Abstract
The relationship between virus evolution and recombination in species B human enteroviruses was investigated through large-scale genetic analysis of echovirus type 9 (E9) and E11 isolates (n = 85 and 116) from 16 European, African, and Asian countries between 1995 and 2008. Cluster 1 E9 isolates and genotype D5 and A E11 isolates showed evidence of frequent recombination between the VP1 and 3Dpol regions, the latter falling into 23 (E9) and 43 (E11) clades interspersed phylogenetically with 46 3Dpol clades of E30 and with those of other species B serotypes. Remarkably, only 2 of the 112 3Dpol clades were shared by more than one serotype (E11 and E30), demonstrating an extremely large and genetically heterogeneous recombination pool of species B nonstructural-region variants. The likelihood of recombination increased with geographical separation and time, and both were correlated with VP1 divergence, whose substitution rates allowed recombination half-lives of 1.3, 9.8, and 3.1 years, respectively, for E9, E11, and E30 to be calculated. These marked differences in recombination dynamics matched epidemiological patterns of periodic epidemic cycles of 2 to 3 (E9) and 5 to 6 (E30) years and the longer-term endemic pattern of E11 infections. Phylotemporal analysis using a Bayesian Markov chain Monte Carlo method, which placed recombination events within the evolutionary reconstruction of VP1, showed a close relationship with VP1 lineage expansion, with defined recombination events that correlated with their epidemiological periodicity. Whether recombination events contribute directly to changes in transmissibility that drive epidemic behavior or occur stochastically during periodic population bottlenecks is an unresolved issue vital to future understanding of enterovirus molecular epidemiology and pathogenesis.
Human enteroviruses (HEV) are single-stranded, positive-sense RNA viruses in the virus family Picornaviridae. Infections with these viruses are enterically transmitted and normally cause subclinical or mild symptoms, but they are also capable of causing a wide array of often severe disease presentations, including aseptic meningitis, encephalitis, and acute flaccid paralysis. Echovirus type 9 (E9) and E11 are serotypes of species B enteroviruses and are among the most common etiological agents of aseptic meningitis (14). The first epidemiological study of E9 investigated isolates obtained from sick children in 1995 (24). E9 is chiefly associated with mild infections affecting children over the age of 5 years and teenagers, although it can cause more severe syndromes in neonates and immunosuppressed patients (14). E11 infections, which predominantly affect infants less than 1 year old, have been associated with large outbreaks of uveitis in infants (15, 21), sepsis, and other neonatal systemic illnesses with high mortality rates (14, 23).
Circulating strains and genotypes of E9 (2, 8, 11, 14) and E11 (4, 7, 21) have been characterized in many different countries through analysis of the structural genome regions, principally VP1. In common with other mammalian RNA viruses, both E9 and E11 show rapid accumulation of nucleotide substitutions over time, but their epidemiologies are distinct. E9 is associated with widespread, large-scale seasonal outbreaks and displays a regular epidemic pattern of outbreaks occurring approximately every 3 years (8, 14). Outbreaks of E11 are less common, occur irregularly, and often last for several years (14, 25).
In contrast to time-related diversification observed in the capsid-encoding regions of the HEV genome, nonstructural region genes are subject to frequent recombination (7, 18, 19, 26, 30), leading to the concept of separate, modular evolution of the structural and nonstructural regions of the HEV genome (20). Recombination in the nonstructural gene region of HEV is limited to members of the same species (27). In a recent large-scale investigation of echovirus type 30 (E30) molecular epidemiology, phylogenetic analysis of the 3Dpol region revealed the existence of a number of discrete, bootstrap-supported clades, allowing circulating E30 variants to be classified into a series of distinct recombinant forms (RFs) (22). Individual RFs became very rapidly disseminated over large geographical distances through repeated cycles of emergence, dominance, and disappearance in a 3- to 5-year cycle.
In the current study, we carried out parallel investigations of the molecular epidemiology and evolution of clinically presenting isolates of E9 and E11 collected in 16 countries in Europe, central Asia, Southeast Asia, and West Africa over a 14-year period. By comprehensive sequencing of these isolates in the structural (VP1) and nonstructural (3Dpol) genome regions, it was possible to explore the dynamics of sequence diversification and recombination. The turnover of individual recombinant groups and the phylogeographical correlates of recombination were determined and compared to those of E30, providing new insights into the nature of the global spread of these important viral pathogens and the role of recombination in enteroviral evolution.
MATERIALS AND METHODS
Samples.
E9 (n = 85) and E11 (n = 119) isolates from 16 countries collected between the years 1995 and 2008 (Table 1) were obtained from internationally distributed referral centers. The following convention was used to name isolates: E9- or E11-two-letter country code plus isolate number/two-letter city or region abbreviation/3Dpol clade/year of collection (e.g., E11-ES13/Ma/DU/03 for an E11 isolate number 13 referred from Madrid in Spain, isolated in 2003, and belonging to the 3Dpol clade DU). Sequences obtained in the current study were supplemented with published sequences of complete genomes. These comprised the E9 prototype strain Hill (X84981), the E9 type-specific strain Barty (AF524866), an E9 isolate from The Netherlands (AF524867), the E11 prototype strain Gregory (X80059), and E11 isolates from India (AJ276224), Russia (AY167104 to AY167106), Finland (AJ577590), Romania (AJ577594), and Hungary (AJ577589).
TABLE 1.
Sources and dates of isolation of survey specimens
| Country | Code | No. of isolates |
Yr of isolation | |
|---|---|---|---|---|
| E9 | E11 | |||
| Azerbaijan | AZ | 0 | 3 | 2004, 2006 |
| Belarus | BY | 0 | 1 | 2008 |
| Finland | FI | 1 | 8 | 2004-2007 |
| Georgia | GE | 0 | 1 | 2002 |
| Ghana | GH | 0 | 3 | 2005 |
| Great Britain | GB | 26 | 0 | 2007-2008 |
| Iceland | IS | 5 | 0 | 2002, 2004 |
| Kazakhstan | KZ | 0 | 1 | 2001 |
| Kyrgyzstan | KG | 1 | 1 | 2004-2008 |
| Netherlands | NL | 10 | 27 | 1995-2008 |
| Russia | RU | 7 | 29 | 2001-2008 |
| Spain | ES | 35 | 37 | 2000-2007 |
| Turkmenistan | TM | 0 | 2 | 2004, 2007 |
| Ukraine | UA | 0 | 2 | 2001, 2007 |
| Uzbekistan | UZ | 0 | 2 | 2002, 2006 |
| Vietnam | VN | 0 | 2 | 2005 |
| Total | 85 | 119 | 1995-2008 | |
Amplification of VP1 and 3Dpol regions and nucleotide sequencing.
RNA extraction and nested reverse transcription (RT)-PCRs were performed as previously described (22), using species B-specific enterovirus primers designed to amplify a 526-bp region of the VP1 gene (16) and a 549-bp region of the 3Dpol gene (22). The amplicons were directly sequenced using BigDye (ABI), and the inner sense or antisense primer and nucleotide sequences were aligned with the Simmonic sequence package version 1.6 (29; http://www.virus-evolution.org).
Phylogenetic analysis.
Phylogenetic trees were constructed by neighbor joining from 1,000 samplings of maximum composite likelihood (MCL) distances using the MEGA4.0 software package (Molecular Evolution Genetic Analysis) (32) with pairwise deletion for missing data and a gamma distribution value of 0.8. Regression analysis and investigation of geographical and temporal aspects of recombination were performed using MCL distances (calculated by MEGA) between sequences. A Bayesian Markov chain Monte Carlo (MCMC) method implemented in the BEAST package version 1.4 (9) was used to estimate temporal phylogenies and rates of evolution (10). Two independent runs for each set were analyzed using the SRD06 model of substitution (28), with chain lengths of 50 million and a relaxed molecular-clock model that allows evolutionary rates to vary among lineages. All other parameters were optimized during the burn-in period. Output from BEAST was analyzed using the program TRACER (http://beast.bio.ed.ac.uk/Tracer), and the results of each duplicate, checked to ensure convergence, were combined after excluding 10% as burn-in.
Analysis of RF succession in evolutionary processes.
The role played by RF succession in the temporal evolution of echoviruses was examined. A Bayesian phylogeographic model XML file for reconstruction of phylogeographical histories of viruses using a standard continuous-time Markov chain (CTMC) implemented in the program BEAST (17) was directly modified to recognize RF elements. The RF states of individual nodes were annotated in Tree-Annotator, and the resultant maximum clade credibility (MCC) tree was visualized in FigTree.
Nucleotide sequence accession numbers.
The sequences obtained in this study have been submitted to GenBank and have been assigned accession numbers GU393543 to GU393949 and GU473268.
RESULTS
Phylogenetic relationships of E9 and E11.
A total of 85 E9 isolates from 7 countries and 119 E11 isolates from 14 countries (Table 1) were analyzed in the current study, together with sequences from prototype isolates and previously published sequences. The sequenced amplicons were analyzed in the VP1 region between positions 2423 and 2904 for E9 (numbering based on the Barty prototype strain) and positions 2412 and 2916 for E11 (based on the Gregory prototype strain).
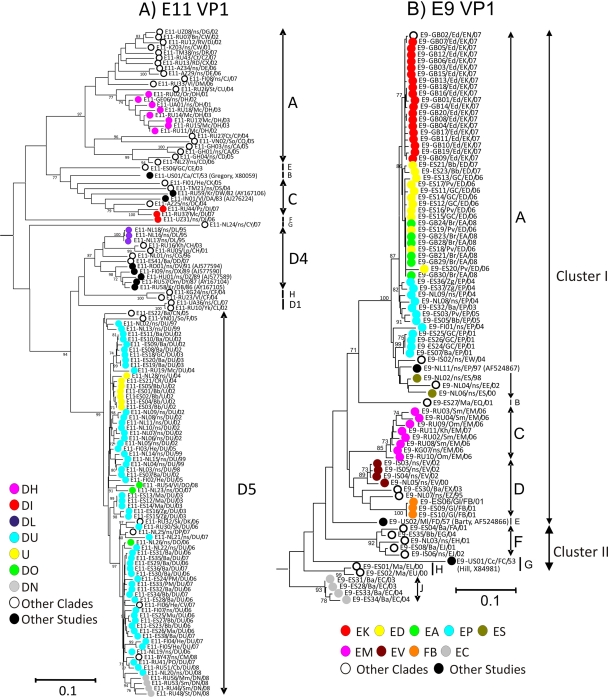
Most E9 isolates (79/85) in the current study belonged to the previously described cluster I lineage and were more closely related to the prototype strain Barty than to strain Hill (11). The isolates assembled into nine genogroups (Fig. 1B), two of which contained the prototype strains as single members. These genogroups were designated A to H and J, with the majority of samples from this study (57/85) belonging to genogroup A, all of which originated from Western Europe. All eight of the Commonwealth of Independent States (CIS) isolates in this study belonged to a single genogroup, C.
FIG. 1.
Neighbor-joining tree of MCL-corrected pairwise distances of VP1 sequences of E11 (A) and E9 (B). The colored labels indicate the 3Dpol group to which the sequences belong, with the exception of sporadic 3Dpol groups (shown with unfilled circles) and database-derived sequences (filled circles). Genogroups are indicated by the appropriate letters and arrows on the right side of each tree. The scale bar depicts an evolutionary distance of 0.1. Only bootstrap resampling values of ≥70% are shown.
The majority of E11 isolates clustered within two of the previously assigned genogroups (25), D5 (74/119) and A (26/119) (Fig. 1A). Of the 26 members of genogroup A, 20 originated from the CIS. The 3 Ghanaian isolates also clustered within genogroup A, as did one of the isolates from Vietnam. The other Vietnamese isolate belonged to genogroup D5, a group which contained most (62/72) of the isolates from Western Europe. A further three new genogroups (designated E to G) and one new subgenogroup (designated D6) that contained a total of five isolates were identified. No sequences within this study clustered within the previously described (25) D2 and D3 lineages.
Sequence divergence of VP1 genome regions.
In the VP1 genome region, variability was predominantly restricted to synonymous sites, with nonsynonymous to synonymous substitution (dN/dS) ratios of 0.013 for E9 and 0.064 for E11, suggesting that most sequence change occurred through neutral drift. Regression analysis of divergence from the earliest isolated sequence for each serotype (Hill prototype for E9 and Gregory prototype for E11, both isolated in 1953) revealed correlation with the date of isolation for E9 (R = 0.389) but poor correlation for E11 (R = 0.089). Based on this regression analysis, the estimated rate of evolution for E9 was 1.6 × 10−3 substitutions per site per year and for E11, 1.1 × 10−3 substitutions per site per year. The same data sets were analyzed by the Bayesian MCMC method (10) using a relaxed molecular clock, and the estimated rate of evolution was greater: 5.8 × 10−3 substitutions per site per year (high-probability distribution [HPD] range, 3.7 × 10−3 to 8.1 × 10−3) for E9 and 4.8 × 10−3 substitutions per site per year (HPD range, 3.6 × 10−3 to 6.1 × 10−3) for E11. The predicted date of the most recent common ancestor (MRCA) for E9 was 1917 (HPD, 1877 to 1947) and for E11 was 1883 (HPD, 1833 to 1927).
The greater intraserotypic sequence divergence and dN/dS ratio and the earlier predicted date of the MRCA of the E11 VP1 region compared to that of E9, along with the poor correlation between sequence divergence and isolation date and obvious tree bifurcation (Fig. 1A), suggested it might be more meaningful to perform regression analysis on individual genogroups containing less divergent sequences. E11 genogroups A, C, D4, and D5 displayed mean within-genogroup sequence divergences of 21.3%, 25.1%, 5.0%, and 4.9%, respectively, with dN/dS ratios of 0.014, 0.024, 0.036, and 0.069. Regression and MCMC analyses of E11-D5 sequences revealed a strong correlation between sequence divergence and the date of isolation (R = 0.895), with an estimated rate of evolution (6.8 × 10−3 substitutions per site per year) greater than that calculated for E11 overall. Using the Bayesian MCMC method, the rate of evolution was estimated to be 8.5 × 10−3 substitutions per site per year (HPD range, 6.3 × 10−3 to 1.09 × 10−2), and the predicted date of the MRCA was 1995 (HPD, 1991 to 1997). However, no correlation was found between sequence diversity and isolation date for the other E11 genogroups analyzed, either individually or together (data not shown).
Sequence divergence in the 3Dpol region.
E9 and E11 sequences were analyzed in the 3Dpol region between positions 6018 and 6567 (numbering based on the type 3 poliovirus prototype strain Leon; accession number K01392). The dN/dS ratios of E9 and E11 were low (0.015 and 0.023, respectively), similar to that observed in VP1. However, sequences formed discrete clusters which were interspersed with E30 sequences from a previous study (22) and with those of other HEV species B serotypes (http://www.virus-evolution.org/downloads/jvi00783-10/supplementaryfigures.doc, Fig. 1). This is in contrast to the VP1 genome region, which exhibited monophyletic groups of HEV according to serotype. Analysis of the distribution of pairwise distances of the 3Dpol region indicated that the threshold between intra- and interclade distances was 0.09 (boundary zone, 0.07 to 0.12 [http://www.virus-evolution.org/downloads/jvi00783-10/supplementaryfigures.doc, Fig. 2]). Using this cutoff value and by phylogeny comparisons, the combined collection of newly determined and previously published E9 and E11 sequences could be consistently assigned to a total of 23 and 43 clades, respectively.
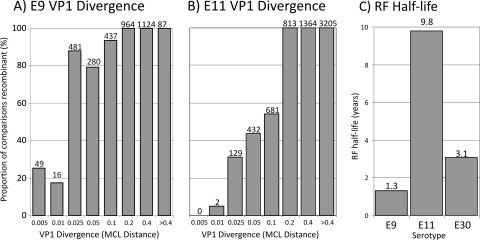
FIG. 2.
(A and B) Association between VP1 sequence divergence (the maximum value is shown for each bar on the x axis) and proportion of recombinant comparisons (i.e., belonging to different 3Dpol clades) for E9 (A) and E11 (B). (C) Comparison of mean RF half-lives of E9, E11, and E30.
Monophyletic clusters in the 3Dpol region consisted of either major RFs (82% and 66% of all E9 and E11 sequences, respectively) or sporadic RFs, defined as those containing two or fewer members. Despite sampling E9, E11, and E30 from the same geographical areas over the same collection period, remarkably few interserotype recombinants were detected; each 3Dpol group was associated with isolates of a single serotype. Of the two exceptions, clade U was initially identified as a single sporadic E30 RF (22) and in seven E11 isolates in the current study. The E30 RF-U sequence was isolated in Bilbao, Spain, in 2002, as were five of the E11 sequences, and shared 99.6 to 99.8% identity (1 or 2 nucleotide changes) with these sequences, suggesting a common and recent parental source for this genome region in both serotypes. The second exception was RF-F, which comprised three E30 sequences isolated from Malaysia in 2004 and one E11 sequence isolated from Vietnam in 2005.
Sequence divergence within RF groups.
To estimate the dates of the recombination events, substitution rates were calculated for the 3Dpol genome regions for several of the larger RF groups, along with their corresponding VP1 genome region. For each recombinant form, substitution rates and estimated times to the most recent common ancestors (TMRCAs) were highly similar in VP1 and 3Dpol genome regions when MCMC methods were used (Table 2). Furthermore, substitution rates for individual RFs in VP1 (such as 6.1 × 10−3 for E9 RF-EP) closely matched the substitution rate for all E9 isolates (5.8 × 10−3). Similarly, the E11 RF-DU RF showed a substitution rate of 9.3 × 10−3, closely matching that of E11 genotype D5 (8.5 × 10−3). Two of the major RF groups of E30 (P and Q) were also analyzed, with data from a previous study (22), using the same 3Dpol region but a different VP1 region from that of the current study (positions 3062 to 3334, with numbering based on the Bastianni prototype strain, accession number AF311938). The substitution rates and TMRCAs of these RF groups were similar between the VP1 and 3Dpol regions and displayed rates of evolution in the VP1 region similar to that of the E30 serotype overall (8.8 × 10−3). Overall, both genome regions analyzed provided consistent estimates for the TMRCAs of each individual RF group for all three enterovirus serotypes analyzed and therefore provide robust estimates for the dates of the recombination events that created each RF.
TABLE 2.
Rates of sequence change by regression and BEAST analysis
| Serotype | Clade | nd | Yr range | Divergence |
dN/dS |
Regression (R) |
MCMC (BEAST)a |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rate of evolution (10−3)b |
TMRCAc |
||||||||||||
| VP1 | 3Dpol | VP1 | 3Dpol | VP1 | 3Dpol | VP1 | 3Dpol | VP1 | 3Dpol | ||||
| E9 | 85 | 14 | 0.136 | 0.274 | 0.013 | 0.015 | 0.389 | 0.077 | 5.8 (3.7-8.1) | ND | 91.3 (61.1-130.8) | ND | |
| E11 | 119 | 14 | 0.259 | 0.254 | 0.064 | 0.023 | 0.089 | 0.138 | 4.8 (3.6-6.1) | ND | 126.2 (81.2-177.4) | ND | |
| E11-D5 | 74 | 12 | 0.049 | 0.139 | 0.069 | 0.022 | 0.895 | 0.293 | 8.5 (6.3-10.9) | ND | 13.4 (11.1-16.6) | ND | |
| E9 | EP | 13 | 8 | 0.042 | 0.044 | 0.030 | 0.035 | 0.933 | 0.502 | 9.1 (4.1-14.6) | 8.4 (3.4-14.2) | 9.3 (8.0-11.7) | 10.0 (8.0-13.8) |
| E11 | DU | 54 | 11 | 0.044 | 0.040 | 0.072 | 0.018 | 0.895 | 0.896 | 9.3 (6.2-12.6) | 8.7 (6.3-11.3) | 12.7 (11.0-14.9) | 12.8 (11.0-15.2) |
| E30 | P | 92 | 6 | 0.024 | 0.022 | 0.047 | 0.036 | 0.534 | 0.564 | 5.9 (3.0-9.1) | 6.6 (3.6-10.4) | 10.0 (6.2-15.6) | 9.1 (6.0-13.9) |
| E30 | Z | 49 | 6 | 0.034 | 0.032 | 0.041 | 0.073 | 0.563 | 0.696 | 8.6 (4.6-12.8) | 7.0 (4.0-10.2) | 10.1 (6.9-15.1) | 8.6 (6.7-11.2) |
The mean value is given, with the HPD interval in parentheses. ND, not determined.
Number of substitutions per site per year.
Within each clade.
Number of sequences in each clade analyzed.
Temporal and geographical aspects of recombination.
Geographical and temporal aspects of E9 and E11 recombination were analyzed as described previously for E30 isolates (22). Sequences assigned to specific RFs according to their clustering patterns in the 3Dpol region were visualized using colored labels to highlight the phylogenetic clustering of members of the major RF groups within the VP1 region (Fig. 1). Individual 3Dpol RF clades predominantly formed monophyletic groups in the VP1 region (Fig. 1). Some exceptions were noted, whereby isolates assembled into VP1 lineages distant from other members of the same 3Dpol group. For example one of the E11 sequences assigned to RF-DO (NL26/ns/DO/06) was found in a VP1 lineage distant from that of the other two members of that RF group.
The relationship between VP1 sequence divergence (as measured by MCL pairwise distances) and the likelihood of recombination was determined. For each pairwise comparison of isolates, whether the two variants belonged to the same or different 3Dpol groups, their divergence in VP1 and their geographical separation and difference in isolation dates were recorded. For both E9 and E11, the proportion of comparisons where isolates had different 3Dpol groups increased with increasing VP1 divergence (Fig. 2A and B). From this, the RF half-lives of E9 and E11 were calculated by determining the mean sequence divergence in VP1 at the 50% recombinant frequency threshold (0.015 and 0.094 for E9 and E11, respectively). Using the rate of evolution determined for VP1 (E9, 5.8 × 10−3; E11, 4.8 × 10−3 [Table 2]), the chronological time to achieve this degree of divergence was calculated. These estimates were compared with an equivalent calculation performed on the previously published E30 data set (22). The RF half-life for E9 was 1.3 years, whereas that of E11 was much greater (9.8 years), with the E30 RF half-life (3.1 years) intermediate between these two values.
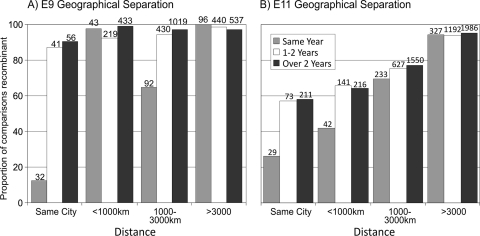
A different recombination dynamic between E9 and E11 was also revealed by analyzing the relationship between the recombination frequency and the geographical distance of separation (Fig. 3). For E11 sequences isolated in the same city, a time relationship with recombination was revealed; only 26% of the sequences isolated within the same year belonged to different 3Dpol groups, whereas >57% of the sequences isolated more than 1 year apart were recombinants. A similar pattern was observed for sequences isolated less than 1,000 km apart but not from the same city: 42% of the sequences isolated in the same year were recombinants, and >64% of the sequences isolated more than 1 year apart were recombinants. There was little difference between the numbers of recombinants for sequences isolated >1,000 km apart, regardless of the time separating the isolation events. A similar analysis of E9 isolates revealed more rapid recombination turnover. Sequences isolated in the same city and in the same year were recombinants in only 12% of pairwise comparisons, but with greater than 1 year separation in times of isolation, this figure increased to over 87%. For sequences that were isolated in the same year and in different cities but less than 1,000 km apart, almost all (98%) fell into different recombination groups.
FIG. 3.
Association between recombination and geographical or temporal separation for E9 (A) and E11 (B). The proportion of recombinant comparisons is shown on the y axis.
Recombinant form succession and capsid evolution.
To more formally reconstruct the succession dynamics of individual RFs and its relationship with diversification in the capsid region, data sets of selected VP1 lineages of E9, E11, and E30 were analyzed using a phylogeographical Bayesian framework implemented in BEAST (17) and adapted to discriminate between RFs. All of the serotypes displayed strikingly different patterns of RF succession and periodicity, which impacted the overall capsid evolution.
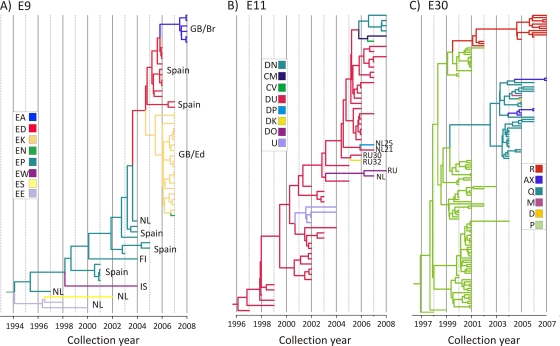
The RF succession and capsid evolutionary dynamics of the RF-P VP1 lineage of E30 were examined. Four major sublineages were identified, each of which contained samples belonging to the RF-P group (Fig. 4C) isolated between 2001 and 2004. Two of these sublineages were wholly comprised of RF-P samples and became extinct by 2004. The other two sublineages contained RF-P sequences isolated between 2000 and 2003 and samples isolated subsequently that belonged to other RF groups (Q, R, AX, D, and M), which extended the life spans of these sublineages to the conclusion of the study period in 2007. The date when recombination events occurred was estimated to lie between the MRCA of the original RF and the new RF and the isolation date of the first clinical sample of the new RF. Thus, it could be estimated that the emergence of RF-R occurred between 1999 and 2002 and that of RF-Q occurred between 1999 and 2003. Several sporadic RFs emerged from the major RF-Q group sharing significant VP1 sequence identity, including a single RF-D sample isolated in the same city on the same day as the closest RF-Q sample (98.5% sequence homology). A general trend was for new RFs to be initially isolated in Spain or France and to spread into northern Europe over the following 1 to 3 years. This was exemplified by RF-R; the earliest samples of this RF were isolated in France and Spain in 2002, one isolate was detected in Croatia in 2005, and by 2006 and 2007, RF-R was detected for the first time in The Netherlands, Great Britain, and Finland. With the exception of RF-AX, which was detected in two relatively divergent VP1 sublineages (93% to 95% sequence similarity), all other RFs were monophyletic and likely originated from a single, datable recombination event.
FIG. 4.
MCMC tree of the main Northern/Western European clades of VP1 sequences of E9 (genotype A), E11 (D5), and E30 visualized in FigTree and plotted on a temporal y axis scale using their sampling dates. The branches are color coded based on the recombination groups of individual sequences and their reconstructed ancestors.
In contrast to E30, a single RF (DU) of E11 (genotype D5) circulated throughout the study period over more than a decade (Fig. 4B). New sublineages of RF-DU and new recombinant groups were highly restricted in distribution and, in contrast to E30, did not replace the parental virus population. For example, between 2007 and 2008, five new RF groups were detected, each remaining a minor and short-lived component of the circulating virus population. Evolution in E9 genogroup A was characterized by large-scale, short-term, geographically limited outbreaks immediately preceded by a recombination event (Fig. 4A). Single outbreaks, with only one exception, were associated with a single RF. Variants within the genogroup A VP1 clade were comprised initially of the RF group EP, detected in three epidemic waves between 1997 and 2005 in The Netherlands and Spain. One direct descendant of this lineage recombined with an unknown donor strain to create the RF group ED, the cause of an outbreak in Spain in 2006 and 2007, which in turn recombined to create the RF group EA seen in the 2008 outbreak in England. The second documented descendant of the original EP group recombined to form the RF group EK, which we detected in an outbreak in Scotland in 2007, and one of these isolates recombined to form a sporadic RF group, EN. The average period of RF quiescence between the MRCA and the first sample isolated was 1 to 2 years, much shorter than that of E30 (3 to 5 years).
DISCUSSION
This study provides a series of novel insights into the global circulation and evolution of E9 and E11. These are two of the most frequently detected HEV serotypes in cases of aseptic meningitis, as well as rarer, more severe diseases. In nontemperate zones, HEV are thought to be endemic, such as E11 in Tunisia (4) and India (12). In contrast, HEV are considered epidemic viruses in temperate zones (14), although the extents and patterns differ between serotypes and countries. In the current study, we found that E9 and E11 exhibited evolutionary dynamics in the VP1 genome region quite distinct from each other and from that of E30, analyzed in a previous study from comparable study populations (22). E9 displayed an epidemic pattern of circulation composed of large-scale, geographically restricted outbreaks occurring over short periods, similar to those demonstrated in previous studies (8, 14). Most sequences isolated from the same region in the same year belonged to the same lineage, as exemplified by an outbreak in Edinburgh, Scotland, over a 6-month period in 2007 in which VP1 nucleotide diversity was only 0.5%. The epidemiology of E11 appears more complex than that of E9. Epidemics caused by E11 have been shown to occur at irregular intervals and to last for a number of years (14), and in the United States, this serotype has been largely quiescent since 1992 (14). In our study, as in others (25), multiple E11 lineages were isolated in the same geographical region at the same time, and concomitantly, a monophyletic genogroup (D5 in the current study) persisted for over a decade over a wide geographical range.
There were marked differences in the patterns of sequence change between VP1 and 3Dpol regions. Bayesian MCMC methods of analysis predicted rates of evolution in the VP1 region for E9 and E11 of 5.8 × 10−3 and 4.8 × 10−3 substitutions per site per year, respectively, similar to those observed previously for E30 (22) and other HEV species (5, 13, 31). However, in the nonstructural region, sequence change was punctuated by frequent recombination events and led to the acquisition of novel 3Dpol sequences interspersed with sequences of other HEV species B isolates. This contrasts dramatically with the monophyletic groupings of serotypes in the VP1 structural genome region.
Analysis of the whole E9 and E11 data sets revealed no discernible relationship between the time of isolation and sequence divergence of E9 and E11 3Dpol sequences (Table 2) (for E30, see reference 22). However, over the much shorter period of sequence diversification within RF groups, there was a strong correlation between the time of isolation and 3Dpol sequence divergence (R values, 0.5 to 0.9) (Table 2). This allowed the rate of evolution in this region to be calculated (which proved to be remarkably similar to those in VP1) and allowed reconstruction of the likely date of the recombination event generating each RF. TMRCAs for individual RFs corresponded well with RF half-lives, for example, E11 RF-DU, with an estimated TMRCA of 12.8 years and an RF half-life for E11 genotype D5 (principally composed of RF-DU isolates) of 5.6 years.
Despite our ability to reconstruct the times when recombination events occurred, the actual serotype identities of recombination partners generating novel RFs in each of the studied echovirus serotypes remain elusive. Collectively E9, E11, and E30 represent approximately 33 to 50% of all species B serotypes isolated by diagnostic laboratories over the past decade in Europe. Given their high prevalence and geographical and temporal cocirculation, it is therefore remarkable how few RFs detected in our current and previous studies could be identified as recombinants between these serotypes. Presumably, recombination has occurred with the larger circulating population of species B serotypes, perhaps with those that do not necessarily present clinically with meningitis or neonatal disease that triggers diagnostic testing and which are therefore not represented in clinical collections of isolates.
Geographical and temporal correlates of recombination.
Classification of E9 and E11 variants into recombinant forms allowed geographical and temporal correlates of recombination to be analyzed. There was a strong direct relationship between recombination and VP1 sequence divergence for both serotypes. From these data and calculated substitution rates, the RF half-lives of 3Dpol groups were determined and indicated that the half-lives of E9 isolates were much shorter than that of E11, with an intermediate life span for E30 3Dpol groups. Previous analyses of parechoviruses have indicated that genotypes 1 and 3 similarly show different half-lives (4.1 and 22.1 years, respectively [6]). The temporal nature of RF turnover in Western Europe was also investigated. The frequencies of shared RF groups were found to be related to geographical distances, with sequences isolated from the same city at the same time predominantly sharing the same RF group. There was no difference between the substitution rates and TMRCAs calculated for the VP1 and 3Dpol regions of E9, E11, and E30 RF groups, suggesting that these regions may emerge simultaneously and that the dynamics of evolution in the structural and nonstructural regions of the genome are interrelated.
To reconstruct more accurately the time scale for the occurrence of specific recombination events in some of the better-sampled evolutionary lineages of E9, E11, and E30, we adapted a Bayesian method for analyzing temporal and geographical correlates of recombination events (17). There was striking evidence that for two of the serotypes examined, E9 and E30, RF succession was specifically linked to the pattern of diversification in the capsid region and to changes in the circulation of each serotype. Most strikingly, combining surveillance data from Spain (33), France (1), Germany (http://www.nlga.niedersachsen.de/master/C40056060_N14377305_L20_D0_I5800417.html), and Finland (3) over the past decade, major peaks of incidence occurred approximately contemporaneously in 2000, 2006, and recently in 2008/2009, coinciding with the emergence of RF-P (which became extinct after 2001) and RF-Q in 2005 and 2006 and the subsequent more rapid emergence of RF-R underlying the outbreaks across Europe in 2007 and 2008. Similarly, outbreaks of E9 between 2001 and 2005 were associated with RF-EP, while its more recent appearance in 2006 originated from descendants of a single VP1 lineage and three rapidly successive RF groups, ED, EK, and EA. In marked contrast, E11, less typically associated with periodic changes in incidence in both the United States and Europe, was comprised predominantly of the EP recombinant, a variant with a reconstructed evolutionary history spanning from 1996 or earlier right through to the end of the study period in 2008. The key question remaining from these reconstructions is whether it is the recombination event that generates a virus whose greater fitness and/or transmissibility drives its emergence and dispersal. Alternatively, might the periodicity in the circulation of E30, E9, and potentially other enterovirus serotypes lead to regular population bottlenecks (such as between 2001 and 2006 for E30) that favor random replacement of virus populations? Resolving this requires detailed phenotypic characterization of RFs that succeed each other, using an infection model capable of detecting subtle differences in transmissibility that may underlie the observed population replacements in nature.
In summary, we have investigated the molecular epidemiology and evolution of clinically presenting isolates of E9 and E11 collected over the past 14 years worldwide. The nonstructural sequences formed distinct clades and, through recombination, created a series of recombinant forms of each serotype interspersed with other species B HEV serotypes. Sequence divergence in the structural region was strongly correlated with recombination frequency and geographical distances. Marked differences in recombination dynamics between serotypes may contribute to the variable patterns of endemicity and cycles of emergence among enteroviruses. This study refines the postulated concept of modular evolution of the structural and nonstructural genes of enteroviruses (20), suggesting that the two genome regions are subject to periods of independent, as well as interdependent, evolution.
Acknowledgments
We acknowledge the collaboration and expertise of another member of the Enterovirus Epidemiology Collaborative Group, Juan Cristina, Centro de Investigaciones Nucleares, Universidad de la República, Montevideo, Uruguay. We thank staff at the Department of Virology, University of Turku, Turku, Finland; Gurutze Rubio, Cruces Hospital, Bilbao, Spain; Manuel Omeñaca, Miguel Servet Hospital, Zaragoza, Spain; Nuria Rabella, Santa Cruz y San Pablo Hospital, Barcelona, Spain; Carmen Perez, Dr. Negrin Hospital, Las Palmas de Gran Canaria, Spain; and T. P. Eremeeva, M. P. Chumakov Institute of Poliomyelitis and Viral Encephalitides, Moscow, Russia.
T. P. Eremeeva was supported in part by the Polio Eradication Initiative through the European Office of the World Health Organization and RFBR grant 08-04-01419-a) for technical assistance with virus isolation. This study was funded by the Wellcome Trust.
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Antona, D., N. Leveque, J. J. Chomel, S. Dubrou, D. Levy-Bruhl, and B. Lina. 2007. Surveillance of enteroviruses in France, 2000-2004. Eur. J. Clin. Microbiol. Infect. Dis. 26:403-412. [DOI] [PubMed] [Google Scholar]
- 2.Ashwell, M. J., D. W. Smith, P. A. Phillips, and I. L. Rouse. 1996. Viral meningitis due to echovirus types 6 and 9: epidemiological data from Western Australia. Epidemiol. Infect. 117:507-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomqvist, S., A. Paananen, C. Savolainen-Kopra, T. Hovi, and M. Roivainen. 2008. Eight years of experience with molecular identification of human enteroviruses. J. Clin. Microbiol. 46:2410-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouslama, L., D. Rezig, A. Ben Yahia, M. Aouni, and H. Triki. 2007. Phylogenetic analysis of echovirus 11 in the 3′ end of the VP1. Intervirology 50:108-114. [DOI] [PubMed] [Google Scholar]
- 5.Brown, B. A., M. S. Oberste, J. P. Alexander, Jr., M. L. Kennett, and M. A. Pallansch. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969-9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvert, J., T. Chieochansin, K. Benschop, E. C. McWilliam-Leitch, J. F. Drexler, K. Grywna, H. da Costa Ribeiro, C. Drosten, H. Harvala, Y. Poovorawan, K. Wolthers, and P. Simmonds. 2010. Recombination dynamics of human parechoviruses: investigation of type-specific differences in frequency and epidemiological correlates. J. Gen. Virol. 91:1229-1238. [DOI] [PubMed] [Google Scholar]
- 7.Chevaliez, S., A. Szendroi, V. Caro, J. Balanant, S. Guillot, G. Berencsi, and F. Delpeyroux. 2004. Molecular comparison of echovirus 11 strains circulating in Europe during an epidemic of multisystem hemorrhagic disease of infants indicates that evolution generally occurs by recombination. Virology 325:56-70. [DOI] [PubMed] [Google Scholar]
- 8.Dalwai, A., S. Ahmad, A. Pacsa, and W. Al Nakib. 2009. Echovirus type 9 is an important cause of viral encephalitis among infants and young children in Kuwait. J. Clin. Virol. 44:48-51. [DOI] [PubMed] [Google Scholar]
- 9.Drummond, A. J., S. Y. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed phylogenetics and dating with confidence. PLoS. Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara, K., T. Kashiwagi, Y. Ohtsu, K. Masunaga, Y. Akasu-Tsuji, N. Tsumura, H. Kato, J. Iwahashi, N. Hamada, M. Toyoda, and T. Toyoda. 2001. Molecular evolution of human echovirus 9 isolated from patients with aseptic meningitis in northern Kyushu during the summer of 1997. Microbiol. Immunol. 45:717-720. [DOI] [PubMed] [Google Scholar]
- 12.Kapoor, A., D. D. Patel, S. Kumar, A. Ayyagari, and T. N. Dhole. 2004. Molecular characterization of echovirus 11 isolates from India. Indian J. Med. Res. 119:149-156. [PubMed] [Google Scholar]
- 13.Kew, O., M. N. Mulders, G. Y. Lipskaya, E. E. da Silva, and M. A. Pallansch. 1995. Molecular epidemiology of polioviruses. Semin. Virol. 6:401-414. [Google Scholar]
- 14.Khetsuriani, N., A. Lamonte-Fowlkes, S. Oberst, and M. A. Pallansch. 2006. Enterovirus surveillance—United States, 1970-2005. MMWR Surveill. Summ. 55:1-20. [PubMed] [Google Scholar]
- 15.Lashkevich, V. A., G. A. Koroleva, A. N. Lukashev, E. V. Denisova, and L. A. Katargina. 2004. Enterovirus uveitis. Rev. Med. Virol. 14:241-254. [DOI] [PubMed] [Google Scholar]
- 16.Leitch, E. C., H. Harvala, I. Robertson, I. Ubillos, K. Templeton, and P. Simmonds. 2009. Direct identification of human enterovirus serotypes in cerebrospinal fluid by amplification and sequencing of the VP1 region. J. Clin. Virol. 44:119-124. [DOI] [PubMed] [Google Scholar]
- 17.Lemey, P., A. Rambaut, A. J. Drummond, and M. A. Suchard. 2009. Bayesian phylogeography finds its roots. PLoS. Comput. Biol. 5:e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindberg, A. M., P. Andersson, C. Savolainen, M. N. Mulders, and T. Hovi. 2003. Evolution of the genome of human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 84:1223-1235. [DOI] [PubMed] [Google Scholar]
- 19.Lukashev, A. N., V. A. Lashkevich, O. E. Ivanova, G. A. Koroleva, A. E. Hinkkanen, and J. Ilonen. 2003. Recombination in circulating enteroviruses. J. Virol. 77:10423-10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukashev, A. N., V. A. Lashkevich, O. E. Ivanova, G. A. Koroleva, A. E. Hinkkanen, and J. Ilonen. 2005. Recombination in circulating human enterovirus B: independent evolution of structural and non-structural genome regions. J. Gen. Virol. 86:3281-3290. [DOI] [PubMed] [Google Scholar]
- 21.Lukashev, A. N., V. A. Lashkevich, G. A. Koroleva, J. Ilonen, G. G. Karganova, V. I. Reznik, and A. E. Hinkkanen. 2003. Molecular epidemiology of enteroviruses causing uveitis and multisystem hemorrhagic disease of infants. Virology 307:45-53. [DOI] [PubMed] [Google Scholar]
- 22.McWilliam Leitch, E. C., J. Bendig, M. Cabrerizo, J. Cardosa, T. Hyypia, O. E. Ivanova, A. Kelly, A. C. Kroes, A. Lukashev, A. Macadam, P. McMinn, M. Roivainen, G. Trallero, D. J. Evans, and P. Simmonds. 2009. Transmission networks and population turnover of echovirus 30. J. Virol. 83:2109-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modlin, J. F. 1996. Update on enterovirus infections in infants and children. Adv. Pediatr. Infect. Dis. 12:155-180. [PubMed] [Google Scholar]
- 24.Nelsen-Salz, B., O. Schildgen, M. Klein, D. Hadaschik, H. J. Eggers, and H. Zimmermann. 1999. Determinants of pathogenicity of echovirus 9 in man: significance of a functional RGD-motif. Zentrabl. Bakteriol. 289:347-354. [DOI] [PubMed] [Google Scholar]
- 25.Oberste, M. S., W. A. Nix, D. R. Kilpatrick, M. R. Flemister, and M. A. Pallansch. 2003. Molecular epidemiology and type-specific detection of echovirus 11 isolates from the Americas, Europe, Africa, Australia, southern Asia and the Middle East. Virus Res. 91:241-248. [DOI] [PubMed] [Google Scholar]
- 26.Oprisan, G., M. Combiescu, S. Guillot, V. Caro, A. Combiescu, F. Delpeyroux, and R. Crainic. 2002. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 83:2193-2200. [DOI] [PubMed] [Google Scholar]
- 27.Santti, J., T. Hyypia, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro, B., A. Rambaut, and A. J. Drummond. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23:7-9. [DOI] [PubMed] [Google Scholar]
- 29.Simmonds, P., and D. B. Smith. 1999. Structural constraints on RNA virus evolution. J. Virol. 73:5787-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simmonds, P., and J. Welch. 2006. Frequency and dynamics of recombination within different species of human enteroviruses. J. Virol. 80:483-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeda, N., M. Tanimura, and K. Miyamura. 1994. Molecular evolution of the major capsid protein VP1 of enterovirus 70. J. Virol. 68:854-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 33.Trallero, G., A. Avellon, A. Otero, T. de Miguel, C. Perez, N. Rabella, G. Rubio, J. E. Echevarria, and M. Cabrerizo. 2010. Enteroviruses in Spain over the decade 1998-2007: virological and epidemiological studies. J. Clin. Virol. 47:170-176. [DOI] [PubMed] [Google Scholar]