Abstract
Recently, we found that several commercial pet vaccines were contaminated with an infectious endogenous retrovirus, RD-114-related virus. Here, we determined the entire nucleotide sequences of RD-114-related viruses isolated from CRFK cells and a vaccine manufactured using CRFK cells. These RD-114-related viruses were nearly identical to the authentic RD-114 virus.
RD-114 virus is a retrovirus isolated in 1971 from a human tumor cell line (RD-114 cells) derived from a human rhabdomyosarcoma after passage through fetal cats (9). Subsequent studies revealed that the virus was an endogenous retrovirus of feline origin but not a human exogenous retrovirus (6, 8, 13). All domestic cats have the RD-114 viral genome, and several feline cell lines produce RD-114-related viruses (2, 6, 13). In particular, Baumann et al. reported that a widely used feline cell line, Crandell-Rees feline kidney (CRFK) cells (3), produced a retrovirus which interfered with the authentic RD-114 virus and was antigenically related to the virus (2). At the time of that research, there was no information about the nucleotide sequence of the original RD-114 virus; therefore, the RD-114-related virus derived from CRFK cells was designated CrLE virus. No further studies related to the CrLE virus have been reported since then.
Several live attenuated vaccines for dogs and cats are produced by using feline cell lines, including CRFK cells. Recently, we found that some live vaccines for dogs and cats contained infectious RD-114-related viruses (10, 11). Moreover, the Japanese regulatory authority (National Veterinary Assay Laboratory, Kokubunji, Tokyo, Japan) confirmed that certain feline live vaccines contained infectious RD-114-related viruses (12); however, there has been no information about the entire nucleotide sequences of the RD-114-related viruses produced from CRFK cells and contaminated vaccines. In this study, we constructed an infectious molecular clone of the RD-114-related virus produced from CRFK cells and compared it with the original RD-114 virus. Furthermore, we characterized an RD-114-related virus contained in a vaccine manufactured using CRFK cells.
CRFK cells (ATCC CCL-94) (3), TE671 cells (a human rhabdomyosarcoma cell line; ATCC CRL-8805), and TE671 cells transduced with the nlsLacZ gene [TE671(LacZ) cells] (15) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, penicillin (100 units/ml), and streptomycin (100 μg/ml).
An infectious molecular clone of the original RD-114 virus, termed pSc3c, was kindly provided by Roger Reeves (Johns Hopkins University, Baltimore, MD) through Massimo Palmarini (Glasgow University, Glasgow, United Kingdom). To amplify the 5′ and 3′ halves of RD-114-related viruses, we designed primers based on the nucleotide sequence of pSc3c deposited in GenBank (accession number NC009889) (Fig. 1 A). The PCR primers used to amplify the 5′ and 3′ halves of the virus were 5′-TGAGAAGTCAGAACCCCCCACC-3′ (forward) and 5′-GCACTATGGCCTCTAAAGCATGCGG-3′ (reverse) and 5′-CCGCATGCTTTAGAGGCCATAGTGC-3′ (forward) and 5′-TGTTAGGAGCCAAACTCCTAGGCC-3′ (reverse), respectively.
FIG. 1.
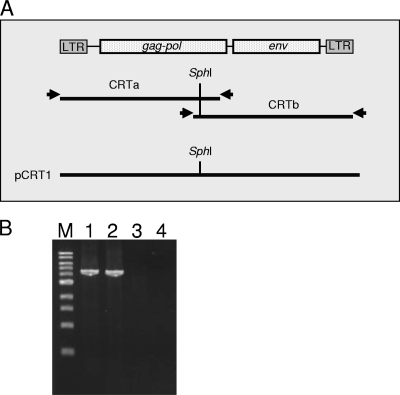
Cloning of RD-114-related virus derived from CRFK cells. (A) Primer positions for long-range PCR and cloning strategy of RD-114-related viruses. (B) Long-range PCR used to clone the 5′ and 3′ halves of an RD-114-related virus derived from CRFK cells. Long-range PCR using genomic DNA isolated from TE671/CRsup (lanes 1 and 2) and TE671 cells (lanes 3 and 4) was conducted to amplify the 5′ (lanes 1 and 3) and 3′ (lanes 2 and 4) halves of the RD-114-related virus. Lane M, 1-kb-ladder molecular weight marker.
We performed long-range PCR using genomic DNA isolated from TE671 cells inoculated with culture supernatant of CRFK (designated TE671/CRsup cells). The PCR conditions were as follows. The reaction mixture (total, 25 μl) consisted of 1 μl DNA template (100 ng), 1 μl PrimeSTAR GXL polymerase (TaKaRa, Ohtsu, Shiga, Japan), 5 μl of 5× buffer containing 5 mM MgCl2 (PrimeSTAR GXL buffer), 2 μl of 2.5 mM deoxynucleotide triphosphates, 0.5 μl of each primer (100 pmol/μl), and 16.5 μl distilled water. The amplification conditions were 98°C for 3 min, followed by 30 cycles of amplification, consisting of denaturation at 98°C for 10 s, annealing at 64°C for 15 s, and extension at 68°C for 40 s, and then a final extension at 68°C for 5 min. PCR was carried out in 200 μl thin-walled tubes using a C1000 thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA). By electrophoresis of the amplified products in a 1% agarose gel, we detected positive bands with estimated sizes (Fig. 1B). The amplicons were designated fragments CRTa and CRTb for the 5′ and 3′ halves of the viral genome, respectively (Fig. 1A).
To reconstitute fragments CRTa and CRTb as a proviral form, we used the SphI restriction enzyme digestion site present in the middle of the RD-114 genome (Fig. 1A). Fragment CRTb was digested with SphI and cloned into the SphI/EcoRV sites of the pSP73 vector (Promega, Madison, WI) to produce pSP73/CRTb. Fragment CRTa was also digested with SphI and inserted into the SphI/PvuI sites of pSP73/CRTb. We designated the reconstituted plasmid pCRT1 (Fig. 1A).
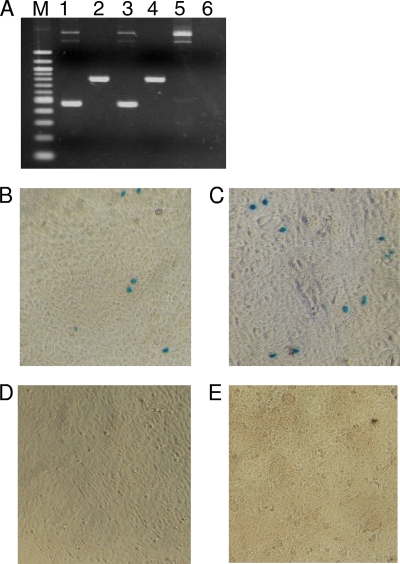
To determine whether the reconstituted plasmid is an infectious molecular clone of RD-114-related virus, either pCRT1 or pSc3c was transfected into TE671 cells. Eight days after transfection, culture supernatants were collected and then inoculated into TE671(LacZ) cells. As a positive control, the culture supernatant of CRFK cells was inoculated into TE671(LacZ) cells. These inocula were treated with DNase I prior to inoculation, and we confirmed that no proviral DNA was contaminated in the inocula by PCR. Nine days later, we isolated genomic DNA from the inoculated TE671(LacZ) cells, and then PCR was performed using specific primers for the partial env and pol genes of RD-114 virus. The primers for the partial pol and env genes, designed based on the sequence of the RD-114 virus, were 5′-GAGACCCTTACTAAATTGAC-3′ (forward) and 5′-AGTTTCTGGTCCAGGGGTTT-3′ (reverse) and 5′-CCCTCGATACTAAGAGAGTG-3′ (forward) and 5′-ACTTCAGCTAACGAGTCTAC-3′ (reverse), respectively. The PCR was carried out using Ex Taq polymerase (TaKaRa) instead of PrimeSTAR GXL polymerase. The PCR conditions were 94°C for 10 min, followed by 30 cycles of amplification, consisting of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 90 s, and then a final extension at 72°C for 10 min. Consequently, we detected specific bands in TE671(LacZ) cells inoculated with viruses prepared from both pCRT1 and pSc3c (Fig. 2 A). The infectivity of the viruses was also confirmed by the LacZ marker rescue assay, conducted as described previously (11, 15) (Fig. 2B). The virus derived from pCRT1 interfered well with RD-114 virus derived from clone pSc3c (Fig. 2D). From these data, we concluded that pCRT1 is an infectious molecular clone of the RD-114-related virus.
FIG. 2.
Infectivity of the virus derived from pCRT1. (A) PCR was conducted to amplify the partial pol (lanes 1, 3, and 5) and env (lanes 2, 4, and 6) genes of RD-114 virus from genomic DNA isolated from TE671(LacZ) cells inoculated with viruses derived from pCRT1 (lanes 1 and 2) and pSc3c (lanes 3 and 4). PCR was also conducted using genomic DNA isolated from mock-infected TE(LacZ) cells (lanes 5 and 6) as a negative control. Lane M, 100-bp-ladder molecular weight marker. (B to D) LacZ marker rescue assay. TE671(LacZ) cells were inoculated with viruses derived from pCRT1 and pSc3c. Nine days after inoculation, 10-fold serially diluted culture supernatants collected from infected cells were inoculated into naïve TE671 cells. Two days after inoculation, the cells were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). TE671 cells inoculated with 10-fold-diluted culture supernatants derived from TE671(LacZ) cells infected with viruses derived from pCRT1 (B) and pSc3c (C) are shown. (D) TE671 cells inoculated with the culture supernatant of mock-infected TE671(LacZ) cells are shown as a negative control. (E) Interference assay using TE671 cells chronically infected with RD-114 virus derived from pSc3c (TE/RD cells). Culture supernatant from TE671(LacZ) cells inoculated with RD-114-related virus derived from pCRT1 was inoculated into TE/RD cells. Two days after inoculation, the cells were stained with X-Gal.
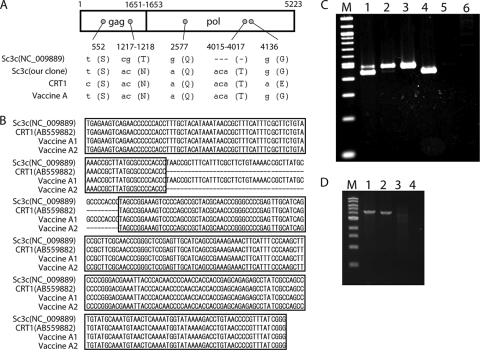
Then, we determined the entire nucleotide sequence of pCRT1 (GenBank accession number AB559882) and compared it with that of pSc3c deposited in GenBank (accession number NC_009889). In pCRT1, we found 3 and 2 nucleotide substitutions in the gag and pol genes, respectively. When the first nucleotide for the initiation codon of the Gag-Pol precursor polyprotein was defined as nucleotide position +1, nucleotide substitutions were found at positions 552, 1217 to 1218, 2577, and 4136 (Fig. 3 A). There was no nucleotide substitution in the env gene. The nucleotide substitutions at positions 1217 to 1218 and 4136 were nonsynonymous, and the others were synonymous. We also found a successive 3-base-pair (bp) deletion (1-amino-acid deletion) in pSc3c in the pol gene at nucleotide positions 4015 to 4017 (Fig. 3A). When we searched for the pol gene sequence in the cat genome database (http://www.ncbi.nlm.nih.gov/projects/genome/guide/cat/), we could not find this 3-bp deletion. Therefore, this deletion seems to be unique to pSc3c, as noted in a previous study (7); however, by our own sequencing analysis of the pSc3c plasmid obtained, we could not detect the 3-bp deletion and the nucleotides at positions 1217 to 1218 and 2577 were identical to those of pCRT1 (Fig. 3A). Besides the coding regions, pCRT1 has a successive 47-bp deletion in the U3 region of the long terminal repeat (LTR) in comparison with the sequence of pSc3c (7, 16) (Fig. 3B). This 47-bp sequence is a direct repeat present in the LTR of exogenous RD-114 virus, which was reported previously (16). When we searched for the LTR sequence in the cat genome database, only the deleted type of LTR was found in the cat genome. To determine whether the inserted type of LTR is present in the virus derived from CRFK cells, we performed PCR to amplify the entire LTR of the virus by use of genomic DNA from TE671/CRsup cells. The primers designed based on the sequence of the RD-114 virus were 5′-TGAGAAGTCAGAACCCCCCACC-3′ (forward) and 5′-TGTTAGGAGCCAAACTCCTAGGCC-3′ (reverse). PCR was carried out using PrimeSTAR GXL polymerase (TaKaRa). The amplification conditions were 98°C for 3 min, followed by 30 cycles of amplification, consisting of denaturation at 98°C for 10 s, annealing at 62°C for 15 s, and extension at 68°C for 30 s, and then a final extension at 68°C for 5 min. As a result, we detected two sizes of LTR amplicons (Fig. 3C, lane 1). The sequence of the larger band was identical to that of pSc3c, suggesting that the virus derived from CRFK cells has two types of LTR. Although we cloned the 5′ and 3′ halves of the RD-114 clone independently by PCR, the sequences of the 5′ and 3′ LTRs of pCRT1 were identical to each other. These data indicate that pCRT1 should be derived from the same proviral integrant in CRFK cells.
FIG. 3.
Comparison of RD-114 virus and RD-114-related viruses. (A) Comparison of nucleotide sequences of the partial gag-pol gene of RD-114 virus (clone pSc3c) and RD-114-related viruses derived from CRFK cells and vaccine A. The first nucleotide for the initiation codon of the Gag-Pol precursor polyprotein of pCRT1 is defined as nucleotide position +1. Small letters and dashes indicate nucleotides and deletions, respectively. Capital letters in parentheses indicate amino acids. (B) Comparison of nucleotides of the U3 region of the LTR of RD-114 virus (clone pSc3c) and RD-114-related viruses derived from CRFK cells and vaccine A. (C) Comparison of the sizes of the entire LTRs of RD-114 virus and RD-114-related viruses. PCR for amplification of the entire LTR was conducted using genomic DNA of TE671/CRsup (lane 1) and TE671(LacZ)/VA (lane 2) cells and mock-infected TE671 (lane 5) and TE671(LacZ) (lane 6) cells. As positive controls, pSc3c (lane 3) and pCRT1 (lane 4) were used. Lane M, 100-bp-ladder molecular weight marker. (D) Long-range PCR used to clone the 5′ and 3′ halves of RD-114-related virus derived from vaccine A. The 5′ (lanes 1 and 3) and 3′ (lanes 2 and 4) halves of the RD-114-related virus were amplified by PCR from TE671(LacZ)/VA cells (lanes 1 and 2) and TE671(LacZ) cells (lanes 3 and 4). Lane M, 1-kb-ladder molecular weight marker.
Next, we attempted to characterize the RD-114-related virus from a live trivalent vaccine (here called vaccine A), which was manufactured using CRFK cells and is commercially available in Japan (10). TE671(LacZ) cells were inoculated with 1 ml vaccine A, and then the inoculated cells [designated TE671(LacZ)/VA cells] were cultured for 3 weeks.
The 5′ and 3′ halves of RD-114-related virus were amplified from genomic DNA of TE671(LacZ)/VA cells by long-range PCR, as described above (Fig. 3D). Both amplicons were cloned, and three independent clones from each half were sequenced. The consensus sequence of the clones (sequences matched in two or three clones out of three sequenced) in the gag, pol, and env genes was compared with that of pCRT1. We found one nucleotide substitution each in the gag and pol genes at nucleotide positions 552 and 4136 (Fig. 3A). There was no nucleotide substitution in the env gene. The nucleotide substitution at position 4136 was nonsynonymous. The 3-bp deletion in the pol gene, which is specific to the already deposited sequence of pSc3c (GenBank accession number NC_009889), was not present in the clone. As with the RD-114-related virus from CRFK cells, we found two types of LTR in the clones (Fig. 3B). We confirmed the types of LTR by PCR, amplifying the entire LTR from genomic DNA of TE671(LacZ)/VA cells (Fig. 3C, lane 2).
In this study, we characterized RD-114-related viruses derived from CRFK cells and a commercial vaccine. RD-114 virus was first isolated from “human” RD-114 cells, and RD-114-related viruses from “feline” cells were distinguished from the original RD-114 virus (2, 6, 8). In this study, we determined the entire nucleotide sequence of an infectious RD-114-related virus derived from CRFK cells for the first time. Viruses from both CRFK cells and vaccine A were nearly identical to the authentic RD-114 virus, clone pSc3c, which was cloned in 1984 (14); therefore, we refer to the virus derived from CRFK cells as RD-114 virus, not RD-114-related virus or CrLE virus, hereafter. The reason for the presence of two types of LTR in RD-114 virus is unknown at present; however, we considered that the 47-bp insertion (a direct repeat) may have been introduced during multiplication of the virus in CRFK or TE671 cells, because we could not find the sequence of the inserted type of LTR in the cat genome. Duplications in the LTR are often associated with increased replication, as described for porcine endogenous retroviruses (4) as well as feline and murine leukemia viruses (1, 5, 17). The 47-bp repeat seemed not to confer prominent positive effects in viral growth in TE671 cells, because the virus derived from pCRT1 lacking the repeat grew as efficiently as the virus derived from pSc3c (Fig. 2B and C), although we have not compared the viral growths of the clones in CRFK cells. Further studies are needed to elucidate the significance of the 47-bp repeat in viral growth in various cell lines.
Acknowledgments
We are grateful to Roger Reeves (Johns Hopkins University) and Massimo Palmarini (Glasgow University) for providing pSc3c.
This work was supported by grants from the Japan Society for the Promotion of Science and from the Bio-Oriented Technology Research Advancement Institution in Japan.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Athas, G. B., P. Lobelle-Rich, and L. S. Levy. 1995. Function of a unique sequence motif in the long terminal repeat of feline leukemia virus isolated from an unusual set of naturally occurring tumors. J. Virol. 69:3324-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, J. G., W. H. Gunzburg, and B. Salmons. 1998. CrFK feline kidney cells produce an RD114-like endogenous virus that can package murine leukemia virus-based vectors. J. Virol. 72:7685-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crandell, R. A., C. G. Fabricant, and W. A. Nelson-Rees. 1973. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK). In Vitro 9:176-185. [DOI] [PubMed] [Google Scholar]
- 4.Denner, J., V. Specke, U. Thiesen, A. Karlas, and R. Kurth. 2003. Genetic alterations of the long terminal repeat of an ecotropic porcine endogenous retrovirus during passage in human cells. Virology 314:125-133. [DOI] [PubMed] [Google Scholar]
- 5.DesGroseillers, L., and P. Jolicoeur. 1984. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J. Virol. 52:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischinger, P. J., P. T. Peebles, S. Nomura, and D. K. Haapala. 1973. Isolation of an RD-114-like oncornavirus from a cat cell line. J. Virol. 11:978-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghani, K., and S. Cottin. 2009. Characterization of an alternative packaging system derived from the cat RD114 retrovirus for gene delivery. J. Gene Med. 11:664-669. [DOI] [PubMed] [Google Scholar]
- 8.Livingston, D. M., and G. J. Todaro. 1973. Endogenous type C virus from a cat cell clone with properties distinct from previously described feline type C virus. Virology 53:142-151. [DOI] [PubMed] [Google Scholar]
- 9.McAllister, R. M., M. Nicolson, M. B. Gardner, R. W. Rongey, S. Rasheed, P. S. Sarma, R. J. Huebner, M. Hatanaka, S. Oroszlan, R. V. Gilden, A. Kabigting, and L. Vernon. 1972. C-type virus released from cultured human rhabdomyosarcoma cells. Nat. New Biol. 235:3-6. [DOI] [PubMed] [Google Scholar]
- 10.Miyazawa, T., R. Yoshikawa, M. Golder, M. Okada, H. Stewart, and M. Palmarini. 2010. Isolation of an infectious endogenous retrovirus in a proportion of live attenuated vaccines for pets. J. Virol. 84:3690-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazawa, T. 2010. Endogenous retroviruses as potential hazards for vaccines. Biologicals 38:371-376. [DOI] [PubMed] [Google Scholar]
- 12.Narushima, R., T. Usui, T. Ogawa, and T. Shimazaki. Detection of infectious RD114 virus in feline live multivalent vaccines and provisional safety evaluation in cats. J. Jpn. Vet. Med. Assoc., in press. (In Japanese with English summary.)
- 13.Okabe, H., R. V. Gilden, and M. Hatanaka. 1973. RD 114 virus-specific sequences in feline cellular RNA: detection and characterization. J. Virol. 12:984-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves, R. H., and S. J. O'Brien. 1984. Molecular genetic characterization of the RD-114 gene family of endogenous feline retrovirus sequences. J. Virol. 52:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakaguchi, S., M. Okada, T. Shojima, K. Baba, and T. Miyazawa. 2008. Establishment of a LacZ marker rescue assay to detect infectious RD114 virus. J. Vet. Med. Sci. 70:785-790. [DOI] [PubMed] [Google Scholar]
- 16.Spodick, D. A., A. K. Ghosh, S. Parimoo, and P. Roy-Burman. 1988. The long terminal repeat of feline endogenous RD-114 retroviral DNAs: analysis of transcription regulatory activity and nucleotide sequence. Virus Res. 9:263-283. [DOI] [PubMed] [Google Scholar]
- 17.Stoye, J. P., C. Moroni, and J. M. Coffin. 1991. Virological events leading to spontaneous AKR thymomas. J. Virol. 65:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]