Abstract
Alpha interferon (IFN-α) is an approved medication for chronic hepatitis B. Gamma interferon (IFN-γ) is a key mediator of host antiviral immunity against hepatitis B virus (HBV) infection in vivo. However, the molecular mechanism by which these antiviral cytokines suppress HBV replication remains elusive. Using an immortalized murine hepatocyte (AML12)-derived cell line supporting tetracycline-inducible HBV replication, we show in this report that both IFN-α and IFN-γ efficiently reduce the amount of intracellular HBV nucleocapsids. Furthermore, we provide evidence suggesting that the IFN-induced cellular antiviral response is able to distinguish and selectively accelerate the decay of HBV replication-competent nucleocapsids but not empty capsids in a proteasome-dependent manner. Our findings thus reveal a novel antiviral mechanism of IFNs and provide a basis for a better understanding of HBV pathobiology.
Hepatitis B virus (HBV) is a noncytopathic hepatotropic DNA virus which belongs to the family Hepadnaviridae (11, 44). Despite the fact that most adulthood HBV infections are transient, approximately 5 to 10% of infected adults and more than 90% of infected neonates fail to clear the virus and develop a lifelong persistent infection, which may progress to chronic hepatitis, cirrhosis, and primary hepatocellular carcinoma (4, 33, 34). It has been shown by several research groups that resolution of HBV and other animal hepadnavirus infection in vivo depends on both killing of infected hepatocytes by viral antigen-specific cytotoxic T lymphocytes and noncytolytic suppression of viral replication, which is most likely mediated by inflammatory cytokines, such as gamma interferon (IFN-γ) and tumor necrosis factor α (TNF-α) (10, 12, 15, 20, 26, 27, 48). Moreover, together with five nucleoside or nucleotide analogs that inhibit HBV DNA polymerase, alpha IFN (IFN-α) and pegylated IFN-α are currently available antiviral medications for the management of chronic hepatitis B. Compared to the viral DNA polymerase inhibitors, the advantages of IFN-α therapy include a lack of drug resistance, a finite and defined treatment course, and an increased likelihood for hepatitis B virus surface antigen (HBsAg) clearance (8, 39). However, only approximately 30% of treated patients achieve a sustained virological response to a standard 48-month pegylated IFN-α therapy (6, 32). Thus far, the antiviral mechanism of IFN-α and IFN-γ and the parameters determining the success or failure of IFN-α therapy in chronic hepatitis B remain elusive. Elucidation of the mechanism by which the cytokines suppress HBV replication represents an important step toward understanding the pathobiology of HBV infection and the molecular basis of IFN-α therapy of chronic hepatitis B.
Considering the mechanism by which IFNs noncytolytically control HBV infection in vivo, it is possible that the cytokines either induce an antiviral response in hepatocytes to directly limit HBV replication or modulate the host antiviral immune response to indirectly inhibit the virus infection. However, due to the fact that IFN-α and -γ do not inhibit or only modestly inhibit HBV replication in human hepatoma-derived cell lines (5, 22, 23, 30), the direct antiviral effects of the cytokines and their antiviral mechanism against HBV have been studied with either an immortalized hepatocyte cell line derived from HBV transgenic mice or duck hepatitis B virus (DHBV) infection of primary duck hepatocytes (37, 53). While these studies revealed that IFN treatment significantly reduced the amount of encapsidated viral pregenomic RNA (pgRNA) in both mouse and duck hepatocytes, further mechanistic analyses suggested that IFN-α inhibited the formation of pgRNA-containing nucleocapsids in murine hepatocytes (52) but shortened the half-life of encapsidated pgRNA in DHBV-replicating chicken hepatoma cells (21). Moreover, the fate of viral DNA replication intermediates or nucleocapsids in the IFN-treated hepatocytes was not investigated in the previous studies.
To further define the target(s) of IFN-α and -γ in the HBV life cycle and to create a robust cell culture system for the identification of IFN-stimulated genes (ISGs) that mediate the antiviral response of the cytokines (25), we established an immortalized murine hepatocyte (AML-12)-derived stable cell line that supported a high level of HBV replication in a tetracycline-inducible manner. Consistent with previous reports, we show that both IFN-α and IFN-γ potently inhibited HBV replication in murine hepatocytes (37, 40). With the help of small molecules that inhibit HBV capsid assembly (Bay-4109) (7, 47) and prevent the incorporation of pgRNA into nucleocapsids (AT-61) (9, 29), we obtained evidence suggesting that the IFN-induced cellular antiviral response is able to distinguish and selectively accelerate the decay of HBV replication-competent nucleocapsids but not empty capsids in a proteasome-dependent manner. Our findings provide a basis for further studies toward better understanding of IFN′s antiviral mechanism, which might ultimately lead to the development of strategies to improve the efficacy of IFN therapy of chronic hepatitis B.
MATERIALS AND METHODS
Establishment of murine hepatocyte-derived cell line that supports high-level HBV replication.
AML12 cells (a gift of Chen Liu at Florida State University, Jacksonville, FL) were maintained in Dulbecco's modified Eagle medium, nutrient mixture F12 (DMEM/F12) (Invitrogen), supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. To obtain cell lines that support tetracycline-inducible HBV replication, the AML12 cells were cotransfected with the plasmid pTet-off (Clontech), which expresses a tetracycline (Tet)-responsive transcriptional activator (tTA), and the plasmid pTREHBVDES, in which HBV pgRNA expression is controlled by a cytomegalovirus early promoter with a tetracycline-responsive element (17). Transfected cells were selected with 500 μg/ml G418 in the presence of 1 μg/ml tetracycline. G418-resistant colonies were picked and expanded into cell lines. HBV replication was induced by culturing cells in tetracycline-free medium, and the levels of viral DNA replicative intermediates were determined by Southern blot hybridization. One of the cell lines with the highest level of HBV replication, designated AML12HBV10, was chosen for this study.
Viral DNA and RNA analysis.
Intracellular viral core DNA was extracted as described previously (19). One-half of the DNA sample from each well of 12-well plates was resolved by electrophoresis into a 1.5% agarose gel and transferred onto a Hybond-XL membrane. For viral RNA analysis, total cellular RNA was extracted with TRIzol reagents (Invitrogen). Five micrograms of total RNA was resolved in a 1.5% agarose gel containing 2.2 M formaldehyde and transferred onto a Hybond-XL membrane in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Encapsidated HBV RNA was extracted as previously described (21). One-half of the RNA sample from each well of 12-well plates was resolved in 1.5% agarose gel containing 2.2 M formaldehyde and transferred onto Hybond-XL membrane in 20× SSC buffer. For the detection of HBV DNA and RNA, membranes were probed with either an [α-32P]UTP (800 Ci/mmol; Perkin Elmer)-labeled minus-strand- or plus-strand-specific full-length HBV riboprobe as described previously. The membrane was exposed to a phosphorimager screen, and hybridization signals were quantified with the QuantityOne software program (Bio-Rad).
Viral nucleocapsid analysis.
HBV capsids (cores) and associated viral DNA were analyzed essentially as previously described, with minor modifications (16, 17). Briefly, AML12HBV10 cells were lysed by addition of 300 μl buffer containing 10 mM Tris-HCl (pH 7.6), 100 mM NaCl, 1 mM EDTA, and 0.1% NP-40 to each well of a 12-well plate. Cell debris was removed by centrifugation at 5,000 × g for 10 min. Ten microliters of the clarified cell lysates were fractionated by electrophoresis through nondenaturing 1% agarose gels and transferred to a nitrocellulose filter by blotting with TNE buffer (10 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 1 mM EDTA). HBV capsids were detected by probing the membrane with an antibody against the HBV core protein (Dako). Bound antibody was revealed by IRDye secondary antibodies and visualized by the Li-COR Odyssey system. To detect capsid-associated HBV DNA, the membranes were treated with buffer containing 0.5 N NaOH and 1.5 M NaCl for 5 min, followed by neutralization with buffer containing 1 M TRIS-HCl and 1.5 M NaCl for 5 min. The viral DNA was detected by hybridization with a [α-32P]UTP (800 Ci/mmol; Perkin Elmer)-labeled minus-strand-specific full-length HBV riboprobe.
Viral core protein analysis.
AML12HBV10 cells were lysed with 1× Laemmli buffer. A fraction of cell lysate was separated on sodium dodecyl sulfate-12% polyacrylamide gels and electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). Membranes were blocked with phosphate-buffered saline (PBS) containing 5% nonfat dry milk and probed with antibodies against HBcAg (18) or β-actin (Chemicon International). Bound antibody was revealed by IRDye secondary antibodies and visualized and quantified by the Li-COR Odyssey system.
IFN and chemicals.
Recombinant murine IFN-α, IFN-γ, TNF-α, interleukin-1 (IL-1), and IL-6 were purchased from PBL Interferon Source and PeproTech. AT-61 and Bay-4109 were synthesized in-house by following the published procedures (7, 29). Lamivudine is a gift from William S. Mason, Fox Chase Cancer Center, Philadelphia, PA.
RESULTS
IFN-α and IFN-γ potently inhibit HBV replication in murine hepatocytes.
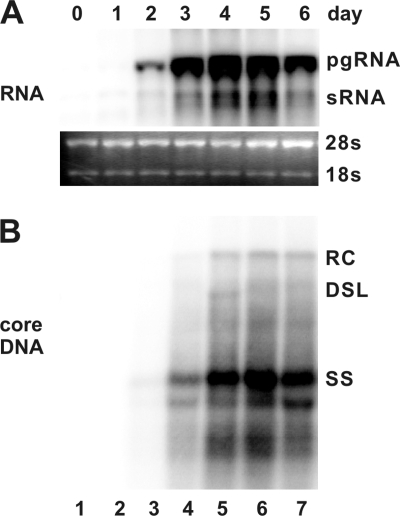
To investigate the antiviral effects and illustrate the antiviral mechanism of IFNs and other inflammatory cytokines, we first established an immortalized mouse hepatocyte (AML12)-derived cell line, designated AML12HBV10, that allows tetracycline (Tet)-inducible transcription of HBV pgRNA and viral DNA replication. In the presence of Tet, the transcription of viral pgRNA is suppressed. However, upon removal of Tet from cultural media, HBV mRNAs are transcribed and accumulated to a detectable level at 24 h, reaching the peak level at 96 h (Fig. 1, upper panel). As expected, following pgRNA transcription, HBV DNA replication intermediates were detected at 48 h and reached the steady-state level at 96 h after Tet removal (Fig. 1, lower panel).
FIG. 1.
Kinetics of HBV RNA transcription and DNA replication in a murine hepatocyte-derived cell line. AML12HBV10 cells were maintained in the medium containing 1 μg/ml of tetracycline to inhibit viral pgRNA transcription and then cultured in the medium without the antibiotic for the indicated number of days. Total cellular RNA and cytoplasmic core DNA were extracted and analyzed by Northern and Southern blot hybridization, respectively. (A) For viral RNA analysis, each lane was loaded with 5 μg of total RNA. pgRNA, pregenomic RNA; sRNA, mRNAs specifying the three envelope proteins. rRNA (28S and 18S) served as loading controls. (B) For core DNA analysis, each lane represents the amount of viral DNA extracted from one-half of cells in a well of a 12-well plate. RC, relaxed circular DNA; DSL, double-stranded linear DNA; SS, single-stranded DNA.
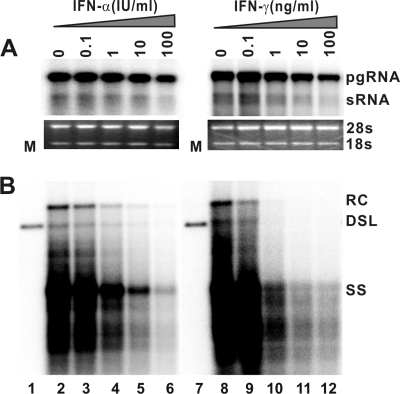
To determine the antiviral effects of IFN-α, IFN-γ, and three other inflammatory cytokines that are induced in the liver during HBV infection and potentially play a role in noncytopathic suppression of HBV replication (20, 24), AML12HBV10 cells were cultured in the absence of Tet and presence of the indicated concentrations of murine IFN-α, IFN-γ, TNF-α, IL-1, or IL-6 for 4 days. The results show that while IL-1 and IL-6 do not affect the levels of intracellular HBV DNA, a modest reduction of HBV DNA replication intermediates can be observed in cells treated with higher concentrations of TNF-α (data not shown). However, in marked contrast, treatment of the cells with either IFN-α or IFN-γ dramatically reduces the levels of intracellular HBV DNA replication intermediates in a dose-dependent manner (Fig. 2 B). Moreover, under the concentrations at which HBV replication is inhibited, the levels of viral pgRNA are not affected or are only slightly reduced (Fig. 2A). Hence, the results imply that the type I and type II IFNs inhibit one or several posttranscriptional steps of HBV replication in the murine hepatocytes.
FIG. 2.
IFN-α and IFN-γ treatment potently reduce the level of HBV DNA. AML12HBV10 cells were cultured in the absence of tetracycline and in the presence of the indicated concentrations of murine IFN-α or IFN-γ for 4 days. Total cellular RNA and cytoplasmic core DNA were extracted and analyzed by Northern and Southern blot hybridization, respectively. (A) For viral RNA analysis, each lane was loaded with 5 μg of total RNA. rRNAs (28S and 18S) serve as loading controls. (B) For core DNA analysis, each lane represents the amount of viral DNA extracted from one-half of cells in a well of 12-well plate.
IFN-α and IFN-γ reduce the level of intracellular HBV nucleocapsids.
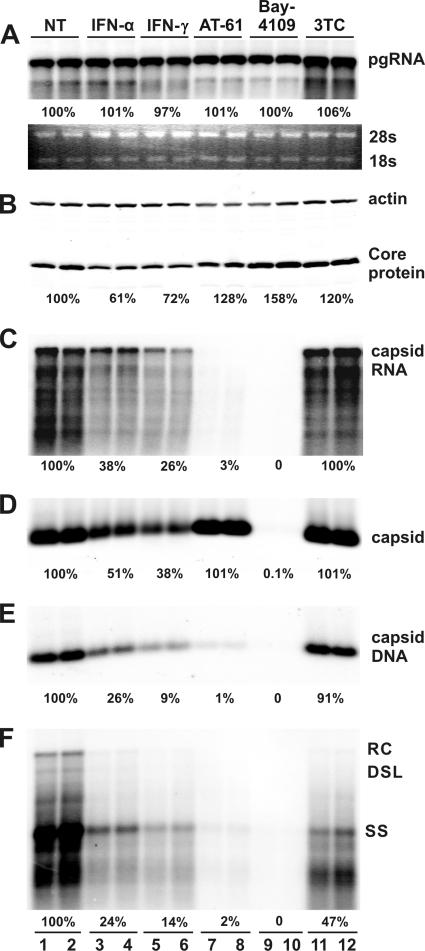
To map the replication steps that are inhibited by IFNs, the levels of viral gene products (pgRNA and core protein) and replication intermediates (capsids, encapsidated pgRNA, capsid-associated DNA, and DNA replication intermediates) were determined in cells that were either left untreated or treated with the indicated concentrations of IFN-α, IFN-γ, Bay-4109, AT-61, or lamivudine (3TC) for 4 days. The three antiviral compounds inhibited HBV replication in a dose-dependent manner in the murine hepatocytes (data not shown). Because each of these antiviral compounds inhibits a well-defined step of HBV replication, they are used in this study as references to help dissect the antiviral mechanism of IFNs.
Our results from this experiment revealed the following. First, none of the treatments affected the levels of pgRNA (Fig. 3 A). Second, none of the three antiviral compounds affected the levels of the HBV core protein, but the amount of the core protein is modestly reduced in cells treated with either IFN-α or IFN-γ (Fig. 3B). Third, in accordance with the inhibition of HBV DNA polymerase, lamivudine treatment did not affect pgRNA encapsidation (Fig. 3C) and capsid formation (Fig. 3D) but reduced the levels of capsid-associated viral DNA replication intermediates (Fig. 3E and F). The observed poor antiviral activity of lamivudine in this cell line is most likely due to its inefficient phosphorylation in rodent hepatocytes (57). Fourth, treatment of cells with Bay-4109, a capsid assembly inhibitor (7, 47), completely inhibited capsid formation, as revealed by the particle gel assay which detects intact nucleocapsids (17) (Fig. 3D). As expected, pgRNA encapsidation (Fig. 3C) and HBV DNA replication (Fig. 3E and F) did not occur in Bay-4109-treated cells. Fifth, consistent with the previous reports (9, 29), treatment of cells with an HBV pgRNA encapsidation inhibitor, AT-61, did not affect capsid formation (Fig. 3D) but eliminated encapsidated pgRNA (Fig. 3C). As a consequence, viral DNA replication was also inhibited (Fig. 3E and F). Finally, unlike all the three antiviral compounds, treatment of cells with either IFN-α or IFN-γ significantly reduced the levels of HBV capsids (Fig. 3D). Consistently, the levels of encapsidated pgRNA (Fig. 3C) and capsid-associated viral DNA replication intermediates (Fig. 3E and F) were also proportionally reduced. Taken together, the distinct profile of HBV core protein and capsid accumulation in IFN-α- or IFN-γ-treated cells suggests that the cytokines may inhibit HBV capsid assembly, which results in the degradation of the unassembled core protein and reduced pgRNA encapsidation and DNA replication. Alternatively, IFNs may promote the degradation of nucleocapsids.
FIG. 3.
IFN-α and IFN-γ treatment reduce the level of intracellular HBV nucleocapsids. AML12HBV10 cells were cultured in the absence of tetracycline and left untreated (control) or treated with 10 IU/ml IFN-α, 5 ng/ml IFN-γ, 25 μM AT-61, 2 μM Bay-4109, or 50 μM lamivudine (3TC) for 4 days, and cells were then harvested. (A) Intracellular viral RNA was determined by Northern blot hybridization. (B) HBV core protein was determined by a Western blot assay with an antibody recognizing the carboxyl-terminal 14 amino acid residues of HBcAg. (D) The amounts of nucleocapsids were determined by a particle gel assay (see Materials and Methods for details) that measures intact nucleocapsids. (E) Nucleocapsid-associated HBV DNA was quantified by alkaline treatment of nucleocapsids on the membrane following the particle gel assay and hybridized with a full-length HBV riboprobe that anneals to the minus strand of HBV DNA. Encapsidated pgRNA (C) and HBV DNA replication intermediates (F) were extracted and determined by Northern blot and Southern blot hybridizations, respectively.
IFN promotes the decay of intracellular HBV nucleocapsids in cells in which pgRNA transcription is inhibited.
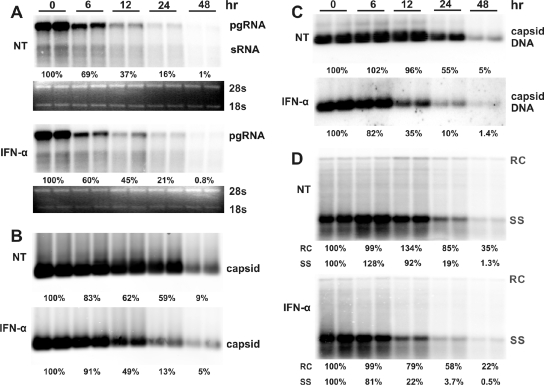
To distinguish the two possibilities described above, we took advantage of our Tet-inducible replication system to determine whether IFN treatment shortens the half-life of HBV nucleocapsids. To this end, AML12HBV10 cells were cultured in the absence of Tet for 4 days to allow the accumulation of nucleocapsids and replication intermediates. At this time (0 h), Tet was added back to culture medium to stop pgRNA transcription and thus the assembly of nucleocapsids. As shown in Fig. 4 A, upon inhibition of de novo pgRNA transcription, the level of preexisting pgRNA quickly declined and IFN-α treatment apparently did not accelerate the rate of pgRNA decrease. Interestingly, treatment of the cells with IFN-α reduced the half-lives of nucleocapsids and capsid-associated viral DNA from approximately 24 h to less than 12 h (Fig. 4B and C). To further determine whether IFN treatment has a differential effect on the decay of nucleocapsids containing different DNA replication intermediates, we performed a Southern blot analysis on the intracellular core-associated HBV DNA and found that IFN treatment drastically accelerated the rate of decay of single-stranded DNA containing nucleocapsids but only modestly promoted the decay of nucleocapsids containing mature relaxed circular (rc) DNA (Fig. 4D). Similar results were obtained with IFN-γ treatment (data not shown).
FIG. 4.
IFN-α promotes the decay of intracellular HBV nucleocapsids in cells in which pgRNA transcription is blocked. AML12HBV10 cells were cultured in the absence of tetracycline for 4 days to allow HBV replication, and tetracycline was then added back to stop pgRNA transcription from viral transgenes integrated in the cellular chromosome. At the same time, cells were left untreated (control) or treated with 10 IU/ml IFN-α. Cells were harvested at the indicated period of time post-IFN treatment. Intracellular viral RNA (A), nucleocapsid (B), core-associated HBV DNA (C), and HBV DNA replication intermediates (D) were determined as described in Materials and Methods.
IFN accelerates the decay of viral DNA-containing nucleocapsids in cells in which pgRNA encapsidation is inhibited.
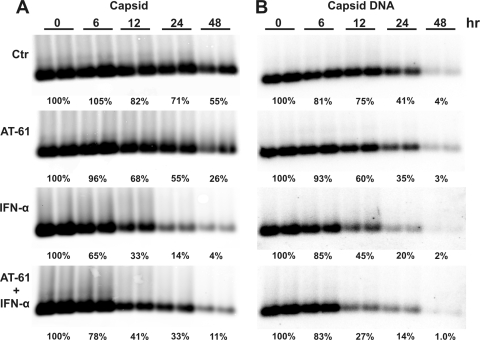
Although the amount of preexisting pgRNA declined quickly upon addition of Tet to culture medium (Fig. 4A), pgRNA-containing capsid formation might not stop promptly. Therefore, the apparently accelerated decay of nucleocapsids observed in the above experiment could be alternatively explained as a result of IFN-induced inhibition of pgRNA-containing capsid formation. To rule out this possibility, the effect of IFN-α on nucleocapsid decay kinetics was further examined in the presence of AT-61, which inhibits the formation of pgRNA-containing nucleocapsids but not empty capsids (9). Briefly, AML12HBV10 cells were cultured in the absence of Tet for 4 days to allow the accumulation of nucleocapsids and replication intermediates. At this time (0 h), Tet was added to culture medium to stop pgRNA transcription. At the same time, the cells were either left untreated or treated with IFN-α in the absence or presence of AT-61. The results showed that compared with untreated controls, AT-61 treatment only modestly increased the rates of capsid and capsid-associated HBV DNA decline (Fig. 5 ). However, consistent with the results presented above (Fig. 4B and C), treatment of the cells with IFN-α in the absence of AT-61 resulted in a profound reduction of both capsids and capsid-associated viral DNA (Fig. 5). Similar results were obtained with IFN-γ treatment (data not shown). These results suggest that although assembly of pgRNA-containing nucleocapsids may occur after cessation of pgRNA transcription, it is obvious that inhibition of pgRNA-containing capsid formation under this experimental condition could not account for the profound reduction in HBV nucleocapsids and viral DNA in IFN-treated cells. Hence, our results favor a hypothesis that an IFN-induced antiviral response promotes the decay of HBV nucleocapsids.
FIG. 5.
IFN-α accelerates the decay of DNA-containing capsid in cells in which pgRNA-containing capsid formation is inhibited. AML12HBV10 cells were cultured in the absence of tetracycline for 4 days, and tetracycline was then added back to stop pgRNA transcription. At the same time, cells were left untreated (control) or treated with 25 μM AT-61, 10 IU/ml IFN-α, or a combination of 25 μM AT-61 and 10 IU/ml mouse IFN-α (mIFN-α). Cells were harvested at the indicated periods of time posttreatment. Intracellular viral capsids (left panel) and core-associated HBV DNA (right panel) were determined as described in Materials and Methods.
However, it was rather unexpected that compared with IFN-α monotreatment, treatment of the cells with IFN-α in the presence of AT-61 resulted in a slightly slower decay of capsids but a higher rate of capsid-associated HBV DNA decay (Fig. 5). One possible explanation for this intriguing observation is that whereas HBV nucleic acid-containing capsids or the replication-competent nucleocapsids are sensitive to IFN-induced decay, the empty capsids formed in the presence of AT-61 might be resistant to the cytokine-induced antiviral response.
IFN selectively promotes the decay of HBV nucleocapsids but not empty capsids formed in the presence of AT-61.
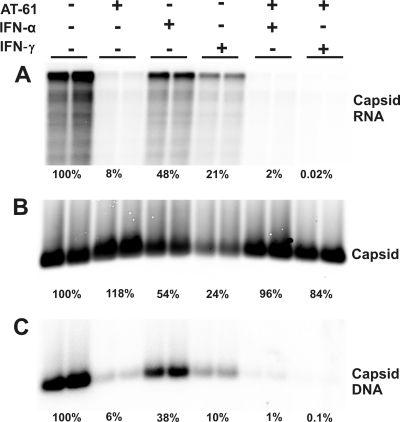
To test this hypothesis, we first investigated the effect of IFN-α and IFN-γ on the accumulation of empty capsids in AT-61-treated cells. Briefly, AML12HBV10 cells were cultured in the absence of Tet and left untreated or treated with appropriate concentrations of AT-61, IFN-α, or IFN-γ, alone or in combinations, for 4 days. Cells were then harvested, and levels of intracellular capsids, encapsidated pgRNA, and capsid-associated HBV DNA were determined. As shown in Fig. 6, consistent with the results presented in Fig. 3, AT-61 treatment did not affect capsid formation but efficiently inhibited pgRNA encapsidation and thus DNA synthesis. Moreover, treatment of the cells with either IFN-α or IFN-γ reduced the levels of capsids, encapsidated pgRNA, and HBV DNA. However, as predicted by the hypothesis, neither IFN-α nor IFN-γ significantly reduced the levels of capsids in AT-61-treated cells. In contrast, the residual amounts of encapsidated pgRNA (8%) and capsid-associated DNA (6%) in AT-61-treated cells were further reduced by the cytokine treatment.
FIG. 6.
Effects of combinational treatment of AT-61 and IFN on HBV nucleocapsids. AML12HBV10 cells were left untreated or treated with 10 IU/ml IFN-α, 5 ng/ml IFN-γ, or a combination of 25 μM AT-61 and 10 IU/ml IFN-α or 5 ng/ml IFN-γ for 4 days. Cells were then harvested, and encapsidated viral RNA (A), capsids (B), and core-associated HBV DNA (C) were determined as described in Materials and Methods.
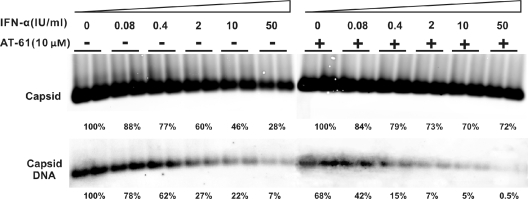
To further determine the differential sensitivities of replication-competent nucleocapsids and empty capsids to the IFN-induced antiviral response, AML12HBV10 cells were cultured in the absence of Tet and left untreated or treated with serial concentrations of IFN-α in the absence or presence of 10 μM AT-61. Under these concentrations, AT-61 only partially inhibits pgRNA-containing capsid formation and reduces core-associated HBV DNA by approximately 40% (Fig. 7). In support of our hypothesis, the results (Fig. 7) showed that compared with IFN-α treatment alone, in the presence of AT-61, IFN-α induced a much less profound reduction of total capsids but a more profound reduction of capsid-associated HBV DNA in a dose-dependent manner. Furthermore, as observed in AT-61-treated AMLHBV10 cells, treatment of a stable AML12-derived cell line expressing the HBV core protein with either IFN-α or IFN-γ did not apparently alter the level of intracellular empty capsids, as revealed by a particle gel assay (data not shown). The results presented above thus imply that the IFN-induced antiviral response is able to distinguish HBV nucleic acid-containing capsids (nucleocapsids) from empty capsids and selectively promotes the decay of the former but not the latter.
FIG. 7.
Effects of combinational treatment of AT-61 and IFN-α on HBV nucleocapsids. AML12HBV10 cells were left untreated or treated with 10 IU/ml IFN-α in the absence or presence of 10 μM AT-61 for 4 days. Cells were then harvested, and encapsidated viral RNA, capsids, and core-associated HBV DNA were determined as described in Materials and Methods.
IFN-induced decay of HBV nucleocapsids, at least in part, depends on proteasome activity.
Expression of IFN-inducible proteasome catalytic and regulatory subunits correlates with the IFN-α- and IFN-γ-mediated noncytopathic inhibition of HBV in transgenic mice and murine hepatocytes and with clearance of the virus in acutely infected chimpanzees (50, 54). It was also demonstrated previously that proteasome inhibitors were able to abrogate the antiviral activity of IFN-β and IFN-γ against HBV in immortalized HBV transgenic mouse hepatocytes (41). It is therefore possible that the accelerated decay of HBV DNA-containing nucleocapsids by IFN-α and IFN-γ in AML12HBV10 cells is mediated by cellular proteasome activity.
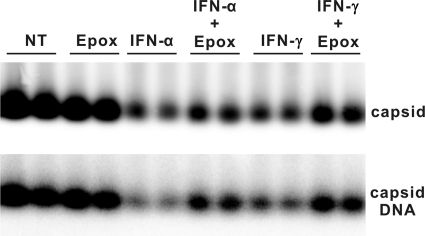
To test this hypothesis, we examined the effect of epoxomicin, an irreversible proteasome inhibitor, on the IFN-induced reduction of nucleocapsids. Because treatment of AML12 cells with epoxomicin for more than 8 h induces apoptosis (data not shown), the cells were treated for only 6 h with the inhibitor. Briefly, AML12HBV10 cells were cultured in the absence of Tet for 4 days to allow the accumulation of HBV DNA-containing nucleocapsids. The cells were then left untreated or treated with 50 IU/ml of IFN-α or 10 ng/ml of IFN-γ. Six hours later, epoxomicin was added to the culture medium and incubated for an additional 6 h. As shown in Fig. 8, treatment of the cells with epoxomicin for only 6 h after IFN treatment attenuated the cytokine-induced reduction of HBV capsids and core-associated viral DNA, suggesting that IFN-induced decay of HBV nucleocapsids, at least in part, depends on cellular proteasome activity.
FIG. 8.
Epoxomicin attenuates IFN-induced decay of HBV nucleocapsids. AML12HBV10 cells were cultured in the absence of tetracycline for 4 days, and tetracycline was then added back to stop pgRNA transcription. At the same time, cells were left untreated (control [NT]) or treated with 50 IU/ml IFN-α. Six hours later, epoxomicin (Epox) was added to culture medium to a final concentration of 0.75 μM. Cells were harvested 6 h after addition of epoxomicin. Capsids and core-associated HBV DNA were determined as described in Materials and Methods.
DISCUSSION
IFN-α and IFN-γ are key mediators of host innate and adaptive immunity against viral infection (45). In the case of HBV infection, it has been demonstrated that induction of IFN-γ and the cytokine-induced cellular gene expression in the livers of HBV-infected chimpanzees is correlated with suppression of HBV replication and ultimately clearance of the virus infection (15, 50). In contrast, unlike the case with hepatitis C virus (HCV) infection, induction of type I IFN-stimulated gene (ISG) expression was not detectable in the livers of chimpanzees during the initial phase of HBV infection, suggesting that HBV may have evolved a strategy to either evade the detection or inhibit the activation of the type I IFN-mediated innate defense system (51). However, IFN-α is an approved antiviral medication for chronic hepatitis B and is able to significantly reduce the viral load in 30% of hepatitis B virus e antigen (HBeAg)-positive patients and 40% of HBeAg-negative patients after a standard 48-month pegylated IFN-α therapy (8, 39). Although it was shown that IFN-α and IFN-γ were able to modestly reduce the levels of HBV DNA at concentrations higher than 100 IU/ml in HBV genome-stably transfected human hepatoma cell lines, the relatively poor antiviral activity of the cytokines in these cell lines prevented the further dissection of their antiviral mechanism (5, 22).
The AML12 cell line consists of well-differentiated and nontumorigenic murine hepatocytes derived from the liver of a transgenic mouse overexpressing human transforming growth factor alpha (55). Because this immortalized murine hepatocyte cell line has typical features of normal hepatocytes and expresses high levels of hepatocyte-specific proteins, such as albumin, α1-antitrypsin, and transferrin (55), it has been used as a cell culture model to study hepatocellular metabolism and effects of the HBV X protein and other exogenous or host cellular proteins on the growth and differentiation of hepatocytes (46, 49). Taking advantages of this well-differentiated murine hepatocyte cell line, we created a stable cell line expressing HBV pgRNA in a Tet-inducible manner to study HBV replication and its inhibition by IFN and other inflammatory cytokines. Our results show that in spite of robust HBV DNA replication, the murine hepatocytes fail to support covalently closed circular DNA (cccDNA) formation (data not shown), which is consistent with that observed in HBV transgenic mice in vivo (13). Moreover, in agreement with a previous report suggesting that both type I and type II IFNs, but not TNF-α, inhibit HBV replication in an HBV transgenic mouse hepatocyte-derived cell line (37), we found that IFN-α and IFN-γ potently inhibit HBV replication via a posttranscriptional mechanism. Furthermore, although the possibility that the cytokines inhibit the pgRNA-containing nucleocapsid formation, as suggested by others (52), could not be directly investigated in our cell culture system, we obtained compelling evidence supporting the notion that the IFN-induced antiviral response accelerates the decay of HBV nucleocapsids (as depicted in Fig. 9), which is consistent with our previous observation that IFN-α treatment shortens the half-life of encapsidated DHBV pgRNA in a chicken hepatoma-derived cell line (dstet5) (21). Furthermore, we provide evidence suggesting that the accelerated decay of HBV nucleocapsids by IFNs requires cellular proteasome activity.
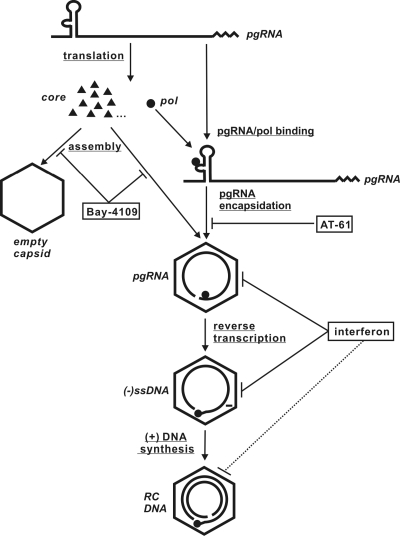
FIG. 9.
Graphic representation of the antiviral mechanisms of IFNs and two key antiviral compounds used in this study. Bay-4109 inhibits HBV capsid assembly. AT-61 does not affect capsid assembly but abrogates pgRNA encapsidation. As revealed in this study, IFNs promote the decay of nucleocapsids (especially unmatured nucleocapsids) but not empty capsids.
Interestingly, we observed in this study that the IFN-induced antiviral response is able to distinguish the replication-competent (DNA-containing) nucleocapsids from empty capsids and selectively promote the decay of viral nucleic acid-containing but not empty capsids. Furthermore, we also observed that single (minus)-stranded DNA-containing capsids are more prone to IFN-induced degradation than mature double-stranded (rc) DNA-containing capsids (Fig. 4D and 9). These intriguing observations imply that the distinct structural features displayed by the different capsids might be differentially recognized by the innate immune response and targeted for degradation. This hypothesis is supported by recent structural and genetic analyses showing that nucleic acid-containing and empty HBV capsids display distinct structural features on their surfaces (36, 42). Alternatively, the phosphorylation status of the HBV core protein, the structural component of capsids, has been shown to affect capsid stability, pgRNA encapsidation, and HBV DNA synthesis (2, 31, 35, 56) and is dynamically regulated during viral capsid maturation (3, 38). It is therefore possible that the differential phosphorylation or dephosphorylation of the core protein on replication-competent nucleocapsids could serve as a molecular tag for recognition by IFN-induced antiviral proteins.
Moreover, our finding that the IFN-induced antiviral response selectively purges replication-competent but not empty capsids may actually bear important biological significance. For example, early electron microscopic studies suggested that the majority of capsids in HBV-infected livers do not contain viral DNA (43) and DNA-negative “empty” Dane particles are predominant in sera (1, 28). Importantly, it was reported by Chisari's group in a chimpanzee study that during the recovery phase of an acute HBV infection, while the levels of HBV DNA in the livers were reduced by more than 10-fold, the HBV core antigen (HBcAg)-positive hepatocytes remains at the same levels during this period of time and started to decline only at later time points (15). Accordingly, these authors concluded that viral DNA was more sensitive to cytokine-induced noncytolytic clearance mechanisms than HBcAg, suggesting that the turnover of HBcAg was slower than that of viral DNA in infected cells (15). Because HBV DNA is synthesized exclusively in nucleocapsids, the results imply that in HBV-infected livers, DNA-containing capsids are differentially eliminated early during the noncytolytic clearance phase, which is believed to be mediated by inflammatory cytokines, including IFNs (14, 15, 53).
Nevertheless, elucidation of the molecular mechanism by which IFN accelerates the decay of HBV nucleocapsids and identification of IFN-induced cellular proteins that mediate the robust antiviral response in murine hepatocytes will ultimately lead to a better understanding of the IFN antiviral mechanism against HBV in human hepatocytes. Such knowledge should be helpful for development of novel therapeutic strategies to improve the antiviral efficacy of IFN-α and cure chronic HBV infections.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (AI061441) and by the Hepatitis B Foundation through an appropriation of the Commonwealth of Pennsylvania. Ju-Tao Guo is the Bruce Witte Scholar of the Hepatitis B Foundation.
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Alberti, A., S. Diana, G. H. Scullard, W. F. Eddleston, and R. Williams. 1978. Full and empty Dane particles in chronic hepatitis B virus infection: relation to hepatitis B e antigen and presence of liver damage. Gastroenterology 75:869-874. [PubMed] [Google Scholar]
- 2.Barrasa, M. I., J. T. Guo, J. Saputelli, W. S. Mason, and C. Seeger. 2001. Does a cdc2 kinase-like recognition motif on the core protein of hepadnaviruses regulate assembly and disintegration of capsids? J. Virol. 75:2024-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basagoudanavar, S. H., D. H. Perlman, and J. Hu. 2007. Regulation of hepadnavirus reverse transcription by dynamic nucleocapsid phosphorylation. J. Virol. 81:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Block, T. M., H. Guo, and J. T. Guo. 2007. Molecular virology of hepatitis B virus for clinicians. Clin. Liver Dis. 11:685-706, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caselmann, W. H., M. Meyer, S. Scholz, P. H. Hofschneider, and R. Koshy. 1992. Type I interferons inhibit hepatitis B virus replication and induce hepatocellular gene expression in cultured liver cells. J. Infect. Dis. 166:966-971. [DOI] [PubMed] [Google Scholar]
- 6.Cooksley, W. G., T. Piratvisuth, S. D. Lee, V. Mahachai, Y. C. Chao, T. Tanwandee, A. Chutaputti, W. Y. Chang, F. E. Zahm, and N. Pluck. 2003. Peginterferon alpha-2a (40 kDa): an advance in the treatment of hepatitis B e antigen-positive chronic hepatitis B. J. Viral Hepat. 10:298-305. [DOI] [PubMed] [Google Scholar]
- 7.Deres, K., C. H. Schroder, A. Paessens, S. Goldmann, H. J. Hacker, O. Weber, T. Kramer, U. Niewohner, U. Pleiss, J. Stoltefuss, E. Graef, D. Koletzki, R. N. Masantschek, A. Reimann, R. Jaeger, R. Gross, B. Beckermann, K. H. Schlemmer, D. Haebich, and H. Rubsamen-Waigmann. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299:893-896. [DOI] [PubMed] [Google Scholar]
- 8.Dienstag, J. L. 2009. Benefits and risks of nucleoside analog therapy for hepatitis B. Hepatology 49:S112-S121. [DOI] [PubMed] [Google Scholar]
- 9.Feld, J. J., D. Colledge, V. Sozzi, R. Edwards, M. Littlejohn, and S. A. Locarnini. 2007. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res. 76:168-177. [DOI] [PubMed] [Google Scholar]
- 10.Guidotti, L. G., K. Ando, M. V. Hobbs, T. Ishikawa, L. Runkel, R. D. Schreiber, and F. V. Chisari. 1994. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 91:3764-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidotti, L. G., and F. V. Chisari. 2006. Immunobiology and pathogenesis of viral hepatitis. Annu. Rev. Pathol. 1:23-61. [DOI] [PubMed] [Google Scholar]
- 12.Guidotti, L. G., T. Ishikawa, M. V. Hobbs, B. Matzke, R. Schreiber, and F. V. Chisari. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25-36. [DOI] [PubMed] [Google Scholar]
- 13.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidotti, L. G., A. Morris, H. Mendez, R. Koch, R. H. Silverman, B. R. Williams, and F. V. Chisari. 2002. Interferon-regulated pathways that control hepatitis B virus replication in transgenic mice. J. Virol. 76:2617-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidotti, L. G., R. Rochford, J. Chung, M. Shapiro, R. Purcell, and F. V. Chisari. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825-829. [DOI] [PubMed] [Google Scholar]
- 16.Guo, H., C. E. Aldrich, J. Saputelli, C. Xu, and W. S. Mason. 2006. The insertion domain of the duck hepatitis B virus core protein plays a role in nucleocapsid assembly. Virology 353:443-450. [DOI] [PubMed] [Google Scholar]
- 17.Guo, H., D. Jiang, T. Zhou, A. Cuconati, T. M. Block, and J. T. Guo. 2007. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J. Virol. 81:12472-12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, H., R. Mao, T. M. Block, and J. T. Guo. 2010. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J. Virol. 84:387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, H., W. S. Mason, C. E. Aldrich, J. R. Saputelli, D. S. Miller, A. R. Jilbert, and J. E. Newbold. 2005. Identification and characterization of avihepadnaviruses isolated from exotic anseriformes maintained in captivity. J. Virol. 79:2729-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, J.-T., H. Zhou, C. Liu, C. Aldrich, J. Saputelli, T. Whitaker, M. I. Barrasa, W. S. Mason, and C. Seeger. 2000. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infection. J. Virol. 74:1495-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, J. T., M. Pryce, X. Wang, M. I. Barrasa, J. Hu, and C. Seeger. 2003. Conditional replication of duck hepatitis B virus in hepatoma cells. J. Virol. 77:1885-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi, Y., and K. Koike. 1989. Interferon inhibits hepatitis B virus replication in a stable expression system of transfected viral DNA. J. Virol. 63:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiraga, N., M. Imamura, T. Hatakeyama, S. Kitamura, F. Mitsui, S. Tanaka, M. Tsuge, S. Takahashi, H. Abe, T. Maekawa, H. Ochi, C. Tateno, K. Yoshizato, T. Wakita, and K. Chayama. 2009. Absence of viral interference and different susceptibility to interferon between hepatitis B virus and hepatitis C virus in human hepatocyte chimeric mice. J. Hepatol. 51:1046-1054. [DOI] [PubMed] [Google Scholar]
- 24.Hosel, M., M. Quasdorff, K. Wiegmann, D. Webb, U. Zedler, M. Broxtermann, R. Tedjokusumo, K. Esser, S. Arzberger, C. J. Kirschning, A. Langenkamp, C. Falk, H. Buning, S. Rose-John, and U. Protzer. 2009. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology 50:1773-1782. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, D., H. Guo, C. Xu, J. Chang, B. Gu, L. Wang, T. M. Block, and J. T. Guo. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jilbert, A. R., T. T. Wu, J. M. England, P. M. Hall, N. Z. Carp, A. P. O'Connell, and W. S. Mason. 1992. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. J. Virol. 66:1377-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kajino, K., A. R. Jilbert, J. Saputelli, C. E. Aldrich, J. Cullen, and W. S. Mason. 1994. Woodchuck hepatitis virus infections: very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. J. Virol. 68:5792-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura, T., N. Ohno, N. Terada, A. Rokuhara, A. Matsumoto, S. Yagi, E. Tanaka, K. Kiyosawa, S. Ohno, and N. Maki. 2005. Hepatitis B virus DNA-negative Dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J. Biol. Chem. 280:21713-21719. [DOI] [PubMed] [Google Scholar]
- 29.King, R. W., S. K. Ladner, T. J. Miller, K. Zaifert, R. B. Perni, S. C. Conway, and M. J. Otto. 1998. Inhibition of human hepatitis B virus replication by AT-61, a phenylpropenamide derivative, alone and in combination with (-)beta-L-2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 42:3179-3186. (Erratum, 43:726.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ladner, S. K., M. J. Otto, C. S. Barker, K. Zaifert, G. H. Wang, J. T. Guo, C. Seeger, and R. W. King. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 41:1715-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan, Y. T., J. Li, W. Liao, and J. Ou. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342-348. [DOI] [PubMed] [Google Scholar]
- 32.Lau, G. K., T. Piratvisuth, K. X. Luo, P. Marcellin, S. Thongsawat, G. Cooksley, E. Gane, M. W. Fried, W. C. Chow, S. W. Paik, W. Y. Chang, T. Berg, R. Flisiak, P. McCloud, and N. Pluck. 2005. Peginterferon alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 352:2682-2695. [DOI] [PubMed] [Google Scholar]
- 33.Liang, T. J. 2009. Hepatitis B: the virus and disease. Hepatology 49:S13-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahon, B. J. 2005. Epidemiology and natural history of hepatitis B. Semin. Liver Dis. 25(Suppl. 1):3-8. [DOI] [PubMed] [Google Scholar]
- 35.Melegari, M., S. K. Wolf, and R. J. Schneider. 2005. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J. Virol. 79:9810-9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pairan, A., and V. Bruss. 2009. Functional surfaces of the hepatitis B virus capsid. J. Virol. 83:11616-11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasquetto, V., S. F. Wieland, S. L. Uprichard, M. Tripodi, and F. V. Chisari. 2002. Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J. Virol. 76:5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perlman, D. H., E. A. Berg, P. B. O'Connor, C. E. Costello, and J. Hu. 2005. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 102:9020-9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perrillo, R. 2009. Benefits and risks of interferon therapy for hepatitis B. Hepatology 49:S103-S111. [DOI] [PubMed] [Google Scholar]
- 40.Robek, M. D., B. S. Boyd, S. F. Wieland, and F. V. Chisari. 2004. Signal transduction pathways that inhibit hepatitis B virus replication. Proc. Natl. Acad. Sci. U. S. A. 101:1743-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robek, M. D., S. F. Wieland, and F. V. Chisari. 2002. Inhibition of hepatitis B virus replication by interferon requires proteasome activity. J. Virol. 76:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roseman, A. M., J. A. Berriman, S. A. Wynne, P. J. Butler, and R. A. Crowther. 2005. A structural model for maturation of the hepatitis B virus core. Proc. Natl. Acad. Sci. U. S. A. 102:15821-15826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakamoto, Y., G. Yamada, M. Mizuno, T. Nishihara, S. Kinoyama, T. Kobayashi, T. Takahashi, and H. Nagashima. 1983. Full and empty particles of hepatitis B virus in hepatocytes from patients with HBsAg-positive chronic active hepatitis. Lab. Invest. 48:678-682. [PubMed] [Google Scholar]
- 44.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 46.Shang, X. Z., H. Zhu, K. Lin, Z. Tu, J. Chen, D. R. Nelson, and C. Liu. 2004. Stabilized beta-catenin promotes hepatocyte proliferation and inhibits TNFalpha-induced apoptosis. Lab. Invest. 84:332-341. [DOI] [PubMed] [Google Scholar]
- 47.Stray, S. J., and A. Zlotnick. 2006. BAY 41-4109 has multiple effects on hepatitis B virus capsid assembly. J. Mol. Recognit. 19:542-548. [DOI] [PubMed] [Google Scholar]
- 48.Summers, J., A. R. Jilbert, W. Yang, C. E. Aldrich, J. Saputelli, S. Litwin, E. Toll, and W. S. Mason. 2003. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc. Natl. Acad. Sci. U. S. A. 100:11652-11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarn, C., L. Zou, R. L. Hullinger, and O. M. Andrisani. 2002. Hepatitis B virus X protein activates the p38 mitogen-activated protein kinase pathway in dedifferentiated hepatocytes. J. Virol. 76:9763-9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wieland, S., R. Thimme, R. H. Purcell, and F. V. Chisari. 2004. Genomic analysis of the host response to hepatitis B virus infection. Proc. Natl. Acad. Sci. U. S. A. 101:6669-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wieland, S. F., and F. V. Chisari. 2005. Stealth and cunning: hepatitis B and hepatitis C viruses. J. Virol. 79:9369-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wieland, S. F., A. Eustaquio, C. Whitten-Bauer, B. Boyd, and F. V. Chisari. 2005. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 102:9913-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wieland, S. F., L. G. Guidotti, and F. V. Chisari. 2000. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol. 74:4165-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wieland, S. F., R. G. Vega, R. Muller, C. F. Evans, B. Hilbush, L. G. Guidotti, J. G. Sutcliffe, P. G. Schultz, and F. V. Chisari. 2003. Searching for interferon-induced genes that inhibit hepatitis B virus replication in transgenic mouse hepatocytes. J. Virol. 77:1227-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, J. C., G. Merlino, and N. Fausto. 1994. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. U. S. A. 91:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Y., X. D. Fu, and J. H. Ou. 2005. Suppression of hepatitis B virus replication by SRPK1 and SRPK2 via a pathway independent of the phosphorylation of the viral core protein. Virology 342:150-158. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, T., J. T. Guo, F. A. Nunes, K. L. Molnar-Kimber, J. M. Wilson, C. E. Aldrich, J. Saputelli, S. Litwin, L. D. Condreay, C. Seeger, and W. S. Mason. 2000. Combination therapy with lamivudine and adenovirus causes transient suppression of chronic woodchuck hepatitis virus infections. J. Virol. 74:11754-11763. [DOI] [PMC free article] [PubMed] [Google Scholar]