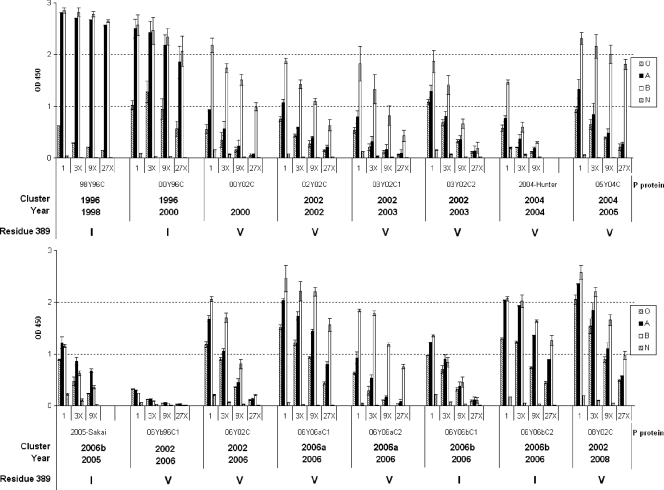
FIG. 2.
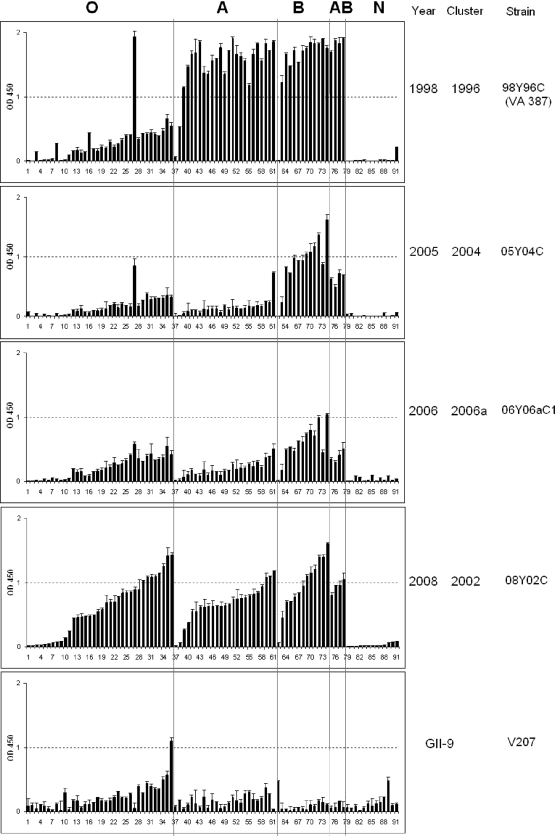
Binding of GII-4 P-domain proteins to human saliva samples. Boiled saliva samples were coated onto 96-well plates prior to the addition of P proteins from 16 strains of GII-4 viruses (98Y96C, 00Y96C, 00Y02C, 02Y02C, 03Y02C1, 03Y02C2, 2004-Hunter, 05Y04C, 2005-Sakai, 06Yb96C1, 06Y02C, 06Y06aC1, 06Y06aC2, 06Y06bC1, 06Y06bC2, and 08Y02C). The P proteins were tested in a series of 3-fold-dilution enzyme-linked immunosorbent assays (ELISA) (optical densities at 450 nm were averaged from at least two independent experiments). “O,” “A,” “B,” and “N” represent the type O (H antigen), A, B, and nonsecretor saliva, respectively. The amino acid (I or V) present at position 389 is indicated for each strain.
P-particle formation and HBGA binding activity.
In our previous studies we have found that the formation of P particles is critical for the HBGA binding property of NVs (59, 60, 65). Using the same approach of an E. coli expression system (54, 59, 67, 68, 69), the expressed P proteins of all 16 GII-4 strains formed P particles, which was demonstrated by gel filtration and nondenaturing SDS-PAGE (data not shown). The formation of P particles was also suggested by their strong binding to the saliva of A, B, and/or O secretors but not to nonsecretors that are known to be specific to GII-4 viruses (see results below). The yields of P particles slightly varied among some strains, even when standard E. coli culture conditions were used. To overcome this problem, a starting material of P protein at a concentration of 0.5 mg/ml was prepared for all strains for the HBGA binding assays. Specific reactions were obtained at this concentration of P particles used (data not shown).
The GII-4 NVs recognize secretor HBGAs.
Thirteen of the first fourteen GII-4 strains exhibited similar patterns of HBGA binding to saliva of the type A, B and O secretors, but not to the nonsecretors (Fig. 2), similar to what was reported for the prototype VA387 and Grimsby viruses (24). To our surprise, the previously reported nonbinding GII-4 strains, 2004-Hunter and 2005-Sakai (43), also revealed specific binding to the secretor saliva. These bindings were confirmed by the oligosaccharide-based binding assay (Fig. 3).
FIG. 3.
Binding of GII-4 P-domain proteins to synthetic oligosaccharides. P proteins from 10 strains of GII-4 viruses (98Y96C, 00Y96C, 02Y02C, 03Y02C1, 05Y04C, 2004-Hunter, 2005-Sakai, 06Y06aC1, 06Y06bC1, and 06Y06bC2) were captured onto 96-well plates coated with pooled rabbit antinoroviruses and incubated with oligosaccharides (H type 1, H type 2, H type 3, A, B, Leb, Ley, Lea, Lex, Sialyl-Lea, or Sialyl-Lex) overnight. The y axis indicates captured HBGAs detected by HRP-avidin (optical densities at 450 nm were averaged from at least two independent experiments). The concentrations of P proteins were 24 ± 9 μg/ml.
Sequence inspection of the only nonbinding strain (06Yb96C1, Fig. 2) identified an amino acid mutation (N373S) in one of the three conserved HBGA binding sites of the viral capsid (68, 69). Since N373 is conserved among 90 of the 93 GII-4 viruses (Fig. 4; see also Fig. S3 in the supplemental material), we hypothesize that the N373S mutation may be responsible for the nonbinding (saliva) or low-binding (oligosaccharides; see result below) phenotype of this strain. It is noticed that this strain was isolated from a stool sample in which another GII-4 strain (06Yb96C2) was identified. Strain 06Yb96C2 did not have the N373S mutation, and its P particles displayed the typical GII-4 binding pattern (Fig. 2). Taken together, these results show that, while the binding intensities to individual carbohydrate epitopes may vary, the ability to recognize the major secretor antigen (α-1,2 fucose) has not changed among different GII-4 variants in the years from 1998 to 2008.
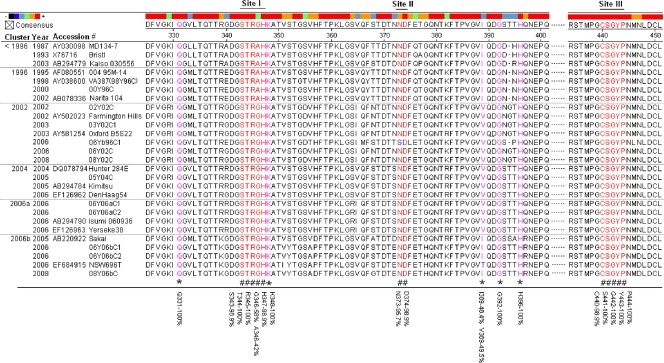
FIG. 4.
Sequence alignment of the HBGA-binding interfaces and the surrounding regions. The P domain sequences of 27 GII-4 viruses selected from different years were aligned using MegAlign. The numbers along the top indicate the position of the residues according to consensus sequence. Amino acids in red (noted by “#” at the bottom) indicate residues that are required for HBGA binding. Pink residues with “*” at the bottom indicate amino acids that affect the binding specificity. Blue letters indicate differences from the consensus sequence within the binding interfaces or surrounding regions. The percent similarity between all 93 strains is indicated at the bottom for key amino acid positions involved in HBGA binding.
The H-related antigens (H-3, Leb, and Ley) play an important role in GII-4 NV recognition.
To further define the HBGA binding specificity of the GII-4 viruses, 10 strains, including the nonbinding strain (06Yb96C1) in the saliva binding assays, were tested with the oligosaccharide-based binding assays. All 10 strains that had typical binding patterns in the saliva binding assay reacted strongly to H-1, H-3, Leb, and Ley (Fig. 3). It was noted that the nonbinding strain (06Yb96C1) in the saliva assays also revealed a low binding activity toward Leb, Ley, and H-3 (Fig. 3). Some of the GII-4 P particles also variably reacted with A and B antigens (see below), but none reacted with the nonsecretor antigens (Lea, Lex, sialyl-Lea, and sialyl-Lex), which is consistent with the results from the saliva-based binding assays.
GII-4 NVs bind to saliva of the majority of the population (A, B, and O secretors).
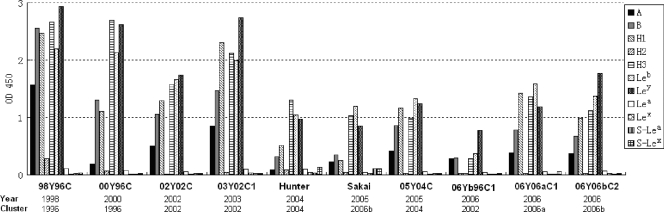
Four strains isolated in 1998, 2005, 2006, and 2008 were tested for their abilities to bind a large panel of saliva samples obtained from 91 individuals with different blood types. Although the binding affinities varied, all strains retained the ability to bind all A, B, and O secretors, but not the nonsecretors (Fig. 5). Strain GII-9 is a nonsecretor binder according to our previous studies (23, 24). It bound nonsecretors as well as type O secretors, with lower binding to type A secretors and the lowest binding to B- and AB-type secretors (Fig. 5), which was expected according to our previous studies (23, 24). These data suggest that the ability of GII-4 viruses to infect the majority (A, B, and O secretors, 80 to 85%) of the general population is unlikely to change.
FIG. 5.
Binding of P particles from representative GII-4 norovirus strains to saliva from 91 individuals of different blood types. Boiled saliva samples were coated onto 96-well plates prior to the addition of 2 μg/ml P proteins from 4 strains of GII-4 viruses (98Y96C, 05Y04C, 06Y06aC1, and 08Y02C) and 1 GII-9 virus (VA207). The y axis indicates the relative binding of the P proteins as detected using ELISA (optical densities at 450 nm were averaged from at least two independent experiments). The saliva samples are grouped by blood types and designated O, A, B, AB, and N representing type O (H antigen), A, B, AB, and nonsecretor saliva, respectively.
The HBGA binding interfaces of the GII-4 NVs are highly conserved.
Sequence alignment of 27 GII-4 strains selected from the 6 clusters described above revealed a high degree of conservation among the amino acids involved in the three HBGA binding sites (S343, T344, R345, D374, S441, G442, and Y443) (Fig. 4) (7, 60, 69) of GII-4 capsids according to the atomic structures of the HBGA binding interfaces (7, 68, 69). A further alignment of all 93 GII-4 sequences supports this conclusion (Fig. 4; see also Fig. S3 in the supplemental material). These data support the phenotypic conservation of the binding pattern of GII-4 NVs to HBGAs. However, residues around the three binding sites and in the rest regions of the P-2 domain displayed increased variability. These data indicate a continual genetic drift that may be related to the potential antigenic variations suggested by others (43).
A single amino acid substitution is associated with binding to the A antigen.
Our previous studies demonstrated that five amino acids (Q331, A346, K348, I389, and S441) in the P-2 domain constitute the A epitope (N-acetylgalactosamine) binding sites (7, 68). Mutation of Q331A, K348A, and I389V reduced binding to the A antigens without affecting the binding to the B antigens (68). Sequence alignment of the 93 GII-4 strains revealed that around half (45/93) of the strains contain an “I” at residue 389, while the remaining half contain a “V” (Fig. 2 and 4; see Fig. S3 in the supplemental material). The relative binding preference for A and B saliva closely correlated with the occurrence of I or V, in which strains with I389 had high binding to both the A and B antigens, while strains with V389 had a lower binding to the A saliva than to the B saliva. These data confirm that residue 389 is responsible for the specificity to the A antigen (Fig. 2 and 4). Interestingly, strains with “I” or “V” segregated among the six genetic clusters and appeared in waves in the past decade. The “I” strains dominated in clusters <1996 and 1996, and then a shift to mostly “V” strains occurred in 2000 (cluster 2002, cluster 2004, and cluster 2006a). Cluster 2006b indicates a shift back to “I” between 2005 and 2008 (Fig. 4). While the A epitope may not affect the binding to the H epitopes and, therefore, would not change the ability to infect the majority of the population, it remains unknown whether the high affinity of the “I389” phenotype to the A antigens has clinical significance compared to the “V389” phenotype.
Cross-reactions of antigenic and HBGA binding activities among epidemic GII-4 variants.
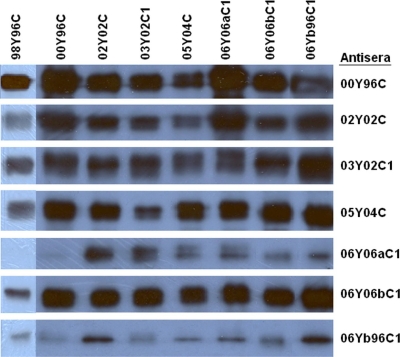
To examine the antigenic variations of GII-4 viruses in the studied period, mice were immunized with P particles of 8 representative strains (98Y96C, 00Y96C, 02Y02C, 03Y02C1, 05Y04C, 06Y06aC1, 06Y06bC1, and 06Yb96C1). To avoid any possible interference from E. coli proteins, which could possibly have contaminated the P-particle preps and could void the results of antibody detection EIA, Western blot assays were performed to confirm the results. Antisera from all mice recognized P particles of homologous strains as well as almost all heterologous strains studied (Fig. 6). We also noticed variations among some strains. For example, antisera of 06Y06aC1 isolated in 2006 resulted in the weakest signal that we observed when reacted against 98Y96C (VA387) that was isolated in 1998. Similarly, the same sera reacted weakly with 00Y96C that was isolated in 2000.
FIG. 6.
Interaction of GII-4 P-domain proteins with mouse hyperimmune sera. P proteins (1.5 μg each) from GII-4 strains 98Y96C, 00Y96C, 02Y02C, 03Y02C1, 05Y04C, 06Y06aC1, 06Y06bC1, and 06Yb96C1 were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and detected using antisera from mice immunized with P-particle proteins from various strains (on the right).
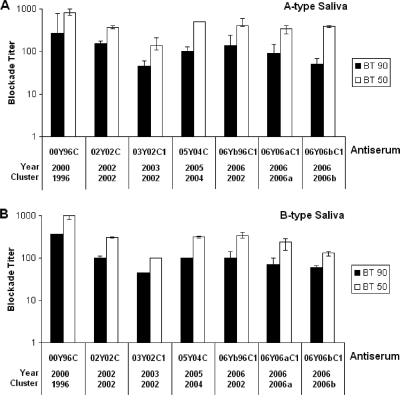
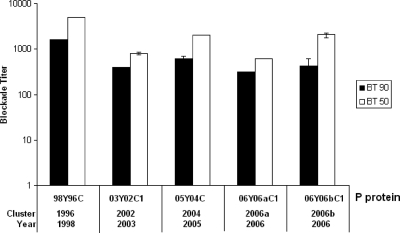
To further characterize the antigenic variations, we examined the ability of antibodies to block the prototype VA387 (98Y96C, 1998) P particles binding to the HBGAs in human saliva. Significant blocking activities of all serum samples against VA387 P-particle binding to type A (Leb+, Ley+, and A+) and/or type B (Leb+, Ley+, and B+) saliva were observed (Fig. 7). No statistical significance was found among the values of BT50 or BT90 across strains (P > 0.05, Kruskal-Wallis test and Ansari-Bradley one-way analysis).
FIG. 7.
The antibodies induced in mice by P-domain protein blocked GII-4 P-particle protein (VA387) binding to A- and B-type saliva. VA387 P proteins were blocked with the mouse antisera against each of 7 different strains (00Y96C, 02Y02C, 03Y02C1, 05Y04C, 06Yb96C1, 06Y06aC1, and 06Y06bC1) for 1 h prior to their addition to either A-type or B-type saliva-coated plates. The y axis indicates the serum dilution (blockade titer) as determined from optical density values. The white bars indicate the antiserum titer at which 50% of binding was blocked (BT50) in comparison to unblocked controls, and the black bars indicate the titer at which 90% of binding was blocked (BT90). The results are averaged from at least two independent experiments.
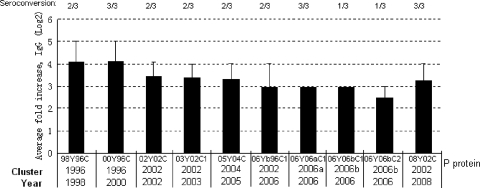
We also examined serum samples from patients infected with the 98Y96C GII-4 (VA387) virus isolated in 1998 in reaction with GII-4 viruses isolated in different years. P particles from 10 strains isolated between 1998 and 2008 were tested, and significant increases in antibody titers of convalescent-phase sera, with no significant differences in the acute-phase sera, were observed for all strains (Fig. 8). A trend of increases of antibody responses against strains that were isolated closer to 1998 (the year the antisera were collected) was observed, indicating a continual accumulation of antigenic variations during the study period.
FIG. 8.
Responses of human serum IgG to GII-4 P-domain proteins. Acute- and convalescent-phase serum samples were collected from individuals infected during a VA387 outbreak in 1998 and incubated with P proteins (98Y96C, 00Y96C, 02Y02C, 03Y02C1, 05Y04C, 06Yb96C1, 06Y06aC1, 06Y06bC1, 06Y06bC2, and 08Y02C) captured onto rabbit anti-NV-coated plates. The y axis indicates the fold increase in convalescent-phase serum IgG over acute-phase IgG titers (results were averaged across the three sample pairs). The number of individual sample pairs exhibiting seroconversion (defined as a fold increase of ≥4) is shown across the top.
We also tested the ability of the paired patient serum samples to block the binding of the GII-4 P particles to synthetic Leb, one of the major antigens of secretor HBGAs (as shown in Fig. 3). The convalescent-phase sera were able to completely block binding of the P particles to Leb at dilutions of ≥1:300 for all strains (Fig. 9), with no significant differences among values of the BT50 and/or BT90 across strains (P > 0.05, Kruskal-Wallis test and Ansari-Bradley one-way analysis). No substantial (≥50%) blocking was observed when the acute antisera were tested (data were not shown).
FIG. 9.
P proteins from 5 strains of GII-4 viruses (98Y96C, 03Y02C, 05Y04C, 06Y06aC1, and 06Y06bC1) were captured onto 96-well plates coated with pooled rabbit antisera against noroviruses and then blocked with a pair of human antiserum samples for 1 h prior to incubation with Leb oligosaccharide. The amount of bound Leb was detected using HRP-avidin. The y axis indicates the serum dilution (blockade titer) as determined from optical density values. The white bars indicate the antiserum titer at which 50% of binding was blocked (BT50) in comparison to unblocked controls, and the black bars indicate the titer at which 90% of binding was blocked (BT90). The results are averaged from at least two independent experiments.
DISCUSSION
The HBGA binding interfaces of NVs are known to be highly conserved among strains within each of the two major genogroups (GI and GII) (41). This finding has led to the hypothesis that the human HBGAs play an important role in NV evolution. In this study, we provide further support for this hypothesis through systematic characterization of the genetic and antigenic variations and HBGA binding patterns of extended GII-4 viruses from GenBank and our collection. Sequence alignments demonstrated that the HBGA binding sites remain highly conserved among GII-4 viruses over time, which is consistent with recent reports by others (1, 41). The amino acid residues essential for HBGA recognition, according to the atomic resolution structures and mutagenesis studies (4, 38), remained virtually unchanged among 93 GII-4 sequences spanning 2 decades from 1987 to 2008. A recent study indicated that the conservation of the HBGA binding site has been traced back to the 1970s (1). Our current study showed that the ability to bind the secretor HBGAs also did not change among 15 selected strains representing all six major genetic subclusters of GII-4 viruses isolated at different time points during the past decade, although certain levels of variation in binding affinity to some HBGAs by several GII-4 variants were noted.
The observed consensus phenotype of GII-4 binding to saliva of the A, B, and O secretors suggests a functional selection pressure on the capsid-ligand interface exerted by the human HBGAs. Structurally, the H epitope (α-1,2-fucose) plays the central role in recognition by the GII-4 viruses, as shown by VA387-HBGA interaction (7). With a conformational multivalent interface, additional carbohydrates around the H epitopes, such as N-acetyl-galactosamine, galactose, and α-1,3/4 fucose, may also participate in GII-4 binding. These epitopes form the A, B, Leb, Ley, H type A, and H type B plus the H-1, H-2, and H-3 molecules which are made in the A, B, and/or O secretors that represent 80 to 85% of the general population (40). According to these results we hypothesize that the persistence of such a broad spectrum of HBGA binding could be the major reason for GII-4 predominance over GI and other GII viruses that generally recognize a narrower spectrum of HBGAs (23, 24).
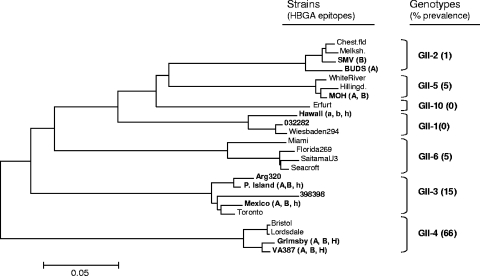
In our previous studies, we have described eight HBGA binding patterns among different GI and GII NVs studied (23, 24). Many of them recognize saliva of only the A and/or B secretors and do not recognize (or bind poorly to) the type O secretors, such as the GII-1, GII-2, GII-3, GII-5, GII-12, and GII-14 strains (23, 24; our unpublished data); thus, it would be logical to speculate why the GII-4 strains are so predominant among NV strains. In another study of NV gastroenteritis in China during 1999 to 2005 (74), we observed a typical gradient prevalence among different GII viruses in the case of acute gastroenteritis in children, which fits well with the gradient spectra of HBGA binding observed in this study (Fig. 10). This result is encouraging because it suggests that we may be able to predict NV prevalence for individual genotypes based on their HBGA binding spectra. Further studies in this direction are necessary.
FIG. 10.
Phylogenetic tree of selected GII-4 and other GII viruses. The receptor binding patterns of strains are shown in the parentheses as HBGA epitopes. The capital letters A, B, and H indicate a strong binding, and the lowercase letters a, b, and h indicate a weak binding. The percentage of prevalence of individual genotypes in epidemics of NV gastroenteritis was estimated based on surveillance of acute gastroenteritis in China from 1999 to 2005 (74).
The observed antigenic variations by different assays using antibodies from mice and GII-4-infected patients suggested that the host immunity continually drives antigenic changes within the GII-4 genotype. However, the high conservation of the HBGA binding interfaces, with variations only in surrounding regions, and the observed cross-blocking activities of HBGA binding among GII-4 from different years indicate that the HBGAs are involved and may act as an important counterbalancing factor in the selection process. This should be emphasized in the hypothesis of GII-4 epochal evolution, in which the carbohydrate variation may be only partially tolerated. Thus, antigenic variants that do not affect the HBGA binding interface and maintain the consensus phenotypes of HBGA binding are likely to be selected, contributing predominantly to the main stream of circulation in the population. On the other hand, antigenic variants with changes in the HBGA interface may have lost their survival advantage and, therefore, may not continue circulating in the population. The only nonbinder of GII-4 seen in this study was found to have an amino acid change in the HBGA binding interface and could be such an example.
It should be noted that our results are in conflict with recently published data of a study in which GII-4 variants with significant HBGA binding changes were reported (43). As a result, an alternative hypothesis, that the variation of HBGA binding is tolerated and that selection via host immunity may result in antigenic variants acquiring new HBGA binding patterns, has been proposed (43). Further studies are required to resolve this discrepancy. We would like to comment that the two studies were performed using different viral particles (VLPs versus P particles) as models. In retrospective analysis of our data, we do not anticipate that the use of P particles would pose a problem because our laboratory has developed over 40 P particles for variable GI and GII NVs and we have not observed significant antigenic and receptor binding changes in P particles compared with VLPs (59; unpublished data). We also obtained the HBGA binding results of additional strains in the Hunter and Sakai clusters, which further confirmed the consensus binding phenotype of GII-4 viruses. Since this issue is fundamentally important, confirmatory studies and side-by-side comparison, as well as testing of additional GII-4 variants in more laboratories, will be necessary.
The antigenic variations described in our study are comparable to those reported in other studies, in which the surrogate neutralization assay to measure HBGA blocking activities was used (6, 43). In general, the blockade of GII-4 VLP/P particles binding to HBGAs was greater with serum samples from patients in outbreaks that occurred near the time of origin of the VLP/P-particle strain (6, 43), indicating a continual antigenic drift (possibly due to a selection by the host immunity). Whether these variants represent new escape mutants responsible for new epidemics or pandemics seen with flu virus remains to be confirmed. According to the epidemiology and transmission mode of NVs, such likelihood may be low. NVs are transmitted by the fecal/oral route, which commonly causes community outbreaks, and may not spread as widely as influenza viruses do. Consequently, NVs may not reach (or reach as quickly) the same level of herd immunity in the general population as that of flu virus. Thus, any sublineages of GII-4 may continue circulating somewhere in the world even if they may die off locally. Their chance to emerge in a new season would also be equal if the herd immunity is low.
The observed variation in A-antigen binding due to the I389V mutation may serve as a good example of the equal chance of circulation (random fluctuation). It is known that the European and North American populations are high A/low B, while the Pacific and Asian populations are low A/high B for their blood antigen types (15, 22). The high or low bindings to the A antigen by GII-4 variants suggest a fitness to either the A or non-A individuals by adaption (selection). Thus, a strain with high affinity for the A antigen is more likely to be derived from the European and North American population, while a low A binder is more likely to come from an Asian population. Since the binding to the A antigen is nonessential for GII-4 viruses, mutants with both phenotypes would have similar survival advantages. The discovery of a single mutation that could be the cause of such epidemic cluster-switching is highly significant.
The present study has several limitations. For example, significant variation in the binding affinities to HBGAs by 16 different GII-4 variants was observed even under strict control of the P particles used in the comparison. At this stage, we do not know whether the difference is due to microvariation of the receptor binding interfaces among the GII-4 variants or variation due to the in vitro expression system used for making the P particles. Regrettably, due to the lack of a reliable in vivo system for assessment, this issue cannot be resolved at this time. In addition, we also observed variation of results between the saliva- and the oligosaccharide-based assays. Such differences were also reported in our previous studies and in other laboratories (19, 20, 23, 24). We believe that both assays are important; while the oligosaccharide assays provide valuable information on the structure and target of the HBGAs, the saliva binding assays are more biologically relevant. Finally, the antigenic characterization described in our study remains preliminary and needs to be improved by using increasingly standardized assay conditions with more defined reagents and more human subjects.
In summary, both host immunity and HBGAs play a role in NV evolution. While the host immunity will continue to drive antigenic change, the HBGAs, as convergent factors, may enforce a functional selection through structural constraint. This may allow only partial tolerance of HBGA binding variation. The diverged lineages (genotypes) of NVs in the two genogroups seen today are the results of such selection by the polymorphic HBGA types. Each lineage may fit one or a combination of a few HBGAs as a stable genetic trait. The GII-4 viruses have the best fit to the secretor types that express the major determinants of the H antigens, which may be the reason for their predominance over other NVs. We hope that our results help to clarify some epidemiology and evolution issues of NVs. If the GII-4 viruses remain in a clone expanding stage and undergo only low levels of antigenic variation (drift), the vaccine strategy used to select vaccine strains for influenza viruses may not be suitable for NVs. It is also possible that a longer time may be needed for a new antigenic variant to emerge for NVs than for flu virus. However, these questions remain to be resolved in future studies.
Supplementary Material
Acknowledgments
The research described in this article was supported by the National Institute of Health, the National Institute of Allergy and Infectious Diseases (R01 AI37093 and R01 AI55649) and National Institute of Child Health (PO1 HD13021), and the Department of Defense (PR033018) to X.J. This work was also supported by a grant from the Translational Research Initiative of Cincinnati Children's Hospital Medical Center (SPR102032) to M.T.
We thank Tatsuya Miyoshi for technical support, Tom Tanaka for the plasmid cDNA of the Sakai GII-4 strain, and Ralph Baric for the plasmid cDNAs for the 2004-Hunter and 2005-Sakai strains.
Footnotes
Published ahead of print on 30 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allen, D. J., J. J. Gray, C. I. Gallimore, J. Xerry, and M. Iturriza-Gomara. 2008. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 3:e1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, D. J., R. Noad, D. Samuel, J. J. Gray, P. Roy, and M. Iturriza-Gomara. 2009. Characterisation of a GII-4 norovirus variant-specific surface-exposed site involved in antibody binding. Virol. J. 6:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., B. Yount, L. Lindesmith, P. R. Harrington, S. R. Greene, F. C. Tseng, N. Davis, R. E. Johnston, D. G. Klapper, and C. L. Moe. 2002. Expression and self-assembly of Norwalk virus capsid protein from Venezuelan equine encephalitis virus replicons. J. Virol. 76:3023-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bok, K., E. J. Abente, M. Realpe-Quintero, T. Mitra, S. V. Sosnovtsev, A. Z. Kapikian, and K. Y. Green. 2009. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83:11890-11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bu, W., A. Mamedova, M. Tan, M. Xia, X. Jiang, and R. S. Hegde. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 82:5340-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon, J. L., L. C. Lindesmith, E. F. Donaldson, L. Saxe, R. S. Baric, and J. Vinje. 2009. Herd immunity to GII.4 noroviruses is supported by outbreak patient sera. J. Virol. 83:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, S., Z. Lou, M. Tan, Y. Chen, Y. Liu, Z. Zhang, X. C. Zhang, X. Jiang, X. Li, and Z. Rao. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 9.Choi, J. M., A. M. Hutson, M. K. Estes, and B. V. Prasad. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175-9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dereeper, A., V. Guignon, G. Blanc, S. Audic, S. Buffet, F. Chevenet, J. F. Dufayard, S. Guindon, V. Lefort, M. Lescot, J. M. Claverie, and O. Gascuel. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465-W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donaldson, E. F., L. C. Lindesmith, A. D. Lobue, and R. S. Baric. 2008. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 225:190-211. [DOI] [PubMed] [Google Scholar]
- 12.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 15.Franco, R. F., B. P. Simoes, and M. A. Zago. 1995. Relative frequencies of the two O alleles of the histo-blood ABH system in different racial groups. Vox Sang. 69:50-52. [DOI] [PubMed] [Google Scholar]
- 16.Graham, D. Y., X. Jiang, T. Tanaka, A. R. Opekun, H. P. Madore, and M. K. Estes. 1994. Norwalk virus infection of volunteers: new insights based on improved assays. J. Infect. Dis. 170:34-43. [DOI] [PubMed] [Google Scholar]
- 17.Green, K. Y., J. F. Lew, X. Jiang, A. Z. Kapikian, and M. K. Estes. 1993. Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J. Clin. Microbiol. 31:2185-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 19.Harrington, P. R., L. Lindesmith, B. Yount, C. L. Moe, and R. S. Baric. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76:12335-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrington, P. R., J. Vinje, C. L. Moe, and R. S. Baric. 2004. Norovirus capture with histo-blood group antigens reveals novel virus-ligand interactions. J. Virol. 78:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hein, J. 1990. Unified approach to alignment and phylogenies. Methods Enzymol. 183:626-645. [DOI] [PubMed] [Google Scholar]
- 22.Holgersson, J., M. E. Breimer, and B. E. Samuelsson. 1992. Basic biochemistry of cell surface carbohydrates and aspects of the tissue distribution of histo-blood group ABH and related glycosphingolipids. APMIS Suppl. 27:18-27. [PubMed] [Google Scholar]
- 23.Huang, P., T. Farkas, S. Marionneau, W. Zhong, N. Ruvoen-Clouet, A. L. Morrow, M. Altaye, L. K. Pickering, D. S. Newburg, J. LePendu, and X. Jiang. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 188:19-31. [DOI] [PubMed] [Google Scholar]
- 24.Huang, P., T. Farkas, W. Zhong, M. Tan, S. Thornton, A. L. Morrow, and X. Jiang. 2005. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 79:6714-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutson, A. M., F. Airaud, J. LePendu, M. K. Estes, and R. L. Atmar. 2005. Norwalk virus infection associates with secretor status genotyped from sera. J. Med. Virol. 77:116-120. [DOI] [PubMed] [Google Scholar]
- 26.Hutson, A. M., R. L. Atmar, and M. K. Estes. 2004. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 12:279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutson, A. M., R. L. Atmar, D. Y. Graham, and M. K. Estes. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 185:1335-1337. [DOI] [PubMed] [Google Scholar]
- 28.Hutson, A. M., R. L. Atmar, D. M. Marcus, and M. K. Estes. 2003. Norwalk virus-like particle hemagglutination by binding to H histo-blood group antigens. J. Virol. 77:405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, X. 2003. Development of serological and molecular tests for the diagnosis of calicivirus infections, p. 505-522. In U. Desselberger and J. Gray (ed.), Viral gastroenteritis, 1st ed. Elsevier Science B. V., Amsterdam, Netherlands.
- 30.Jiang, X., D. Cubitt, J. Hu, X. Dai, J. Treanor, D. O. Matson, and L. K. Pickering. 1995. Development of an ELISA to detect MX virus, a human calicivirus in the Snow Mountain agent genogroup. J. Gen. Virol. 76(11):2739-2747. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, X., D. O. Matson, G. M. Ruiz-Palacios, J. Hu, J. Treanor, and L. K. Pickering. 1995. Expression, self-assembly, and antigenicity of a snow mountain agent-like calicivirus capsid protein. J. Clin. Microbiol. 33:1452-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, X., N. Wilton, W. M. Zhong, T. Farkas, P. W. Huang, E. Barrett, M. Guerrero, G. Ruiz-Palacios, K. Y. Green, J. Green, A. D. Hale, M. K. Estes, L. K. Pickering, and D. O. Matson. 2000. Diagnosis of human caliciviruses by use of enzyme immunoassays. J. Infect. Dis. 181(Suppl. 2):S349-S359. [DOI] [PubMed] [Google Scholar]
- 35.Jin, M., H. P. Xie, Z. J. Duan, N. Liu, Q. Zhang, B. S. Wu, H. Y. Li, W. X. Cheng, S. H. Yang, J. M. Yu, Z. Q. Xu, S. X. Cui, L. Zhu, M. Tan, X. Jiang, and Z. Y. Fang. 2008. Emergence of the GII4/2006b variant and recombinant noroviruses in China. J. Med. Virol. 80:1997-2004. [DOI] [PubMed] [Google Scholar]
- 36.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519. [DOI] [PubMed] [Google Scholar]
- 37.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 38.Le, S. Q., and O. Gascuel. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25:1307-1320. [DOI] [PubMed] [Google Scholar]
- 39.Lee, B. E., J. K. Preiksaitis, N. Chui, L. Chui, and X. L. Pang. 2008. Genetic relatedness of noroviruses identified in sporadic gastroenteritis in children and gastroenteritis outbreaks in northern Alberta. J. Med. Virol. 80:330-337. [DOI] [PubMed] [Google Scholar]
- 40.Le Pendu, J. 2004. Histo-blood group antigen and human milk oligosaccharides: genetic polymorphism and risk of infectious diseases. Adv. Exp. Med. Biol. 554:135-143. [DOI] [PubMed] [Google Scholar]
- 41.Lindesmith, L., C. Moe, J. Lependu, J. A. Frelinger, J. Treanor, and R. S. Baric. 2005. Cellular and humoral immunity following Snow Mountain virus challenge. J. Virol. 79:2900-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindesmith, L., C. Moe, S. Marionneau, N. Ruvoen, X. Jiang, L. Lindblad, P. Stewart, J. LePendu, and R. Baric. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548-553. [DOI] [PubMed] [Google Scholar]
- 43.Lindesmith, L. C., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopman, B. A., M. H. Reacher, Y. Van Duijnhoven, F. X. Hanon, D. Brown, and M. Koopmans. 2003. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg. Infect. Dis. 9:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marionneau, S., N. Ruvoen, B. Le Moullac-Vaidye, M. Clement, A. Cailleau-Thomas, G. Ruiz-Palacois, P. Huang, X. Jiang, and J. Le Pendu. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monroe, S. S., S. E. Stine, X. Jiang, M. K. Estes, and R. I. Glass. 1993. Detection of antibody to recombinant Norwalk virus antigen in specimens from outbreaks of gastroenteritis. J. Clin. Microbiol. 31:2866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Néron, B., H. Menager, C. Maufrais, N. Joly, J. Maupetit, S. Letort, S. Carrere, P. Tuffery, and C. Letondal. 2009. Mobyle: a new full web bioinformatics framework. Bioinformatics 25:3005-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen, T. A., F. Yagyu, M. Okame, T. G. Phan, Q. D. Trinh, H. Yan, K. T. Hoang, A. T. Cao, P. Le Hoang, S. Okitsu, and H. Ushijima. 2007. Diversity of viruses associated with acute gastroenteritis in children hospitalized with diarrhea in Ho Chi Minh City, Vietnam. J. Med. Virol. 79:582-590. [DOI] [PubMed] [Google Scholar]
- 49.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 50.Nordgren, J., E. Kindberg, P. E. Lindgren, A. Matussek, and L. Svensson. 2010. Norovirus gastroenteritis outbreak with a secretor-independent susceptibility pattern, Sweden. Emerg. Infect. Dis. 16:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parker, S., D. Cubitt, J. X. Jiang, and M. Estes. 1993. Efficacy of a recombinant Norwalk virus protein enzyme immunoassay for the diagnosis of infections with Norwalk virus and other human “candidate” caliciviruses. J. Med. Virol. 41:179-184. [DOI] [PubMed] [Google Scholar]
- 52.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 53.Rockx, B. H., H. Vennema, C. J. Hoebe, E. Duizer, and M. P. Koopmans. 2005. Association of histo-blood group antigens and susceptibility to norovirus infections. J. Infect. Dis. 191:749-754. [DOI] [PubMed] [Google Scholar]
- 54.Seto, Y., N. Iritani, H. Kubo, A. Kaida, T. Murakami, K. Haruki, O. Nishio, M. Ayata, and H. Ogura. 2005. Genotyping of norovirus strains detected in outbreaks between April 2002 and March 2003 in Osaka City, Japan. Microbiol. Immunol. 49:275-283. [DOI] [PubMed] [Google Scholar]
- 55.Shirato, H., S. Ogawa, H. Ito, T. Sato, A. Kameyama, H. Narimatsu, Z. Xiaofan, T. Miyamura, T. Wakita, K. Ishii, and N. Takeda. 2008. Noroviruses distinguish between type 1 and type 2 histo-blood group antigens for binding. J. Virol. 82:10756-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siebenga, J. J., H. Vennema, B. Renckens, E. de Bruin, B. van der Veer, R. J. Siezen, and M. Koopmans. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siebenga, J. J., H. Vennema, D. P. Zheng, J. Vinje, B. E. Lee, X. L. Pang, E. C. Ho, W. Lim, A. Choudekar, S. Broor, T. Halperin, N. B. Rasool, J. Hewitt, G. E. Greening, M. Jin, Z. J. Duan, Y. Lucero, M. O'Ryan, M. Hoehne, E. Schreier, R. M. Ratcliff, P. A. White, N. Iritani, G. Reuter, and M. Koopmans. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 200:802-812. [DOI] [PubMed] [Google Scholar]
- 58.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. U. S. A. 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan, M., P. Fang, T. Chachiyo, M. Xia, P. Huang, Z. Fang, W. Jiang, and X. Jiang. 2008. Noroviral P particle: structure, function and applications in virus-host interaction. Virology 382:115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan, M., R. S. Hegde, and X. Jiang. 2004. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J. Virol. 78:6233-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan, M., P. Huang, J. Meller, W. Zhong, T. Farkas, and X. Jiang. 2003. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 77:12562-12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan, M., and X. Jiang. 2008. Association of histo-blood group antigens with susceptibility to norovirus infection may be strain-specific rather than genogroup dependent. J. Infect. Dis. 198:940-941. [DOI] [PubMed] [Google Scholar]
- 63.Tan, M., and X. Jiang. 2007. Norovirus-host interaction: implications for disease control and prevention. Expert Rev. Mol. Med. 9:1-22. [DOI] [PubMed] [Google Scholar]
- 64.Tan, M., and X. Jiang. 2005. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 13:285-293. [DOI] [PubMed] [Google Scholar]
- 65.Tan, M., and X. Jiang. 2005. The P domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J. Virol. 79:14017-14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan, M., M. Jin, H. Xie, Z. Duan, X. Jiang, and Z. Fang. 2008. Outbreak studies of a GII-3 and a GII-4 norovirus revealed an association between HBGA phenotypes and viral infection. J. Med. Virol. 80:1296-1301. [DOI] [PubMed] [Google Scholar]
- 67.Tan, M., J. Meller, and X. Jiang. 2006. C-terminal arginine cluster is essential for receptor binding of norovirus capsid protein. J. Virol. 80:7322-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan, M., M. Xia, S. Cao, P. Huang, T. Farkas, J. Meller, R. S. Hegde, X. Li, Z. Rao, and X. Jiang. 2008. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology 379:324-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan, M., M. Xia, Y. Chen, W. Bu, R. S. Hegde, J. Meller, X. Li, and X. Jiang. 2009. Conservation of carbohydrate binding interfaces: evidence of human HBGA selection in norovirus evolution. PLoS One 4:e5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan, M., W. Zhong, D. Song, S. Thornton, and X. Jiang. 2004. E. coli-expressed recombinant norovirus capsid proteins maintain authentic antigenicity and receptor binding capability. J. Med. Virol. 74:641-649. [DOI] [PubMed] [Google Scholar]
- 71.Victoria, M., M. P. Miagostovich, M. S. Ferreira, C. B. Vieira, J. M. Fioretti, J. P. Leite, R. Colina, and J. Cristina. 2009. Bayesian coalescent inference reveals high evolutionary rates and expansion of norovirus populations. Infect. Genet. Evol. 9:927-932. [DOI] [PubMed] [Google Scholar]
- 72.Xerry, J., C. I. Gallimore, M. Iturriza-Gomara, D. J. Allen, and J. J. Gray. 2008. Transmission events within outbreaks of gastroenteritis determined through analysis of nucleotide sequences of the P2 domain of genogroup II noroviruses. J. Clin. Microbiol. 46:947-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xi, J. N., D. Y. Graham, K. N. Wang, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 74.Zhao, Y. F., H. P. Xie1, H. X. Lv, Q. Zhang, M. Jin, Z. J. Duncan, D. Steele, B. M. Jiang, and X. Jiang. 2007. Investigation of human calicivirus (HuCV) diarrhea among infantile and young children in China, 1999—2005. Chin. J. Virol. 23:9-15. [PubMed] [Google Scholar]
- 75.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.