Abstract
Robinsoniella peoriensis was recently identified as a Gram-positive, spore-forming, anaerobic rod originally isolated from swine manure storage pits. We describe here a case of R. peoriensis bacteremia in a 42-year-old male with pancreatic cancer. The identification was confirmed by unique phenotypic profiles and partial 16S rRNA gene sequences.
CASE REPORT
A 42-year-old male legal clerk presented with jaundice for 2 months. The patient had a 6-year history of chronic viral hepatitis B virus infection with secondary cirrhosis. He had been diagnosed with pancreatic cancer and had undergone surgery 10 months prior to admission. The pancreatic cancer had been diagnosed as adenocarcinoma with medium-to-low differentiation at the pancreatic tail and a tumor size of 2.5 by 2 by 3 cm as determined by biopsy and pathologic examination. The spleen and greater omentum were not involved. Immunohistochemistry staining revealed the following: p170, +; CK7, +++; CK19, +++; Ki-67, + (>75%); vascular endothelial growth factor, +++; p53, −; HER-1, −; HER-2, −. He lived in an urban area of Beijing and denied having traveled or had any animal contact in the past year. Chemotherapy had been started immediately after surgery with gemcitabine and oxaliplatin, but his jaundice continued to progress; therefore, he was admitted to the Chinese PLA General Hospital.
On admission, his physical examination revealed a temperature of 36.8°C (axilla), a pulse of 78 beats/min, a respiration rate of 18 breaths/min, and a blood pressure of 120/75 mmHg. His head, eyes, ears, nose, and throat appeared normal, and his lungs were clear. Several oral ulcers were noticed, and his skin was yellowish, but no lesions were noted. Laboratory values on admission revealed the following: blood hemoglobin level, 104 g/liter; platelet count, 94 × 109/liter; leukocyte count, 7.97 × 109/liter with 71.5% neutrophils. The total bilirubin level was 107.5 μmol/liter, the direct bilirubin level was 83.6 μmol/liter, the total bile acid level was 373.9 μmol/liter, the alanine aminotransferase level was 44.2 U/liter, and the aspartate aminotransferase level was 107.7 U/liter. He was positive for hepatitis B surface antigen, and his D-dimer level was 6.11 μg/ml. Urine and stool analysis results were normal. Several tumor markers, including the carcinoembryonic antigen and cancer antigens CA-125, CA19-9, and CA724, were higher than normal. Computed tomography suggested bone and peritoneal metastases.
On the 5th day after admission, the patient developed a high fever with a temperature of 39.5°C, and one peripheral blood culture was collected by venipuncture. After a 20-h incubation, a Gram-positive, rod-shaped bacterium without obvious spore formation (Fig. 1 A) grew in an anaerobic bottle in the BacT/Alert blood culture system (bioMérieux, Inc., Durham, NC). The organism did not grow in the aerobic bottle. The patient was treated with metronidazole intravenously (0.5 g once a day for 12 days). His temperature returned to normal 5 days after the antibiotic therapy began and stayed between 36.8°C and 37.4°C. However, his cancer status and liver function deteriorated subsequently, and the patient died of multiple organ failure 30 days after hospitalization. No second blood culture was obtained.
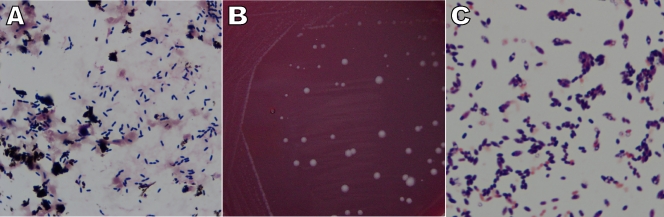
FIG. 1.
Colony and microscopic morphology of the new isolate. Gram staining of the positive anaerobic blood culture bottle showed Gram-positive, rod-shaped bacteria without obvious spore formation (A). Dull white, flat colonies with mixed sizes and smooth edges (B) and Gram-positive, ovoid-shaped spores located subterminally (C) were observed 2 days after subculture on an anaerobic blood plate.
The isolate (0910-06083) was subcultured on a blood plate anaerobically and on a sheep blood agar plate aerobically. Two days later, gray-white, smooth, nonhemolytic colonies with unequal sizes were observed only on the anaerobic blood plate (Fig. 1B). Gram staining of a purified colony revealed a Gram-positive, rod-shaped bacterium with ovoid-shaped spores located subterminally (Fig. 1C). The organism was found to be catalase positive, urease negative, and gelatin positive, with a capability of utilizing glucose, lactose, sucrose, maltose, xylose, mannose, arabinose, raffinose, trehalose, cellobiose, glycerol, and salicin but not sorbitol or mannitol. Identification reactions showed 88.4% and 96% identity with Clostridium clostridioforme by API 20 A and API Rapid ID 32 A (bioMérieux, Inc.), respectively. The key biochemical reactions are listed in Table 1. Susceptibility testing was performed using the Etest (AB Biodisk-V, Solna, Sweden) and was interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (2) and the Etest application sheet. The organism was susceptible to metronidazole, cefoxitin, imipenem, amoxicillin-clavulanic acid, and piperacillin-tazobactam with MICs of 0.064 μg/ml, 8 μg/ml, 1 μg/ml, 0.5 μg/ml, and 6 μg/ml, respectively, and was intermediately resistant to clindamycin with an MIC of 4 μg/ml.
TABLE 1.
Contrast of phenotypic characteristics of the new isolate (0910-06083) and R. peoriensisa
| Phenotypic profile | 0910-06083 | R. peoriensisb |
|---|---|---|
| Growth at: | ||
| 44°C | + | ND |
| 37°C | + | + |
| Hemolysis of sheep blood | − | − |
| Motility | − | − |
| Indole | − | − |
| Urease | − | − |
| Nitrate | − | − |
| Catalase | + | + |
| N-Acetyl-β-glucosaminidase | + | + |
| α-Arabinosidase | + | + |
| α-Galactosidase | + | + |
| β-Galactosidase | + | + |
| α-Glucosidase | + | + |
| β-Glucosidase | + | + |
| β-Glucuronidase | + | + |
| α-Fucosidase | + | + |
| Acid phosphatase | ND | + |
| Esterase | ND | +, weak |
| Esterase lipase | ND | +, weak |
| Utilization of: | ||
| Arabinose | + | + |
| Cellobiose | + | + |
| Fructose | + | + |
| Glucose | + | + |
| Lactose | + | + |
| Sucrose | + | ND |
| Maltose | + | + |
| Raffinose | + | + |
| Salicin | + | ND |
| Trehalose | + | + |
| Xylan | + | + |
| Xylose | + | + |
| Gelatin | + | ND |
| Esculin | + | ND |
| Glycerol | + | ND |
| Mannose | + | ND |
| Sorbitol | − | ND |
| Mannitol | − | ND |
+, positive; −, negative; ND, not done.
Biochemical reactions were recorded from reference 3.
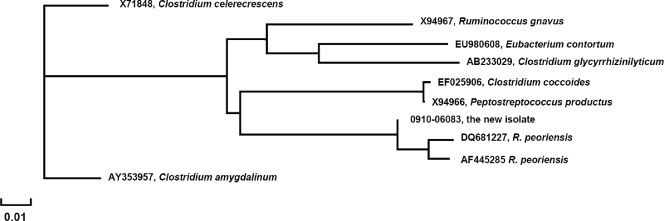
A loopful of the purified isolate was put into 1 ml of distilled water and heated at 95°C for 10 min, the suspension was centrifuged, and 1 μl of supernatant was used for PCR amplification. The first 500 bp of the 16S rRNA gene was amplified by using a primer set (5FPL and 523RPL) spanning the regions of the 16S rRNA gene corresponding to nucleotide positions 5 to 523 of Escherichia coli. The PCR amplification products were bidirectionally sequenced with the same two primers as previously described (6, 7). Neighbor-joining phylogenetic analysis was performed by online analysis at the NCBI Entrez nucleotide database site (http://www.ncbi.nlm.nih.gov/Database/index.html) (Fig. 2). The nucleic acid sequences determined from the isolate were most closely related to the recently identified organism Robinsoniella peoriensis (3), diverging from the published sequences (AF445285, DQ681227) at one or two nucleotide positions (99.02 to 99.22% similarity).
FIG. 2.
Phylogenetic analysis of partial 16S rRNA gene sequences of the isolate and related organisms. X71848, X94967, EU980608, AB233029, EF025906, X94966, 0910-06083, DQ681227, AF445285, and AY353957 are Clostridium celerecrescens, R. gnavus, Eubacterium contortum, Clostridium glycyrrhizinilyticum, C. coccoides, Peptostreptococcus productus, the new isolate, R. peoriensis, R. peoriensis, and Clostridium amygdalinum, respectively. The scale is provided as a measurement of relative phylogenetic distance.
Discussion.
R. peoriensis, which was initially classified in clostridial rRNA cluster XIVa, was recently classified as a novel species named in honor of Isadore M. Robinson (3). Most R. peoriensis strains have been isolated from environmental sources, and the only previously reported human isolate was recovered from a deep wound on the heel of a 79-year-old female according to the description from the Culture Collection of the University of Göteborg, Göteborg, Sweden (3, 4). In this study, we are reporting the isolation of R. peoriensis from a blood culture from a patient with pancreatic cancer. R. peoriensis was the sole organism recovered from this patient during his hospitalization period. The patient responded to intravenous metronidazole therapy, which was consistent with the susceptibility pattern of the R. peoriensis isolate identified in this study. Since the isolate was recovered from the anaerobic bottle of a single two-bottle set, the causal relationship between the isolate and the patient's bacteremia merits further investigation.
In a clinical setting, this microorganism should be distinguished from closely related ovoid spore-forming organisms such as Clostridium coccoides and Ruminococcus gnavus (1, 5). Current phenotypic identification systems, including API 20 A and API Rapid ID 32 A, are unable to identify R. peoriensis. Several unique phenotypic reactions can be used to identify the organism (Table 1) and should be included in the API and other commercial systems to enhance identification accuracy. A genotypic identification method based on 16S rRNA gene sequencing provides rapid and accurate identification. This isolate is susceptible to metronidazole by both the Etest result and the therapeutic response obtained. The Etest seemed to provide reliable antimicrobial susceptibility testing results for this isolate.
To our knowledge, our isolate is the first one recovered from a human with bacteremia. R. peoriensis usually colonizes pig feces and produces a variety of odorous chemicals, which cause contamination of local water supplies and thus result in health problems for both animals and humans (3, 4). In this case, our patient worked in an attorney's office in Beijing without an obvious history of exposure to pigs or pig manure. However, chemotherapy for 10 months undoubtedly compromised his immune function. He had no obvious skin lesions, but an oral ulcer might have been the site of entry for the organism. At this time, no information about risk factors, infection sites, or organ or tissue tropism or other information about pathogenesis is available for this organism in relation to human infections (3).
Nucleotide sequence accession number.
The GenBank accession number for the partial 16S rRNA gene sequences of the R. peoriensis isolate identified in the present study is GU304598.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Chioralia, G., T. Trammer, H. Kampen, and H. M. Seitz. 1998. Relevant criteria for detecting microsporidia in stool specimens. J. Clin. Microbiol. 36:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing of anaerobic bacteria; informational supplement, M11-S1. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Cotta, M. A., T. R. Whitehead, E. Falsen, E. Moore, and P. A. Lawson. 2009. Robinsoniella peoriensis gen. nov., sp. nov., isolated from a swine-manure storage pit and a human clinical source. Int. J. Syst. Evol. Microbiol. 59:150-155. [DOI] [PubMed] [Google Scholar]
- 4.Cotta, M. A., T. R. Whitehead, and R. L. Zeltwanger. 2003. Isolation, characterization and comparison of bacteria from swine faeces and manure storage pits. Environ. Microbiol. 5:737-745. [DOI] [PubMed] [Google Scholar]
- 5.Sakuma, K., M. Kitahara, R. Kibe, M. Sakamoto, and Y. Benno. 2006. Clostridium glycyrrhizinilyticum sp. nov., a glycyrrhizin-hydrolysing bacterium isolated from human faeces. Microbiol. Immunol. 50:481-485. [DOI] [PubMed] [Google Scholar]
- 6.Tang, Y. W., N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 36:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang, Y. W., A. Von Graevenitz, M. G. Waddington, M. K. Hopkins, D. H. Smith, H. Li, C. P. Kolbert, S. O. Montgomery, and D. H. Persing. 2000. Identification of coryneform bacterial isolates by ribosomal DNA sequence analysis. J. Clin. Microbiol. 38:1676-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]