Abstract
Recently it has been shown that the proinflammatory NF-κB pathway promotes efficient influenza virus propagation. Based on these findings, it was suggested that NF-κB blockade may be a promising approach for antiviral intervention. The classical virus-induced activation of the NF-κB pathway requires proteasomal degradation of the inhibitor of NF-κB, IκB. Therefore, we hypothesized that inhibition of proteasomal IκB degradation should impair influenza A virus (IAV) replication. We chose the specific proteasome inhibitor PS-341, which is a clinically approved anticancer drug also known as Bortezomib or Velcade. As expected, PS-341 treatment of infected A549 cells in a concentration range that was not toxic resulted in a significant reduction of progeny virus titers. However, we could not observe the proposed suppression of NF-κB-signaling in vitro. Rather, PS-341 treatment resulted in an induction of IκB degradation and activation of NF-κB as well as the JNK/AP-1 pathway. This coincides with enhanced expression of antiviral genes, such as interleukin-6 and, most importantly, MxA, which is a strong interferon (IFN)-induced suppressor of influenza virus replication. This suggests that PS-341 may act as an antiviral agent via induction of the type I IFN response. Accordingly, PS-341 did not affect virus titers in Vero cells, which lack type I IFN genes, but strongly inhibited replication of vesicular stomatitis virus (VSV), a highly IFN-sensitive pathogen. Thus, we conclude that PS-341 blocks IAV and VSV replication by inducing an antiviral state mediated by the NF-κB-dependent expression of antivirus-acting gene products.
Influenza A viruses are highly contagious pathogens for humans and several animal species. Activation of the proinflammatory nuclear factor κB (NF-κB) pathway is a hallmark of influenza A virus infection (17, 69). This pathway is commonly known to exert antiviral activity because it regulates the expression of inflammatory cytokines, chemokines, and immunoreceptors, as well as beta interferon (IFN-β), one of the most potent antiviral cytokines (61). However, recently it has been shown by us and others that influenza A viruses exploit this signaling pathway for efficient replication (78). Several mechanisms have been proposed to be involved in the virus-supportive action of NF-κB. The NF-κB-dependent expression of the proapoptotic factors FasL and TRAIL has been demonstrated to be crucial for efficient viral propagation, resulting in activation of caspases that in turn promote the nuclear export of the viral ribonucleoprotein complexes (78, 79). Other studies have shown that NF-κB inhibits type I IFN-induced antiviral gene expression either by blocking STAT1 activation via NF-κB-dependent expression of the suppressor of cytokine signaling 3 (SOCS3) (62) or by direct interference with the promoter regions of interferon-induced genes (ISGs) (77). Finally, it has also been suggested that NF-κB directly interferes with influenza virus RNA synthesis (39). These various virus-supportive functions imply that NF-κB might be a suitable target for antiviral intervention (46). Accordingly, it has been shown that inhibition of NF-κB activity by chemical inhibitors or the widely used drug acetylsalicylic acid (ASA) results in impaired influenza virus replication in vitro and in vivo (51, 56).
The classical (or canonical) pathway of NF-κB activation involves release of the NF-κB factors p65 and p50 which, in their latent state, are complexed with their inhibitory proteins, the inhibitors of κB (IκBs), in the cytoplasm. Inducers and activators of the classical NF-κB pathway are cytokines or environmental stress conditions and also bacterial or viral infections (59). A crucial step in NF-κB activation is the initiation of the inhibitor of IκB kinase (IKK) complex, consisting of three isoenzymes. IKK2 is the most crucial kinase for activation of NF-κB, since it phosphorylates IκB at two conserved serine residues in the N-terminal regulatory domain (Ser32 and Ser36 for IκBα) (30, 31). This phosphorylation is a signal for polyubiquitination by the ubiquitin system at conserved lysine residues (9, 31) and leads to degradation of IκB by the 26S proteasome. The consequence of IκB degradation is the exposure of the nuclear localization signal in the p65/p50 complex. Upon further posttranslational modifications of p65 and p50, NF-κB translocates into the nucleus to act as a transcription factor (64).
The proteasome is a self-compartmentalizing, multimeric protease that belongs to the ubiquitin-proteasome system (UPS) for intracellular protein degradation. It exists within all eukaryotic cells and is localized in the cytoplasm and in the nucleus (4, 10, 13, 19). The majority of short-lived but also long-lived proteins are substrates for the proteasome, highlighting the importance of this degradation machinery in many cellular regulatory mechanisms. These include regulation of cell cycle progression, cell growth, and gene expression, as well as termination or activation of certain signal transduction cascades. The substrates are marked with a polyubiquitin chain by the ubiquitin cascade system, which is a signal for subsequent ATP-dependent degradation into small peptide fragments. Three different enzymatic activities, responsible for this degradation, can be distinguished: trypsin like, chymotrypsin like, and caspase like, also known as peptidylglutamyl peptide-hydrolyzing activity (4, 34, 35).
A major breakthrough in the analysis of the function of the proteasome was the identification of proteasome inhibitors (41, 68, 76). Small peptide aldehydes were the first compounds found to inhibit the proteasome (75). Since then many other substances have been identified. It was shown that dipeptidyl boronic acids are reversible, potent, and selective proteasome inhibitors, whereas other classes of proteasome inhibitors also inhibit thiol proteases, such as cathepsins and calpains (1). PS-341, also known as Bortezomib or Velcade, is one of the most prominent dipeptidyl boronic acids that inhibit the proteasome. Adams and colleagues (2) developed PS-341 and showed that it is an agent with effective antitumor activity. Today, PS-341 is approved by the Food and Drug Administration for the therapy of relapsed and refractory multiple myeloma (MM), as well as for the initial treatment of MM (66, 67, 71).
Since efficient influenza A virus (IAV) replication depends on an active NF-κB pathway and since the activity of this pathway is linked to the proteasomal degradation of IκBs, we evaluated the antiviral activity of the clinically approved proteasome inhibitor PS-341. We hypothesized that PS-341 treatment should impair IAV replication as a consequence of preventing NF-κB activity through inhibition of IκBα degradation. This inhibitory effect on the NF-κB pathway has already been shown for other cells, such as human primary umbilical vein endothelial cells (HUVEC), renal carcinoma cell lines, multiple myeloma cell lines, mantle cell lymphoma cell lines, and pancreatic adenocarcinoma cell lines (3, 15, 24, 29, 60, 65). Indeed, we could show that PS-341 leads to reduced viral titers. However, in contrast to our assumption, PS-341 did not prevent IκBα degradation but rather promoted its sustained degradation. The unpredicted hyperactivation of the NF-κB and the JNK/AP-1 pathways by PS-341 pretreatment appears to be linked to a priming of the type I interferon response, which in turn confers an antiviral state to the host cell.
MATERIALS AND METHODS
Viruses, cell lines, and viral infections.
Avian influenza virus A/FPV/Bratislava/79 (H7N7; FPV) and the human influenza virus A/Puerto-Rico/8/34 (H1N1; PR8; Giessen variant) were originally obtained from the virus strain collection of the Institute of Virology (Justus-Liebig-University, Gießen, Germany). The human isolate of the avian influenza virus A/Thailand/KAN-1/2004 (H5N1; KAN-1) was originally isolated at the Siriraj Hospital (Mahidol University, Bangkok, Thailand). Vesicular stomatitis virus strain Indiana (VSV) was a gift from T. Wolff (Robert-Koch Institute, Berlin, Germany). The Madin-Darby canine kidney cell line (MDCK ΙΙ) was grown in minimal essential medium (MEM). The human alveolar epithelial cell line A549, the human embryonic kidney cell line HEK293, and the green monkey epithelial cell line Vero were grown in Dulbecco's minimal essential medium (DMEM), and the human hematopoietic cell line U937 was grown in Roswell Park Memorial Institute Medium (RPMI) 1640. All media were supplemented with 10% heat-inactivated fetal calf serum (FCS) and antibiotics. Primary HUVEC and primary human bronchial epithelial cells (HBEpC) were obtained from Promocell (Heidelberg, Germany) and cultivated in endothelial cell growth medium supplemented with 20 μl/ml FCS, 4 μl/ml endothelial cell growth supplement, 0.1 ng/ml epidermal growth factor, 1 ng/ml basic fibroblast growth factor, 90 μg/ml heparin, and 1 μg/ml hydrocortisone and in airway epithelial growth medium supplemented with 4 μl/ml bovine pituitary extract, 10 ng/ml epidermal growth factor, 5 μg/ml insulin, 0.5 μg/ml hydrocortisone, 0.5 μg/ml epinephrine, 6.7 ng/ml triiodo-l-thyronine, 10 μg/ml transferrin, and 0.1 ng/ml retinoic acid, respectively. All experiments with primary cells were carried out at cell population doubling number 11. All cell lines and cells were cultured at 37°C in a humidified 5% CO2 atmosphere. For infection cells were washed with phosphate-buffered saline (PBS) and infected with the multiplicities of infection (MOIs) indicated in the figure legends. Therefore, virus was diluted in PBS/BA (PBS containing 0.2% bovine serum albumin [BSA], 1 mM MgCl2, 0.9 mM CaCl2, 100 U/ml penicillin, and 0.1 mg/ml streptomycin) and incubated for 30 min at 37°C and 5% CO2. The inoculum was aspirated, and cells were washed and incubated with either MEM, DMEM, or RPMI 1640 containing 0.2% BSA, 1 mM MgCl2, 0.9 mM CaCl2, and antibiotics (MEM/BA, DMEM/BA, or RPMI 1640/BA, respectively) or primary cell growth medium containing common supplements. At the indicated time points supernatants were collected to assess the number of infectious particles by standard plaque assay. Briefly, MDCK II cells grown to 90% confluence in six-well dishes were washed and infected with serial dilutions of the supernatants in PBS/BA for 30 min at 37°C and 5% CO2. The inoculum was aspirated, and cells were overlaid with 2 ml MEM/BA supplemented with 0.6% agar (Oxoid, Hampshire, United Kingdom), 0.3% DEAE-dextran (Amersham Pharmacia Biotech, Freiburg, Germany), and 1.5% NaHCO3 (Gibco Invitrogen, Karlsruhe, Germany). After incubation at 37°C and 5% CO2 for 2 to 3 days virus plaques were visualized by staining with neutral red (Sigma-Aldrich, Munich, Germany).
Treatment of cells with different agents.
For treatment of cells with PS-341 the compound was diluted in the appropriate medium to the indicated concentration. Cells were washed with PBS, medium containing PS-341, or solvent, or no supplement was added, and cells were incubated as indicated in the figure legends. For treatment of cells postinfection PS-341 was diluted in the appropriate medium containing BSA and supplements and added after the inoculum was aspirated or at the time points indicated in the figure legends (time-of-addition kinetics). For activating stimulation of the NF-κB pathway cells were treated with human tumor necrosis factor alpha (TNF-α; 30 ng/ml, 15 min; Sigma-Aldrich, Munich, Germany). TNF-α was directly added into the medium. For proapoptotic stimulation staurosporine (Sigma-Aldrich, Munich, Germany) was added directly into the medium and incubated for the times indicated in the figure legends. Cells were always incubated at 37°C with 5% CO2.
Western blot assays.
For Western blot analysis cells were washed with PBS and lysed in RIPA (25 mM Tris-HCl [pH 8], 137 mM NaCl, 10% glycerol, 0.1% sodium dodecylsulfate, 0.5% sodium deoxycholate, 1% Igepal, 2 mM EDTA [pH 8], 50 mM sodium glycerophosphate, 20 mM sodium pyrophosphate, 1 mM sodium vanadate, 5 μg/ml leupeptin, and 5 mM benzamidine) at 4°C for a minimum of 10 min. Supernatants were cleared by centrifugation in a standard table-top centrifuge (Eppendorf, Hamburg, Germany) at maximum speed at 4°C. Protein content was estimated by employing a protein dye reagent (Bio-Rad Laboratories, Munich, Germany). Equal amounts of total protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted on nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany).
All primary antibodies were diluted 1:1,000 [except for poly(ADP-ribose) polymerase (PARP) antibody (at 1:500) and interferon regulatory factor 3 (IRF-3) antibody (at 1:200)] in TBST buffer (1 M Tris-HCl [pH 7.6], 3.1 M NaCl, 1% Tween 20) containing 5% BSA and 0.02% sodium azide and incubated overnight at 4°C. Secondary antibodies were diluted in TBST buffer (anti-mouse and anti-rabbit at 1:2,500 or anti-goat at 1:5,000) and incubated for 1 h at room temperature.
Native PAGE.
To analyze IRF-3 dimerization, a native PAGE assay was performed according to the methods of Iwamura and colleagues (28). Briefly, cells were scraped from 6-cm dishes in 50 μl lysis buffer (50 nM Tris-HCl [pH 8], 1% NP-40, 150 mM NaCl, 100 μg/ml leupeptin, 5 mM sodium vanadate), and supernatants were cleared by centrifugation in a standard table-top centrifuge (Eppendorf, Hamburg, Germany) at maximum speed at 4°C. Protein content was estimated by employing a protein dye reagent (Bio-Rad Laboratories, Munich, Germany). Equal protein amounts were separated in sample buffer (125 nM Tris-HCl [pH 8], 30% glycerol, bromphenol blue) on a 7.5% polyacrylamide gel with a two-buffer system (upper chamber buffer, 25 mM Tris-HCl [pH 8.4], 192 mM glycine, and 1% sodium deoxycholate; lower chamber buffer, 25 mM Tris-HCl [pH 8.4] and 192 mM glycine) at a constant 20 mA on ice. Afterwards the gel was soaked in SDS-PAGE running buffer (25 mM Tris-HCl [pH 8.4], 250 mM glycine, 0.1% SDS) for 30 min, and the proteins were blotted on nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany).
Inhibitors, antibodies, and reagents.
PS-341 was purchased from Selleck Chemicals LLC and stored as a stock solution of 100 μM dissolved in PBS at −80°C. Staurosporine was obtained from Sigma-Aldrich (Munich, Germany). The monoclonal antibody anti-PARP was purchased from BD Transduction Laboratories (Heidelberg, Germany). Antisera against the influenza virus proteins PB1 and M1 were obtained from Santa Cruz Biotechnology (Heidelberg, Germany). To detect posttranslational modification of NF-κB p65 and to detect general NF-κB p65 expression, a phospho-specific antibody against Ser536 and a pan-antibody (Cell Signaling Technology, Frankfurt am Main, Germany) were used, respectively. IκBα was detected by using a polyclonal antibody from Santa Cruz Biotechnology (Heidelberg, Germany). To determine the activity of the mitogen-activated protein kinase (MAPK) JNK and of its downstream targets ATF-2 and c-Jun, phospho-specific antibodies against phospho-Thr183 and Tyr185 JNK (BD Transduction Laboratories, Heidelberg, Germany) and phospho-Ser63 c-Jun and phospho-Thr71 ATF-2 (Cell Signaling Technology, Frankfurt am Main, Germany) were used. Monomeric and dimeric IRF-3 were detected by using a polyclonal rabbit antibody from Santa Cruz Biotechnology (Heidelberg, Germany). Antisera against cellular extracellular signal-regulated kinase 2 (ERK2), JNK1, α-tubulin, and ATF-2 for loading controls were obtained from Santa Cruz Biotechnology (Heidelberg, Germany), Sigma-Aldrich (Munich, Germany), and Cell Signaling Technology (Frankfurt am Main, Germany).
Flow cytometry analysis.
To determine overall viability of A549 cells after PS-341 treatment, a propidium iodide (PI; Sigma-Aldrich, Munich, Germany) staining procedure was performed. Therefore, A549 cells were incubated with PS-341 as described in the figure legends. For analysis both adherent and detached cells were collected, washed with PBS, and incubated with a PI solution (50 μg/ml PI in PBS) for 15 min at room temperature. Afterwards cells were washed and fluorescence was determined in the FL2 channel of a FACScalibur cytometer (Becton Dickinson, Heidelberg, Germany).
MTT cell proliferation assay.
The pale yellow tetrazolium salt 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide (MTT; Sigma-Aldrich, Munich, Germany) can be a substrate for the mitochondrial succinate dehydrogenase and cleaved to form the dark violet formazan, which accumulates in viable and proliferating cells. An MTT cell proliferation assay was used to determine whether PS-341 has an influence on the proliferative and metabolic capacities of cells. Therefore, cells were treated with PS-341 for the indicated times. Afterwards an MTT-PBS solution (5 mg/ml) was added, and cells were incubated for an additional 2 h at 37°C and 5% CO2. The supernatants were aspired, and cells were subsequently lysed with dimethyl sulfoxide (DMSO). Absorbance was measured at 562 nm by using an Emax Precision microplate reader (Molecular Devices, Sunnyvale, CA), and the untreated control result was arbitrarily set as 100% metabolic active cells.
Proteasome activity assay.
The proteasome activity was measured using the commercially available Proteasome-Glo chymotrypsin-like cell-based assay from Promega (Mannheim, Germany). Briefly, cells were treated with PS-341 and infected with avian FPV. Afterwards Proteasome-Glo cell-based reagent was added, and cells were incubated for an additional 30 min at room temperature. Then, luminescence was measured using the MicroLumatPlus LB 96V luminometer (Berthold Technologies, Bad Wildbad, Germany), and untreated control results were arbitrarily set as 100% of chymotrypsine-like activity of all cellular 26S proteasomes.
Reverse transcription and quantitative real-time PCR.
To analyze the transcription of certain genes, mRNA of differently treated A549 cells was isolated and subjected to reverse transcription as described elsewhere (62). For quantification, real-time PCR was performed using the Mx Pro 3005P cycler (Agilent Technologies/Stratagene, Santa Clara, CA), and changes in the transcription of the gene of interest were ascertained as the difference between the transcription of the housekeeping gene for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and the gene of interest by using the 2−ΔΔCT method (43).
The following human primers were used: GAPDH_fwd, 5′-GCA AAT TTC CAT GGC ACC GT-3′; GAPDH_rev, 5′-GCC CCA CTT GAT TTT GGA GG-3′; IFN-β_fwd, 5′-GGC CAT GAC CAA CAA GTG TCT CCT CC-3′; IFN-β_rev, 5′-GCG CTC AGT TTC GGA GGT AAC CTG T-3′; MxA_fwd, 5′-GTT TCC GAA GTG GAC ATC GCA-3′; MxA_rev, 5′-GAA GGG CAA CTC CTG ACA GT-3′; IL-6_fwd, 5′-AGA GGC ACT GGC AGA AAA CAA C-3′; IL-6_rev, 5′-AGG CAA GTC TCC TCA TTG AAT CC-3′; IL-8_fwd, 5′-CTT GTT CCA CTG TGC CTT GGT T-3′; IL-8_rev, 5′-GCT TCC ACA TGT CCT CAC AAC AT-3′; CCL5_fwd, 5′-CGG CAC GCC TCG CTG TCA TC-3′; CCL5_rev, 5′-GCA AGC AGA AAC AGG CAA AT-3′.
RESULTS
PS-341 has no cytotoxic or proapoptotic effects in cells.
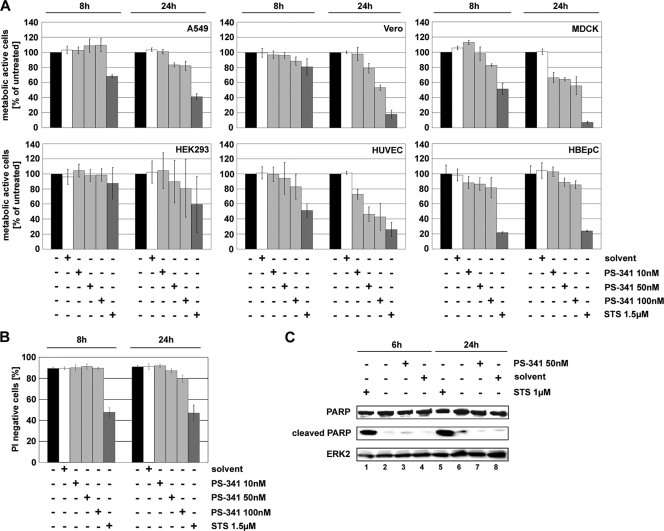
PS-341 is a dipeptidyl boronic acid that inhibits the 26S proteasome. It is clinically approved for the treatment of MM and shows cytotoxic or apoptosis-inducing effects in a variety of transformed and cancer cells. The non-small-cell lung cancer cell line A549 represents the major target tissue of influenza virus infection and is therefore a well-established in vitro model for influenza virus propagation. Since this cell line should be used for infection experiments throughout, it was crucial to find nontoxic concentrations of PS-341 to determine the potential antiviral activity. Thus, it was first determined whether different concentrations of PS-341 would have an influence on the proliferative and metabolic activities by performing an MTT cell proliferation assay. In this assay the metabolic activity of a mitochondrial enzyme (NADH-dependent succinate dehydrogenase) is measured, which is only active in proliferating healthy cells (54). While PS-341 concentrations of 10 nM had no influence on the metabolic activity of A549 cells (Fig. 1A, upper left panel), 50 nM and 100 nM led to a slight reduction of metabolically active cells after a 24-h treatment. However, even at later time points (48 h, 72 h, and 96 h [data not shown]) the proportion of metabolically active cells treated with 50 nM PS-341 remained constant at over 77%. Only at higher concentrations (100 nM) and with longer incubation periods (96 h) was a decrease down to 40% of metabolically active cells observed (data not shown). Similar results were obtained in additional MTT assays using Vero, MDCK II, and HEK293 cell lines, as well as primary HUVEC and primary HBEpC (Fig. 1A). Vero and HEK293 cells and HBEpC exhibited nearly the same sensitivity to 50 nM PS-341 as A549 cells. There was no major impact of the compound on the metabolic activity of these cells. MDCK II and HUVEC are slightly more susceptible to the toxic effects of 50 nM PS-341 than A549 cells. In MDCK II cells the metabolic activity was reduced to about 60% metabolically active cells, and in HUVEC it was reduced to about 45% metabolically active cells. In conclusion, there are only minor differences in PS-341 toxicity for the different cell types used, as measured by MTT assays.
FIG. 1.
A 50 nM concentration of PS-341 is not cytotoxic or proapoptotic in cells for the indicated exposure times. A549 (A and B) and Vero, MDCK II, and HEK293 cells and HUVEC and HBEpC (all shown only in panel A) were treated with different concentrations (100 nM, 50 nM, and 10 nM) of PS-341 for the indicated times or treated with solvent or left untreated. Cells treated with 1.5 μM staurosporine (STS) were used as positive controls for metabolically inactive cells/dead cells. (A) An MTT assay was performed, and metabolic activity of cells was calculated as the percentage of the untreated control. Arrow bars represent standard deviations from four independent experiments. (B) PI staining was performed to measure membrane integrity of cells by fluorescence-activated cell sorter analysis. The diagram shows gated cells which were not stained by PI and therefore had no destruction of the cell membrane. Arrow bars represent standard deviations of three independent experiments. (C) A549 cells were treated with 50 nM PS-341 or solvent or left untreated for the indicated times. Afterwards cells were lysed and Western blot analysis was performed to detect apoptotic cells by cleavage of PARP. As a positive control cells were treated with 1 μM staurosporine. Equal amounts of protein loading were assayed by using the cellular protein ERK2. Shown is one representative blot out of three independent experiments.
Furthermore, under the same experimental conditions the membrane integrity of A549 cells was analyzed by PI staining. PI is a fluorescent molecule that intercalates with nucleic acids but is membrane impermeable, so that it is generally excluded from viable cells. After a 24-h treatment with 50 nM PS-341 no significant differences for PI-positive cells compared to control cells could be detected (Fig. 1B). At later time points (data not shown) the percentage of PI-positive cells only increased slightly at a PS-341 concentration of 50 nM, whereas 100 nM led to a significant increase of up to 85% PI-positive cells.
To analyze potential proapoptotic effects of PS-341 on A549 cells we examined caspase-mediated cleavage of PARP. PARP is a substrate for apoptotic caspases, and its cleavage is a hallmark for apoptosis induction (57). Upon treatment of A549 cells with 50 nM PS-341 for 6 h or 24 h, no induction of PARP cleavage was observed (Fig. 1C). Since these data indicate that a concentration of 50 nM PS-341 has neither a significant cytotoxic nor proapoptotic impact on A549 cells, this concentration was used for further experiments.
PS-341 impairs IAV propagation in a variety of different cells.
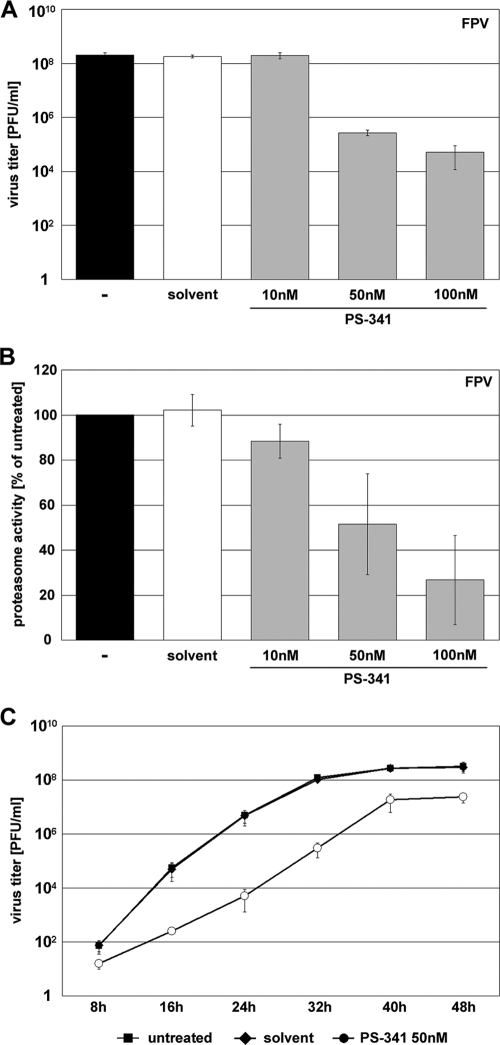
Earlier findings demonstrated that efficient IAV propagation depends on an active NF-κB pathway (51, 56). As PS-341 is a proteasome inhibitor known to prevent proteasomal degradation of IκBα, we hypothesized that it inhibits virus-induced NF-κB activation (14, 35) and that this should finally result in impaired influenza virus propagation. To test this assumption A549 cells were treated with different concentrations of PS-341 1 h prior to IAV infection and throughout the inoculation period of 24 h. Progeny virus titers were determined 24 h postinfection (p.i.). As expected PS-341 treatment resulted in a block of viral replication of the avian influenza virus strain A/FPV/Bratislava/79 (H7N7; FPV) in a concentration-dependent manner. While concentrations of 10 nM had no antiviral effect, 50 nM led to a significant titer reduction of up to 3 orders of magnitude (Fig. 2A). The highest concentration used (100 nM) led to a titer reduction of up to 4 orders of magnitude.
FIG. 2.
PS-341 impairs FPV replication in A549 cells. (A and B) A549 cells were either pretreated for 1 h with different concentrations of PS-341 or with solvent only or were left untreated. Then, cells were infected with FPV at an MOI of 0.001 (A) or 0.05 (B). After virus inoculation cells were posttreated with different concentrations of PS-341. (A) At 24 h p.i. supernatants were obtained and progeny virus titers were measured by standard plaque assay. (B) Proteasome activity and the ability of PS-341 to inhibit the proteasome was determined 24 h p.i. (C) A549 cells were pretreated with 50 nM PS-341 or solvent or left untreated for 1 h. Afterwards cells were infected with FPV at an MOI of 0.0005. Subsequent to virus inoculation cells were posttreated with 50 nM PS-341 or solvent or left untreated. After the indicated times p.i. supernatants were obtained and progeny virus titers were determined by standard plaque assay. Arrow bars in all experiments represent standard deviations of three independent experiments.
These results were also confirmed in a virus growth kinetics study in infected cells that received a single dose of 50 nM PS-341. Virus titers were reduced at every time point analyzed (Fig. 2C). While at early time points titer reductions again were on the order of 3 orders of magnitude, at later time points the antiviral activity was less pronounced, presumably due to a decline of PS-341 activity during the long incubation period.
Since PS-341 is a proteasome inhibitor, we investigated whether the antiviral concentrations of PS-341 may have an inhibitory effect on the 26S proteasome in A549 cells. A concentration-dependent inhibition of the proteasome was observed in FPV-infected A549 cells at 24 h p.i. (Fig. 2B) as well as in uninfected cells (data not shown). While 100 nM PS-341 showed a 70% inhibition of proteasome activity, 50 nM PS-341 led to an inhibition of about 50% compared to untreated controls. Taken together, these data indicate that PS-341 exhibits a strong and sustained antiviral activity at concentrations of 50 nM and 100 nM that do partially block the proteasome in the cell type used.
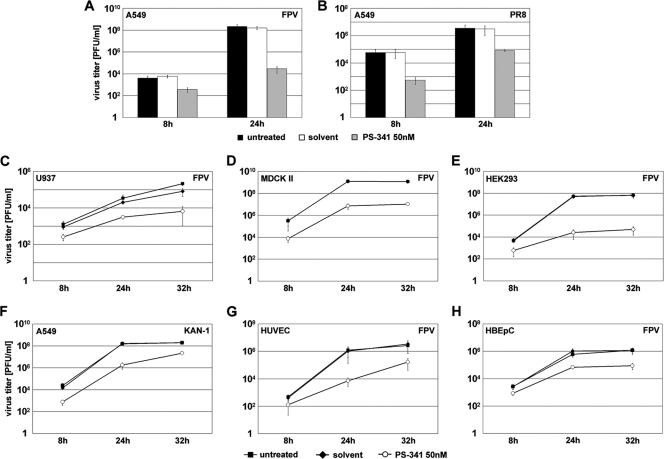
The antiviral activity of PS-341 could also be confirmed for other influenza virus types, including the human H1N1 isolate A/Puerto-Rico/8/34 (PR8) (Fig. 3B) and the human isolate of avian H5N1 strain A/Thailand/KAN-1/2004 (KAN-1) (Fig. 3F), as well as in other cell types, such as the promonocytic cell line U937 (Fig. 3C) and the epithelial cell lines MDCK II and HEK293 (Fig. 3D and E). Furthermore, PS-341 also exhibited antiviral activity in nonimmortalized primary cells such as HUVEC (Fig. 3G) and HBEpC (Fig. 3H).
FIG. 3.
PS-341 impairs viral propagation of different IAV strains in different cell types. A549 (A, B, and F), U937 (C), MDCK II (D), HEK293 (E), HUVEC (G), or HBEpC (H) cells were pretreated for 1 h with 50 nM PS-341 or solvent or left untreated. Then cells were infected with different human and avian IAV strains (FPV MOI of 0.001, PR8 MOI of 0.05, and KAN-1 MOI of 0.01). Subsequent to virus inoculation cells were posttreated with 50 nM PS-341 or solvent or left untreated. After the indicated times p.i. supernatants were obtained and progeny virus titers were determined by standard plaque assay. Arrow bars represent standard deviations of three independent experiments.
PS-341 treatment affects early steps of the viral life cycle.
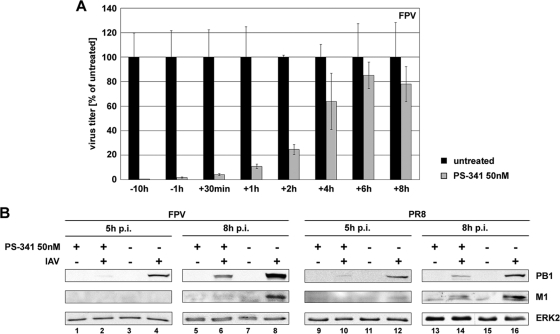
To determine which step in the influenza virus life cycle is affected by PS-341, a time-of-addition kinetics study that spanned the first replication cycle was performed. A549 cells were treated with PS-341 either before or after infection at the time points indicated (Fig. 4A). A strong antiviral activity of PS-341 was observed upon addition of the compound up to 2 h p.i. At time points of 4 h p.i. and later, a dramatic decrease in the antiviral efficacy was observed. This indicates that the event in the viral life cycle that is affected by PS-341 occurs within the first 4 h. Since this correlates with the strong onset of viral gene and protein expression, we analyzed whether viral protein accumulation is affected by PS-341. Indeed, we observed a strong reduction in viral matrix protein (M1) and PB1 polymerase synthesis in FPV- and PR8-infected cells at 5 h and 8 h p.i.
FIG. 4.
Early steps of viral replication within the first replication cycle are affected. (A) For time-of-addition kinetics analysis, A549 cells were either left untreated or were pretreated for 10 h or 1 h with 50 nM PS-341 before infection and additionally posttreated after infection. Cells were infected with FPV at an MOI of 0.005. After virus inoculation cells were posttreated with 50 nM PS-341. Then the proteasome inhibitor was added after virus inoculation (−10 h, −1 h, and +30 min) or it was added at the different times p.i. as indicated (+1 h, +2 h, +4 h, +6 h, and +8 h; cells were not pretreated before infection). At 9 h p.i. supernatants were obtained and progeny virus titers were determined by standard plaque assay. Shown is one representative experiment out of three independent experiments. (B) A549 cells were pretreated with 50 nM PS-341 or left untreated for 1 h. Afterwards cells were infected with avian FPV or human PR8 at an MOI of 1. Subsequent to virus inoculation cells were posttreated with 50 nM PS-341 or left untreated. After the indicated times p.i. cells were lysed and analyzed by Western blotting for accumulation of viral proteins polymerase PB1 and matrix protein M1. Cellular protein ERK2 served as a control to demonstrate equal amounts of protein loading. Shown is one representative blot out of three independent experiments.
While these data are consistent with the time-of-addition kinetics shown in Fig. 4A, the findings are in disagreement with the proposed NF-κB-inhibiting action of PS-341. In earlier studies it was shown that interference with NF-κB activity in infected cells resulted in the nuclear retention of viral ribonucleoprotein (RNP) complexes, while the accumulation of viral proteins was unaffected (78, 79). Thus, we expected that PS-341 would not have an influence on viral protein accumulation within the first replication cycle; however, the contrary was observed. This was a first indication that the antiviral action of PS-341 does not correlate with inhibition of the NF-κB pathway.
PS-341 pretreatment leads to activation of the classical NF-κB pathway and c-Jun NH2-terminal kinase (JNK) pathway.
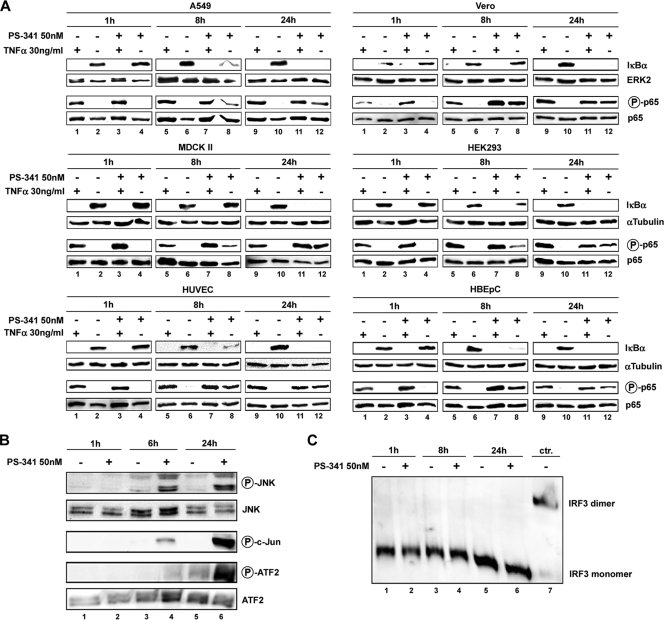
The infection of cells with influenza virus results in activation of the NF-κB pathway (17, 69). However, NF-κB is also induced by other factors, such as proinflammatory cytokines, environmental stress conditions, and different chemical compounds (59). TNF-α is known to be a strong activator of this pathway (27, 58). Effective proteasome inhibitors should inhibit this activation independently of the stimulus. To address the potency of PS-341 to prevent proteasomal degradation of IκBα, TNF-α was used to activate the NF-κB pathway (Fig. 5A). Cells were first treated for the indicated times with PS-341 and subsequently stimulated with TNF-α. A 1-h pretreatment with 50 nM PS-341 did not result in prevention of IκBα degradation in any cell types used (Fig. 5A, lanes 3), indicating that the partial inhibition of the proteasome at this concentration (Fig. 2B) may not be sufficient to block IκBα decay. The same results were obtained when the different cells were pretreated for longer time periods (8 h or 24 h) with PS-341 before TNF-α stimulation (Fig. 5A, lanes 7 and 11). However, at these later time points an additional and unexpected observation was made. We observed effective IκBα degradation in the absence of TNF-α in cells that were only treated with PS-341 (Fig. 5A, lanes 8 and 12). This surprising result was also obtained with 100 nM PS-341 but not with 10 nM, a concentration that also did not show antiviral activity (data not shown).
FIG. 5.
PS-341 treatment leads to an activation of the NF-κB pathway and JNK/AP-1 pathway. (A) A549, Vero, MDCK II, HEK293, HUVEC, and HBEpC cells were treated with 50 nM PS-341 for the indicated times or left untreated. In addition, cells were stimulated with 30 ng/ml TNF-α for 15 min or left unstimulated. Cell lysates were subjected to Western blot analysis against IκBα and phosphorylated p65 (Ser536). p65 and ERK2 served as controls for equal protein amounts. (B and C) A549 cells were treated with PS-341 at 50 nM for the indicated times or left untreated. (B) Western blotting was performed with total cell lysates, using phospho-specific antibodies against JNK and the transcription factors c-Jun and ATF-2 or loading controls, respectively. (C) Native PAGE analysis was performed with total cell lysates to detect IRF-3 dimers. As a control for IRF-3 dimerization (ctr.), A549 cells were transfected for 4 h with 1 μg RNA isolated from FPV-infected A549 cells (MOI of 5; 5 h). In each case one representative blot is shown of three independent experiments.
We now tested whether this PS-341-induced IκBα degradation would also translate into NF-κB activation and induction of NF-κB-dependent gene expression. One indication of activated NF-κB is the phosphorylation of the p65 subunit within the canonical NF-κB pathway. Upon the phosphorylation at Ser536, p65 is stabilized and the nuclear localization and transcriptional activity are enhanced (7, 40, 50, 64, 70, 72). While TNF-α stimulation provoked a phosphorylation of p65 at Ser536 as expected, we also observed p65 phosphorylation upon PS-341 pretreatment in all cell types used (Fig. 5A, lanes 7, 8, 11, and 12). Moreover, in a reporter gene assay, enhanced transcriptional activity from the NF-κB promoter was measured when A549 cells were treated with 50 nM PS-341 (data not shown). Taken together these data demonstrate that treatment of cells with 50 nM PS-341 (or higher doses) results in activation of the classical NF-κB pathway.
Recent studies have shown that proteasome inhibition leads to an activation of the JNK pathway (16, 52, 80). As the JNK pathway is activated upon influenza virus infection and it is involved in antiviral signaling, whether PS-341 affects this signaling pathway in A549 cells has been investigated (44, 45, 48). Therefore, cells were incubated with 50 nM PS-341 for the indicated times (Fig. 5B). After a 6-h treatment with 50 nM PS-341, activation of JNK was detected as evidenced by phosphorylation of the kinase at Thr183 and Tyr185 (Fig. 5B, lane 4). This was even enhanced upon a 24-h treatment (Fig. 5B, lane 6). Downstream substrates of JNK are the AP-1 transcription factors c-Jun and ATF-2, which are activated by JNK-mediated phosphorylation at Ser63 and Thr71, respectively (33). Consistent with JNK activation, we also found c-Jun and ATF-2 were phosphorylated and activated (Fig. 5B, lane 6), leading to the conclusion that, besides NF-κB, the JNK/c-Jun/ATF-2 pathway is also activated in A549 upon PS-341 treatment.
Besides NF-κB and AP-1, IRF-3 is another transcription factor that controls expression of antiviral genes (25). For example, all three factors contribute to IFN-β expression in a complex called the IFN-β enhanceosome (61). Thus, we also investigated the influence of PS-341 on the activation of IRF-3, which can be monitored by dimerization of the factor. As shown in Fig. 5C we could not detect any influence by 50 nM PS-341 on the dimerization of IRF-3, indicating that the potential PS-341-induced activity on the factor is below the detection threshold.
NF-κB- and JNK/AP-1-dependent cytokine and chemokine gene transcription levels are upregulated upon PS-341 treatment.
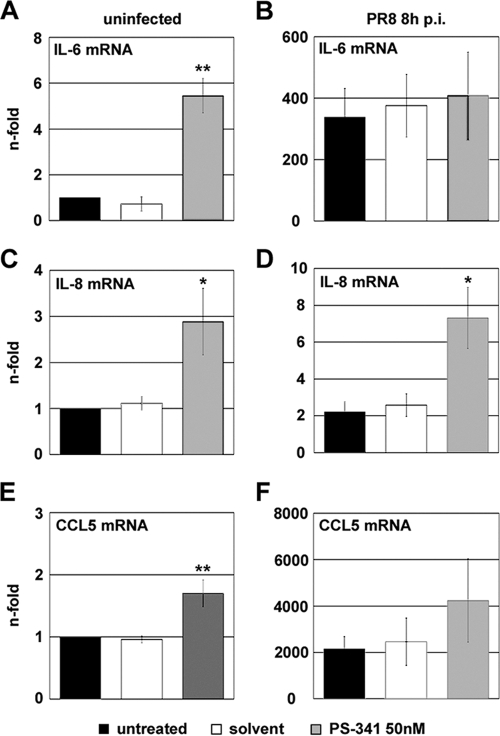
Cytokines and chemokines are small soluble secreted proteins with functions in inflammation and immunity. The expression of many of these proteins depends on the transcription factors NF-κB and AP-1. As we had detected an activation of these two pathways upon PS-341 treatment, we then analyzed transcription levels of certain NF-κB- and AP-1-dependent cytokines and chemokines. The transcription levels of the cytokine interleukin-6 (IL-6) and the chemokine IL-8 are known to be dependent on the NF-κB and JNK/AP-1 pathways (26, 36, 37). The chemokine RANTES (regulated upon activation, normal T cell expressed and secreted), also known as CCL5, was shown to be regulated by the NF-κB, JNK, and p38 MAPK pathways (36, 38). Hence, A549 cells were either treated with 50 nM PS-341 or left untreated and were subsequently infected with PR8 or left uninfected. Upon PS-341 treatment the transcription level of the cytokine IL-6 in uninfected cells was nearly 6-fold elevated compared to control cells (Fig. 6A). No differences in transcription levels in PR8-infected cells were observed between untreated and PS-341-treated samples; however, the effects may have been hidden by a 100-fold-higher transcription level upon infection (Fig. 6B). This was different for the transcription levels of the chemokine IL-8, which are normally not affected upon influenza virus infection. Here, gene expression was increased 3-fold in both the uninfected and PR8-infected cells that were treated with PS-341, compared to controls (Fig. 6C and D). Also, in the case of the chemokine CCL5 (RANTES), treatment with PS-341 resulted in an increase in transcription levels in uninfected and infected cells (Fig. 6E and F). Taken together, these results suggest that upon PS-341 treatment of A549 cells the NF-κB and the JNK/AP-1 pathways are functionally activated and that these activations probably lead to secretion of immune-modulatory factors.
FIG. 6.
NF-κB- and AP-1-dependent gene transcripts are upregulated. A549 cells were pretreated for 1 h with 50 nM PS-341 or solvent or left untreated. Then cells were infected with PR8 at an MOI of 1 or mock infected with solvent containing no virus (uninfected). After virus inoculation cells were posttreated with 50 nM PS-341 or solvent or left untreated. At 8 h p.i. cells were lysed and RNA was subjected to reverse transcription. cDNA was analyzed by quantitative RT-PCR for amounts of transcripts of IL-6 (A and B), IL-8 (C and D), and CCL5 (E and F). Arrow bars represent standard deviations of three independent experiments and are generated as the fold change compared to untreated and mock-infected controls. *, P < 0.05; **, P < 0.01).
PS-341 treatment induces type I IFN response genes, leading to suppression of virus propagation.
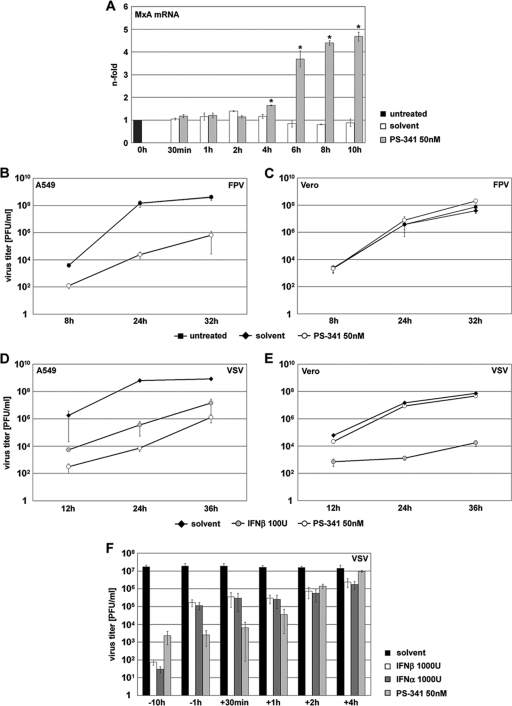
IFNs are a large family of multifunctional proteins, being secreted by virus-infected cells to induce an antiviral state in surrounding cells as a protective mechanism (20, 42, 74). The immediate expression of IFN-β by the virus-induced activation of NF-κB, AP-1, and IRF-3 is the initial step in this antiviral response (61). The finding that PS-341 treatment leads to activation of two of these three transcription factors prompted us to analyze whether IFN signaling is also induced in A549 cells upon PS-341 treatment. While we were unable to detect increased transcription of IFN-β in treated cells, we observed increasing transcription levels of the antiviral IFN-stimulated gene MxA over the observed time course (Fig. 7A). The first significant changes to a nearly 2-fold induction were detected after a 4-h treatment, while a 5-fold increase of transcription levels was observed after a 10-h treatment. Human MxA protein is a GTPase whose expression is strictly dependent on type I IFN and which has been shown to mediate anti-influenza virus activity in vitro and in vivo when expressed in transgenic mice (63). This finding is a strong indication that PS-341 induces a low-level type I IFN response that may be the basis for the antiviral activity of this compound. To address this hypothesis we infected Vero cells in the presence or absence of PS-341. These cells are deficient for type I IFN genes due to genomic deletions (12). Cells were treated with PS-341 and subsequently infected with avian FPV. Indeed, we were able to show that in type I IFN-deficient Vero cells PS-341 did not act antivirally, while in A549 control cells a clear antiviral effect 4 orders of magnitude greater was detectable (Fig. 7C and B, respectively). To exclude that these findings may have been caused by an altered inhibitory action on the 26S proteasome, the activity of the proteasome in the presence of PS-341 was compared in both cell types. There were no significant differences detected between A549 and Vero cells, indicating that the compound inhibits the proteasome to the same level in both cell lines (data not shown). This is also consistent with the results of the toxicity assays shown in Fig. 1A, which also showed responses in a similar range for A549 and Vero cells.
FIG. 7.
PS-341 treatment induces type I IFN response genes, leading to suppression of virus propagation. (A) A549 cells were treated for the indicated times with 50 nM PS-341 or solvent. Then cells were lysed and RNA was subjected to reverse transcription. cDNA was analyzed by quantitative RT-PCR to determine amounts of transcripts of IFN-dependent MxA. Arrow bars represent standard deviations of two independent experiments and are generated as the fold change compared to the untreated control. *, P < 0.05 compared to untreated control. (B to E) A549 cells (B and D) or Vero cells (C and E) were pretreated for 1 h with 50 nM PS-341 (B to E), for 12 h with 100 U of IFN-β (only D and E), solvent, or left untreated. After that cells were infected with avian FPV at an MOI of 0.001 (B and C) or with VSV at an MOI of 0.001 (D) or 0.0001 (E). Subsequent to virus inoculation cells were posttreated with 50 nM PS-341 (B to E), 100 U of IFN-β (D and E only), solvent, or left untreated. After the indicated times p.i. supernatants were obtained and progeny virus titers were determined by standard plaque assay. Arrow bars represent standard deviations of three independent experiments (panel E shows results of one representative experiment out of four). (F) For time-of-addition kinetics analysis, A549 cells were either pretreated for 10 h or 1 h with 50 nM PS-341 or 1,000 U of IFN-β or IFN-α before infection and were additionally posttreated after infection or left untreated. Cells were infected with VSV at an MOI of 0.001. After virus inoculation cells were posttreated with 50 nM PS-341 or 1,000 U of IFN-β or IFN-α. The proteasome inhibitor or IFNs were added after virus inoculation (−10 h, −1 h, or +30 min) or they were added at different times p.i. as indicated (+1 h, +2 h, and +4 h; cells were not pretreated before infection). At 12 h p.i. supernatants were obtained and progeny virus titers were determined by standard plaque assay. Shown is one representative experiment out of three independent experiments.
To further verify an interference of PS-341 with the type I IFN response, we infected cells with VSV, a pathogen which is extremely sensitive to the action of type I IFN. Indeed, treatment of VSV-infected A549 cells with PS-341 resulted in a tremendous drop in virus titers, similar to the IFN-β-treated control (Fig. 7D). In contrast, treatment of VSV-infected Vero cells with PS-341 had no influence on progeny virus titers, most likely because of a lack of type I IFN induction in these cells (Fig. 7E). To evaluate whether the kinetics of PS-341 match that of type I IFN, we treated VSV-infected A459 cells with PS-341, IFN-β, or IFN-α at several time points pre- and postinfection (Fig. 7F). Similar to influenza viruses, VSV replication was only inhibited at early time points postinfection. Most importantly, the treatment with IFNs showed an antiviral efficacy that was similar to PS-341 over the observation period. Taken together, these observations allow the striking conclusion that PS-341 primes the type I IFN response in IFN-competent cells and that this activation is necessary for its antiviral efficacy.
DISCUSSION
Infections with influenza A viruses are still a major problem worldwide. The recent outbreaks of the pandemic Mexican H1N1v swine origin flu (S-OIV) and the ongoing infections of humans with highly pathogenic avian H5N1 viruses in Southeast Asia and Africa demonstrate that there is a continuous threat of novel and maybe more severe pandemics in the future. The S-OIV outbreak has clearly demonstrated that the development and production of vaccines against these viruses takes too long to be an efficient measure against the early phases of a pandemic. This leaves us with a few antiviral compounds to fight such a burden. The increasing incidence of resistance to either the M2 blockers amantadine/rimantadine or the neuraminidase inhibitors oseltamivir and zanamivir shows that antiviral drugs directly targeting viral components are not a long-term option. It has been shown that influenza viruses recruit and manipulate host cell factors for efficient replication (48, 49). These findings suggest that cellular factors which are dispensable for cellular metabolism and survival may be much more promising targets for antiviral therapy (47). Blockade of these factors should provide a broad antiviral activity also against newly emerging strains and the problem of resistance should be minimized, since the virus cannot replace the missing cellular function. In particular, the requirement of the NF-κB signaling pathway and the consequences on viral replication by inhibiting this pathway indicate how useful cellular factors may be as targets for an efficient antiviral therapy (44, 56, 78). The antiviral effect of ASA via its IKK-inhibiting action can be taken as a proof of concept that inhibition of cellular factors such as the NF-κB pathway is well tolerated in cells and organisms (44, 51).
Here, we aimed to interfere with influenza virus replication by pursuing another strategy of NF-κB inhibition. It is well known that the activation of the classical NF-κB pathway depends on the proteasomal degradation of the IκBs (32). Therefore, it was expected that inhibition of the proteasome by a specific proteasome inhibitor would lead to impaired viral replication. PS-341, also known as Bortezomib or Velcade, was chosen, as it is clinically approved for the treatment of MM and well established as a specific proteasome inhibitor (2, 66, 67, 71). Its antitumor activity was predicted to be an effect dependent on the inhibition of NF-κB activity by preventing proteasomal degradation of IκBα and on its general cytotoxic and proapoptotic effects (2, 21, 22). In this study a concentration of PS-341 that was not toxic for the lung epithelial cell line A549 or even primary HBEpC was chosen. Indeed, it could be shown that upon treatment of A549 cells with 50 nM PS-341 influenza A virus replication was impaired up to several orders of magnitude compared to untreated cells. This concentration led to a moderate average inhibition of 50% of all proteasomes in the cell, which might be the reason that we did not observe adverse effects on cell viability and metabolism. The concentration of 50 nM PS-341 only led to a reduction of about 20% in metabolic activity in the A549 cells used in this study (Fig. 1A), and even after a 96-h treatment the percentage of metabolically active cells remained at 77% of active cells (data not shown). This is consistent with the results of Mortenson and colleagues (53), who showed in a clonogenic survival assay that PS-341 treatment of A549 cells over a long time period produced a lower toxicity than expected.
Two other findings illustrate that PS-341 at the concentrations used in our experiments does not have cytotoxic or proapoptotic effects but a real antiviral efficacy. First, we observed a recovery of virus replication in long-term viral growth kinetics in A549 cells which had only received a single dose of the inhibitor (Fig. 2C). Thus, cells are not nonspecifically damaged by PS-341, because otherwise virus replication could not proceed. Furthermore, treatment of Vero cells with PS-341 in concentrations that inhibited the proteasome to the same level as in A549 cells and that had the same effect on the metabolic activity as in A549 cells did not block virus accumulation at all, which in turn indicates that PS-341 does not affect viability of these cells. Finally, the degree of antiviral action of PS-341 seems to be only slightly different for different virus strains and cell types including primary nonimmortalized cells. Therefore, it could be excluded that the observed antiviral activity of PS-341 depends on a possible cytotoxic or proapoptotic effect.
It has been shown that the inhibition of the NF-κB pathway by acetylsalicylic acid has no effect on viral protein accumulation within the first replication cycle of influenza viruses (51). However, here it could be demonstrated that already within the first replication cycle viral protein expression was affected upon PS-341 treatment and that an early treatment of cells parallel to the onset of viral infection was necessary for an efficient antiviral activity of PS-341 (Fig. 3A and B). These findings already indicate that the antiviral action of PS-341 differs from the mechanisms of NF-κB-inhibiting agents. The observation that PS-341 could not prevent IκBα degradation may be attributed to an incomplete inhibition of the proteasome by 50 nM PS-341. However, PS-341 treatment rather led to an activation of the NF-κB pathway in all cell types used and the JNK/AP-1 pathway in A549 cells (Fig. 5), indicating a signal-inducing potency of the drug. While this might appear surprising, our data have recently been confirmed by Hideshima and colleagues (23). While our manuscript was in preparation those authors also showed that PS-341 leads to activation of the NF-κB pathway. This was hypothesized to occur through the direct or indirect activation of IKK2 and the subsequent phosphorylation of IκBα. Furthermore, it was suggested that IκBα is degraded via a proteasome-independent mechanism (23). For MM cells Hideshima and colleagues (23) showed that the NF-κB-activating effect is not only promoted by treatment with up to 20 nM PS-341 for 8 to 12 h (a little less PS-341 than we used) but also by lactacystin and MG-132, and they concluded that NF-κB activation may be a general effect of proteasome inhibitors. This proteasome inhibitor-dependent effect of an induced degradation of IκBα has also been shown by other groups who have used diverse proteasome inhibitors in different cell lines (8, 11, 55). Studies in which PS-341 led to an inhibition of NF-κB signaling were mostly performed in other cell types than the cells used in this study. The treatment with PS-341 was carried out with concentrations from 50 nM up to 10 μM PS-341 for 1 to 24 h (3, 15, 24, 29, 60, 65). In the case of HUVEC an inhibition of IκBα degradation was observed after a 1-h treatment with 10 μM PS-341 (29, 60). In contrast, we treated HUVEC for much longer time periods (8 to 24 h) with much less PS-341 (50 nM) and observed a clear degradation of IκBα (Fig. 5A).
Consistent with an activation of NF-κB and AP-1, we showed that PS-341 upregulates the transcription of antivirus-acting cytokines, which are dependent on these signaling pathways, such as IL-6, IL-8, and CCL5/RANTES (26, 36, 37, 38). Both NF-κB and AP-1 are also known to bind and regulate the IFN-β promoter. While we were not able to detect induced expression of IFN-β itself, we detected upregulation of the strictly IFN-dependent antiviral gene MxA. Such a seemingly contradictory observation has also been made by others (18). For example, in a microarray study of influenza virus-infected cells, a plethora of IFN-induced genes were identified in the absence of detectable levels of IFN-β mRNA. This led to the assumption that IFN-β expression may occur early at low levels in a very transient fashion (18), which might also be the case in our experiments. Another explanation may be that, in contrast to NF-κB and AP-1, IRF-3 is not significantly activated upon PS-341 treatment. IRF-3 is known to provide an exponential boost of expression of IFN-β and IFN-dependent genes, which may be missing here. Thus, PS-341 may only lead to a moderate induction of the response. However, this seems to be still sufficient to prime an antiviral response.
In any case, the strong expression of IFN-dependent MxA suggested that type I IFN may play a crucial role in the antiviral efficacy of PS-341. Accordingly, we showed that in a type I IFN-deficient cell system (Vero cells) no antiviral activity of PS-341 was measured even though the level of proteasomal inhibition in these cells was similar to that in IFN-competent A549 cells. Furthermore, the replication of VSV, a pathogen that is extremely sensitive to type I IFNs and the action of MxA (5, 73), was also significantly decreased by PS-341 in A549 cells but not in Vero cells. The antiviral efficacy of PS-341 at different time points during VSV replication correlated very well with the action of type I IFNs, again suggesting that these cytokines may be the main mediators of the PS-341 effect. This is a second functional indication of an IFN priming action of PS-341.
Taken together our findings are compatible with the following scenario. PS-341 results in activation of NF-κB and the JNK/AP-1 pathway and a low-level IFN-like gene expression response. This seems to confer a certain antiviral state to the cells that may even be boosted upon infection. A similar priming of the antiviral response by active NF-κB was previously observed in the context of Borna disease virus (BDV) infection (6). BDV infection does not lead to an activation of NF-κB and type I IFN expression. However, in cells expressing a constitutive active form of IKK2, causing a preactivated NF-κB pathway, a tremendous drop in virus titers was observed that was most likely due to an early and efficient antiviral induction of type I IFN signaling (6).
The fact that PS-341 is administrated systemically for the treatment of MM and that a partial inhibition of the proteasome by PS-341 in normal cells is well tolerated in treated patients suggests that an antiviral therapy against influenza A viruses with PS-341 is possible at a concentration that may not have adverse side effects. However, as the lung is the primary infected tissue, local administration as an aerosol might be the route of choice to further reduce the likelihood of unknown side effects. Thus, PS-341 may serve as an emergency drug in cases of problematic or fatal infections with virus variants resistant to commonly used drugs like oseltamivir or zanamivir or in case of a pandemic influenza outbreak.
Acknowledgments
This study was in part supported by different grants of the Deutsche Forschungsgemeinschaft (DFG graduate school GRK1409 and DFG Lu477/12-1) and by the German FluResearchNet, a nationwide research network on zoonotic influenza sponsored by the Ministry of Education and Research.
Footnotes
Published ahead of print on 30 June 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Adams, J., M. Behnke, S. Chen, A. A. Cruickshank, L. R. Dick, L. Grenier, J. M. Klunder, Y. T. Ma, L. Plamondon, and R. L. Stein. 1998. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 8:333-338. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J., V. J. Palombella, E. A. Sausville, J. Johnson, A. Destree, D. D. Lazarus, J. Maas, C. S. Pien, S. Prakash, and P. J. Elliott. 1999. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 59:2615-2622. [PubMed] [Google Scholar]
- 3.An, J., Y. Sun, M. Fisher, and M. B. Rettig. 2004. Maximal apoptosis of renal cell carcinoma by the proteasome inhibitor bortezomib is nuclear factor-κB dependent. Mol. Cancer Ther. 3:727-736. [PubMed] [Google Scholar]
- 4.Baumeister, W., J. Walz, F. Zuhl, and E. Seemuller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92:367-380. [DOI] [PubMed] [Google Scholar]
- 5.Belkowski, L. S., and G. C. Sen. 1987. Inhibition of vesicular stomatitis viral mRNA synthesis by interferons. J. Virol. 61:653-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourteele, S., K. Oesterle, S. Pleschka, G. Unterstab, C. Ehrhardt, T. Wolff, S. Ludwig, and O. Planz. 2005. Constitutive activation of the transcription factor NF-κB results in impaired Borna disease virus replication. J. Virol. 79:6043-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buss, H., A. Dorrie, M. L. Schmitz, E. Hoffmann, K. Resch, and M. Kracht. 2004. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-κB at serine 536 is mediated by multiple protein kinases, including IκB kinase (IKK) α, IKKβ, IKKɛ, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 279:55633-55643. [DOI] [PubMed] [Google Scholar]
- 8.Calvaruso, G., M. Giuliano, P. Portanova, A. De Blasio, R. Vento, and G. Tesoriere. 2006. Bortezomib induces in HepG2 cells IκBα degradation mediated by caspase-8. Mol. Cell. Biochem. 287:13-19. [DOI] [PubMed] [Google Scholar]
- 9.Ciechanover, A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17:7151-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coux, O., K. Tanaka, and A. L. Goldberg. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65:801-847. [DOI] [PubMed] [Google Scholar]
- 11.Dolcet, X., D. Llobet, M. Encinas, J. Pallares, A. Cabero, J. A. Schoenenberger, J. X. Comella, and X. Matias-Guiu. 2006. Proteasome inhibitors induce death but activate NF-κB on endometrial carcinoma cell lines and primary culture explants. J. Biol. Chem. 281:22118-22130. [DOI] [PubMed] [Google Scholar]
- 12.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 13.Etlinger, J. D., and A. L. Goldberg. 1977. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc. Natl. Acad. Sci. U. S. A. 74:54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiedler, M. A., K. Wernke-Dollries, and J. M. Stark. 1998. Inhibition of TNF-alpha-induced NF-κB activation and IL-8 release in A549 cells with the proteasome inhibitor MG-132. Am. J. Respir. Cell Mol. Biol. 19:259-268. [DOI] [PubMed] [Google Scholar]
- 15.Fujioka, S., G. M. Sclabas, C. Schmidt, W. A. Frederick, Q. G. Dong, J. L. Abbruzzese, D. B. Evans, C. Baker, and P. J. Chiao. 2003. Function of nuclear factor κB in pancreatic cancer metastasis. Clin. Cancer Res. 9:346-354. [PubMed] [Google Scholar]
- 16.Fujita, T., K. Washio, D. Takabatake, H. Takahashi, S. Yoshitomi, K. Tsukuda, Y. Ishibe, Y. Ogasawara, H. Doihara, and N. Shimizu. 2005. Proteasome inhibitors can alter the signaling pathways and attenuate the P-glycoprotein-mediated multidrug resistance. Int. J. Cancer 117:670-682. [DOI] [PubMed] [Google Scholar]
- 17.Garoufalis, E., I. Kwan, R. Lin, A. Mustafa, N. Pepin, A. Roulston, J. Lacoste, and J. Hiscott. 1994. Viral induction of the human beta interferon promoter: modulation of transcription by NF-kappa B/rel proteins and interferon regulatory factors. J. Virol. 68:4707-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. U. S. A. 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg, A. L., R. Stein, and J. Adams. 1995. New insights into proteasome function: from archaebacteria to drug development. Chem. Biol. 2:503-508. [DOI] [PubMed] [Google Scholar]
- 20.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 21.Hideshima, T., D. Chauhan, P. Richardson, C. Mitsiades, N. Mitsiades, T. Hayashi, N. Munshi, L. Dang, A. Castro, V. Palombella, J. Adams, and K. C. Anderson. 2002. NF-kappa B as a therapeutic target in multiple myeloma. J. Biol. Chem. 277:16639-16647. [DOI] [PubMed] [Google Scholar]
- 22.Hideshima, T., D. Chauhan, R. Schlossman, P. Richardson, and K. C. Anderson. 2001. The role of tumor necrosis factor alpha in the pathophysiology of human multiple myeloma: therapeutic applications. Oncogene 20:4519-4527. [DOI] [PubMed] [Google Scholar]
- 23.Hideshima, T., H. Ikeda, D. Chauhan, Y. Okawa, N. Raje, K. Podar, C. Mitsiades, N. C. Munshi, P. G. Richardson, R. D. Carrasco, and K. C. Anderson. 2009. Bortezomib induces canonical nuclear factor-κB activation in multiple myeloma cells. Blood 114:1046-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hideshima, T., P. Richardson, D. Chauhan, V. J. Palombella, P. J. Elliott, J. Adams, and K. C. Anderson. 2001. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 61:3071-3076. [PubMed] [Google Scholar]
- 25.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19:1-13. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann, E., O. Dittrich-Breiholz, H. Holtmann, and M. Kracht. 2002. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 72:847-855. [PubMed] [Google Scholar]
- 27.Israel, A., B. O. Le, D. Hatat, J. Piette, M. Kieran, F. Logeat, D. Wallach, M. Fellous, and P. Kourilsky. 1989. TNF stimulates expression of mouse MHC class I genes by inducing an NF kappa B-like enhancer binding activity which displaces constitutive factors. EMBO J. 8:3793-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-κB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 29.Kalogeris, T. J., F. S. Laroux, A. Cockrell, H. Ichikawa, N. Okayama, T. J. Phifer, J. S. Alexander, and M. B. Grisham. 1999. Effect of selective proteasome inhibitors on TNF-induced activation of primary and transformed endothelial cells. Am. J. Physiol. 276:C856-C864. [DOI] [PubMed] [Google Scholar]
- 30.Karin, M. 1999. How NF-κB is activated: the role of the IκB kinase (IKK) complex. Oncogene 18:6867-6874. [DOI] [PubMed] [Google Scholar]
- 31.Karin, M. 1999. The beginning of the end: IκB kinase (IKK) and NF-κB activation. J. Biol. Chem. 274:27339-27342. [DOI] [PubMed] [Google Scholar]
- 32.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 33.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 34.Kisselev, A. F., T. N. Akopian, V. Castillo, and A. L. Goldberg. 1999. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol. Cell 4:395-402. [DOI] [PubMed] [Google Scholar]
- 35.Kisselev, A. F., and A. L. Goldberg. 2001. Proteasome inhibitors: from research tools to drug candidates. Chem. Biol. 8:739-758. [DOI] [PubMed] [Google Scholar]
- 36.Kracht, M., and J. Saklatvala. 2002. Transcriptional and post-transcriptional control of gene expression in inflammation. Cytokine 20:91-106. [DOI] [PubMed] [Google Scholar]
- 37.Krause, A., H. Holtmann, S. Eickemeier, R. Winzen, M. Szamel, K. Resch, J. Saklatvala, and M. Kracht. 1998. Stress-activated protein kinase/Jun N-terminal kinase is required for interleukin (IL)-1-induced IL-6 and IL-8 gene expression in the human epidermal carcinoma cell line KB. J. Biol. Chem. 273:23681-23689. [DOI] [PubMed] [Google Scholar]
- 38.Kujime, K., S. Hashimoto, Y. Gon, K. Shimizu, and T. Horie. 2000. p38 mitogen-activated protein kinase and c-jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J. Immunol. 164:3222-3228. [DOI] [PubMed] [Google Scholar]
- 39.Kumar, N., Z. T. Xin, Y. Liang, H. Ly, and Y. Liang. 2008. NF-κB signaling differentially regulates influenza virus RNA synthesis. J. Virol. 82:9880-9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence, T., M. Bebien, G. Y. Liu, V. Nizet, and M. Karin. 2005. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature 434:1138-1143. [DOI] [PubMed] [Google Scholar]
- 41.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 42.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 43.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔCT) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 44.Ludwig, S. 2009. Targeting cell signalling pathways to fight the flu: towards a paradigm change in anti-influenza therapy. J. Antimicrob. Chemother. 64:1-4. [DOI] [PubMed] [Google Scholar]
- 45.Ludwig, S., C. Ehrhardt, E. R. Neumeier, M. Kracht, U. R. Rapp, and S. Pleschka. 2001. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J. Biol. Chem. 276:10990-10998. [PubMed] [Google Scholar]
- 46.Ludwig, S., and O. Planz. 2008. Influenza viruses and the NF-κB signaling pathway: towards a novel concept of antiviral therapy. Biol. Chem. 389:1307-1312. [DOI] [PubMed] [Google Scholar]
- 47.Ludwig, S., O. Planz, S. Pleschka, and T. Wolff. 2003. Influenza-virus-induced signaling cascades: targets for antiviral therapy? Trends Mol. Med. 9:46-52. [DOI] [PubMed] [Google Scholar]
- 48.Ludwig, S., S. Pleschka, O. Planz, and T. Wolff. 2006. Ringing the alarm bells: signalling and apoptosis in influenza virus infected cells. Cell. Microbiol. 8:375-386. [DOI] [PubMed] [Google Scholar]
- 49.Ludwig, S., S. Pleschka, and T. Wolff. 1999. A fatal relationship: influenza virus interactions with the host cell. Viral Immunol. 12:175-196. [DOI] [PubMed] [Google Scholar]
- 50.Mattioli, I., A. Sebald, C. Bucher, R. P. Charles, H. Nakano, T. Doi, M. Kracht, and M. L. Schmitz. 2004. Transient and selective NF-kappa B p65 serine 536 phosphorylation induced by T cell costimulation is mediated by I kappa B kinase beta and controls the kinetics of p65 nuclear import. J. Immunol. 172:6336-6344. [DOI] [PubMed] [Google Scholar]
- 51.Mazur, I., W. J. Wurzer, C. Ehrhardt, S. Pleschka, P. Puthavathana, T. Silberzahn, T. Wolff, O. Planz, and S. Ludwig. 2007. Acetylsalicylic acid (ASA) blocks influenza virus propagation via its NF-κB-inhibiting activity. Cell. Microbiol. 9:1683-1694. [DOI] [PubMed] [Google Scholar]
- 52.Meriin, A. B., V. L. Gabai, J. Yaglom, V. I. Shifrin, and M. Y. Sherman. 1998. Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J. Biol. Chem. 273:6373-6379. [DOI] [PubMed] [Google Scholar]
- 53.Mortenson, M. M., M. G. Schlieman, S. Virudachalam, and R. J. Bold. 2004. Effects of the proteasome inhibitor bortezomib alone and in combination with chemotherapy in the A549 non-small-cell lung cancer cell line. Cancer Chemother. Pharmacol. 54:343-353. [DOI] [PubMed] [Google Scholar]
- 54.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 55.Nemeth, Z. H., H. R. Wong, K. Odoms, E. A. Deitch, C. Szabo, E. S. Vizi, and G. Hasko. 2004. Proteasome inhibitors induce inhibitory kappa B (IκB) kinase activation, IκB alpha degradation, and nuclear factor kappa B activation in HT-29 cells. Mol. Pharmacol. 65:342-349. [DOI] [PubMed] [Google Scholar]
- 56.Nimmerjahn, F., D. Dudziak, U. Dirmeier, G. Hobom, A. Riedel, M. Schlee, L. M. Staudt, A. Rosenwald, U. Behrends, G. W. Bornkamm, and J. Mautner. 2004. Active NF-κB signalling is a prerequisite for influenza virus infection. J. Gen. Virol. 85:2347-2356. [DOI] [PubMed] [Google Scholar]
- 57.Oliver, F. J., R. G. de la Rubia, V. Rolli, M. C. Ruiz-Ruiz, G. de Murcia, and J. M. Murcia. 1998. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J. Biol. Chem. 273:33533-33539. [DOI] [PubMed] [Google Scholar]
- 58.Osborn, L., S. Kunkel, and G. J. Nabel. 1989. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc. Natl. Acad. Sci. U. S. A. 86:2336-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pahl, H. L. 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 60.Palombella, V. J., E. M. Conner, J. W. Fuseler, A. Destree, J. M. Davis, F. S. Laroux, R. E. Wolf, J. Huang, S. Brand, P. J. Elliott, D. Lazarus, T. McCormack, L. Parent, R. Stein, J. Adams, and M. B. Grisham. 1998. Role of the proteasome and NF-κB in streptococcal cell wall-induced polyarthritis. Proc. Natl. Acad. Sci. U. S. A. 95:15671-15676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panne, D. 2008. The enhanceosome. Curr. Opin. Struct. Biol. 18:236-242. [DOI] [PubMed] [Google Scholar]
- 62.Pauli, E. K., M. Schmolke, T. Wolff, D. Viemann, J. Roth, J. G. Bode, and S. Ludwig. 2008. Influenza A virus inhibits type I IFN signaling via NF-κB-dependent induction of SOCS-3 expression. PLoS Pathog. 4:e1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pavlovic, J., H. A. Arzet, H. P. Hefti, M. Frese, D. Rost, B. Ernst, E. Kolb, P. Staeheli, and O. Haller. 1995. Enhanced virus resistance of transgenic mice expressing the human MxA protein. J. Virol. 69:4506-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perkins, N. D. 2006. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene 25:6717-6730. [DOI] [PubMed] [Google Scholar]
- 65.Pham, L. V., A. T. Tamayo, L. C. Yoshimura, P. Lo, and R. J. Ford. 2003. Inhibition of constitutive NF-kappa B activation in mantle cell lymphoma B cells leads to induction of cell cycle arrest and apoptosis. J. Immunol. 171:88-95. [DOI] [PubMed] [Google Scholar]
- 66.Richardson, P. G., B. Barlogie, J. Berenson, S. Singhal, S. Jagannath, D. Irwin, S. V. Rajkumar, G. Srkalovic, M. Alsina, R. Alexanian, D. Siegel, R. Z. Orlowski, D. Kuter, S. A. Limentani, S. Lee, T. Hideshima, D. L. Esseltine, M. Kauffman, J. Adams, D. P. Schenkein, and K. C. Anderson. 2003. A phase 2 study of bortezomib in relapsed, refractory myeloma. N. Engl. J. Med. 348:2609-2617. [DOI] [PubMed] [Google Scholar]
- 67.Richardson, P. G., P. Sonneveld, M. W. Schuster, D. Irwin, E. A. Stadtmauer, T. Facon, J. L. Harousseau, D. Ben-Yehuda, S. Lonial, H. Goldschmidt, D. Reece, J. F. San-Miguel, J. Blade, M. Boccadoro, J. Cavenagh, W. S. Dalton, A. L. Boral, D. L. Esseltine, J. B. Porter, D. Schenkein, and K. C. Anderson. 2005. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N. Engl. J. Med. 352:2487-2498. [DOI] [PubMed] [Google Scholar]
- 68.Rock, K. L., C. Gramm, L. Rothstein, K. Clark, R. Stein, L. Dick, D. Hwang, and A. L. Goldberg. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78:761-771. [DOI] [PubMed] [Google Scholar]
- 69.Ronni, T., S. Matikainen, T. Sareneva, K. Melen, J. Pirhonen, P. Keskinen, and I. Julkunen. 1997. Regulation of IFN-alpha/beta, MxA, 2′,5′-oligoadenylate synthetase, and HLA gene expression in influenza A-infected human lung epithelial cells. J. Immunol. 158:2363-2374. [PubMed] [Google Scholar]
- 70.Sakurai, H., H. Chiba, H. Miyoshi, T. Sugita, and W. Toriumi. 1999. IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J. Biol. Chem. 274:30353-30356. [DOI] [PubMed] [Google Scholar]
- 71.San Miguel, J. F., R. Schlag, N. K. Khuageva, M. A. Dimopoulos, O. Shpilberg, M. Kropff, I. Spicka, M. T. Petrucci, A. Palumbo, O. S. Samoilova, A. Dmoszynska, K. M. Abdulkadyrov, R. Schots, B. Jiang, M. V. Mateos, K. C. Anderson, D. L. Esseltine, K. Liu, A. Cakana, H. van de Velde, P. G. Richardson, et al. 2008. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N. Engl. J. Med. 359:906-917. [DOI] [PubMed] [Google Scholar]
- 72.Sizemore, N., N. Lerner, N. Dombrowski, H. Sakurai, and G. R. Stark. 2002. Distinct roles of the IκB kinase alpha and beta subunits in liberating nuclear factor kappa B (NF-κB) from IκB and in phosphorylating the p65 subunit of NF-κB. J. Biol. Chem. 277:3863-3869. [DOI] [PubMed] [Google Scholar]
- 73.Staeheli, P., and J. Pavlovic. 1991. Inhibition of vesicular stomatitis virus mRNA synthesis by human MxA protein. J. Virol. 65:4498-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 75.Tsubuki, S., H. Kawasaki, Y. Saito, N. Miyashita, M. Inomata, and S. Kawashima. 1993. Purification and characterization of a Z-Leu-Leu-Leu-MCA degrading protease expected to regulate neurite formation: a novel catalytic activity in proteasome. Biochem. Biophys. Res. Commun. 196:1195-1201. [DOI] [PubMed] [Google Scholar]
- 76.Voorhees, P. M., and R. Z. Orlowski. 2006. The proteasome and proteasome inhibitors in cancer therapy. Annu. Rev. Pharmacol. Toxicol. 46:189-213. [DOI] [PubMed] [Google Scholar]
- 77.Wei, L., M. R. Sandbulte, P. G. Thomas, R. J. Webby, R. Homayouni, and L. M. Pfeffer. 2006. NFκB negatively regulates interferon-induced gene expression and anti-influenza activity. J. Biol. Chem. 281:11678-11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wurzer, W. J., C. Ehrhardt, S. Pleschka, F. Berberich-Siebelt, T. Wolff, H. Walczak, O. Planz, and S. Ludwig. 2004. NF-κB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. J. Biol. Chem. 279:30931-30937. [DOI] [PubMed] [Google Scholar]
- 79.Wurzer, W. J., O. Planz, C. Ehrhardt, M. Giner, T. Silberzahn, S. Pleschka, and S. Ludwig. 2003. Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 22:2717-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu, C., B. B. Friday, J. P. Lai, L. Yang, J. Sarkaria, N. E. Kay, C. A. Carter, L. R. Roberts, S. H. Kaufmann, and A. A. Adjei. 2006. Cytotoxic synergy between the multikinase inhibitor sorafenib and the proteasome inhibitor bortezomib in vitro: induction of apoptosis through Akt and c-Jun NH2-terminal kinase pathways. Mol. Cancer Ther. 5:2378-2387. [DOI] [PubMed] [Google Scholar]