Abstract
A multiplex PCR assay for the simultaneous detection of Mycobacterium tuberculosis and Pneumocystis jirovecii was developed using IS6110-based detection for M. tuberculosis and mitochondrial large-subunit (mtLSU) rRNA gene detection for P. jirovecii. Ninety-five pulmonary blinded samples were examined using the developed multiplex PCR assay, and the results were compared with those obtained by the single nested PCRs targeting IS6110 for M. tuberculosis and mtLSU rRNA for P. jirovecii. Of the 95 pulmonary samples tested, the multiplex nested PCR developed here could detect 36 cases of M. tuberculosis infection, 35 cases of P. jirovecii infection, and 17 cases of M. tuberculosis and P. jirovecii coinfections. The sensitivities of the multiplex nested PCR in detecting M. tuberculosis and P. jirovecii were 92.1% and 81.4%, respectively, whereas the specificities in detecting M. tuberculosis and P. jirovecii were 98.2% and 100%, respectively.
Pulmonary tuberculosis (TB) and Pneumocystis jirovecii pneumonia are two of the most common opportunistic infections found in association with AIDS worldwide (4), including Thailand (2, 3, 6, 10). About one-third of the world's population and one-third of people infected with HIV are infected with Mycobacterium tuberculosis. The World Health Organization (WHO) reported that globally 9.2 million new cases of TB and 1.7 million deaths from TB occurred in 2006, and of these, 0.7 million cases and 0.2 million deaths, respectively, were in HIV-positive people (21). At present, Pneumocystis pneumonia, caused by Pneumocystis jirovecii (previously known as P. carinii f. sp. hominis), remains one of the most common AIDS-defining illnesses and is a frequent cause of morbidity and mortality in HIV-infected patients (7). Geographically, TB is the most common respiratory opportunistic infection in people infected with HIV worldwide, especially in the developing world (1, 4, 6, 8, 9, 12), whereas P. jirovecii pneumonia is more prevalent in industrialized countries (4, 5, 13, 16). In Thailand, TB has been the most common opportunistic infection in people with AIDS, whereas P. jirovecii pneumonia has been the second most common (12, 18). The total number of the two infections represents one-half of opportunistic infections in AIDS cases.
TB and P. jirovecii pneumonia can clinically and radiologically mimic each other, including having similar presentations in patients, and they cannot always be diagnosed by clinical presentation or sputum examination. In addition, coinfection in individuals may also occur. Therefore, accurate and rapid diagnosis is required. The molecular means of diagnosis is considered to be a reliable technique, and it is essential that it be developed or improved to simultaneously diagnose TB and P. jirovecii pneumonia. Having a technique for differential diagnosis of the two infections would contribute to the ability to provide immediate treatment, controlling the diseases and decreasing the rates of transmission. The aim of the present study was to develop a multiplex PCR technique for the detection of M. tuberculosis and P. jirovecii simultaneously in clinical samples. In the present study, the development of a multiplex PCR involved selection of the appropriate genes, as well as the optimum PCR mixture and PCR thermal profile. The multiplex PCR was applied to test its sensitivity and specificity with clinical specimens.
MATERIALS AND METHODS
Sample collection.
A total of 95 sputum/bronchoalveolar lavage (BAL) fluid samples obtained from patients of Phramongkutklao Hospital who had signs and symptoms suspected of being TB or P. jirovecii pneumonia were used in this study. M. tuberculosis was detected in these samples using staining for acid-fast bacilli (AFB) and conventional M. tuberculosis culture. P. jirovecii was detected using toluidine blue O stain and immunofluorescence assay (IFA). The results for the samples were confirmed by a PCR specific for each organism (17, 19, 20). Each sample was recorded as a code number and kept at −20°C in order to use in a blind study of the multiplex nested PCR.
DNA extraction.
Prior to DNA extraction, all samples were incubated at 75 to 90°C for 2 to 5 h to inactivate hazardous organisms. They were treated with N-acetyl-l-cysteine and lysis buffer and were subjected to DNA extraction using an automated MagNa Pure Compact extractor system and a MagNa Pure Compact nucleic acid isolation kit (Roche Applied Science, Roche, Switzerland), according to the recommendations of the manufacturer. Final elutions of DNA were made in 100 μl and were stored at −20°C until use.
DNA was extracted from a culture of a BAL fluid sample positive for M. tuberculosis and one positive for P. jirovecii for use as positive controls. The extracted DNA was kept at −20°C and was used as a positive control throughout the study.
Detection of M. tuberculosis and P. jirovecii using single nested PCR.
Each sample was tested using single nested PCR to confirm the presence of M. tuberculosis and P. jirovecii. The PCR products were diluted 1:100 with distilled water, and 1 μl was used as the template in the second-round amplification. The PCR products were analyzed using 2% agarose gel electrophoresis and were visualized under UV light by an UVitec BTS-15.M apparatus. A 100-bp DNA ladder (Promega) was used as a standard marker.
Single nested PCR assay for IS6110 of M. tuberculosis.
PCR amplification was performed using primers described previously (17). The first-round amplification, using primer TB1 as a forward outer primer and TB4 as a reverse outer primer, amplified a 302-bp region of repetitive sequence (Table 1). The reaction mixture consisted of deionized distilled water, Go Taq Flexi buffer, 2 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), 25 pmol of each primer, 1.25 U Taq DNA polymerase (Promega), and 1 μl DNA template. The protocol for IS6110 amplification was modified from that used in a previous study (17) and was initiated with denaturation at 95°C for 5 min, and then the mixture was subjected to 40 cycles of 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s and had a final extension of 72°C for 5 min.
TABLE 1.
Primers used in single nested PCR assay and multiplex nested PCR assay
| Primer | Oligonucleotide sequence (5′-3′) | Size (bp) |
|---|---|---|
| TB1/TB4 | GTGCGGATGGTCGCAGAGAT | 302 |
| CCTGATGATCGGCGATGAAC | ||
| TB2/TB3 | AGCACGATTCGGAGTGGGCA | 141 |
| TCAGCGGATTCTTCGGTCG | ||
| pAZ102-E/pAZ102-H | GATGGCTGTTTCCAAGCCCA | 346 |
| GTGTACGTTGCAAAGTACTC | ||
| pAZ102-X/pAZ102-Y | GTGAAATACAAATCGGACTAGG | 260 |
| TCACTTAATATTAATTGGGGAGC |
In the second-round amplification, the reaction mixture consisted of 25 pmol of each primer and the other reagents used in the first-round amplification. Forward inner primer TB2 and reverse inner primer TB3 amplified a 141-bp fragment (Table 1). The amplification conditions used were the same as those used in the first-round amplification, with the total duration being 25 cycles.
Single nested PCR assay for mtLSU rRNA gene of P. jirovecii.
Amplification of the mitochondrial large-subunit (mtLSU) rRNA gene of P. jirovecii was performed using the oligonucleotide primers published by Wakefield et al. (19, 20), as shown in Table 1. The first-round amplification was performed using primers pAZ102-E and pAZ102-H as the forward and reverse outer primers, respectively, and yielded a 346-bp fragment. The PCR mixture consisted of 1 μl of DNA template, deionized distilled water, Go Taq Flexi buffer, 2 mM MgCl2, 0.2 mM each dNTP, 25 pmol of each primer, and 1.25 U Taq DNA polymerase. The conditions, modified from those from a previous study (18), were as follows: denaturation at 94°C for 5 min and 40 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 1 min, and extension at 72°C for 1 min. A final extension at 72°C for 5 min was performed.
In the second-round amplification, the reaction mixture consisted of 25 pmol of each primer and the other reagents used in the first-round amplification. Forward inner primer pAZ102-X and reverse inner primer pAZ102-Y amplified a 260-bp fragment (Table 1) (20). The amplification conditions used were the same as those used in the first-round amplification, with the total duration being 25 cycles.
Condition optimization of multiplex nested PCR.
Multiplex nested PCR was used to simultaneously amplify the two targets, IS6110 and the mtLSU rRNA gene, for M. tuberculosis and P. jirovecii, respectively. The first- and second-round amplifications of the multiplex nested PCR were performed using primers similar to those used in the single nested PCR for the detection of M. tuberculosis and P. jirovecii. To confirm the absence of cross-amplification between the selected primers, amplification of a mixture of DNA from both organisms was conducted. Optimization to adjust the parameters was performed to obtain a final condition that provided good intensities for both amplicons, as well as to provide no nonspecific bands. Sequence analysis was also performed to confirm that the PCR products of both M. tuberculosis and P. jirovecii matched the theoretical amplicon sizes. Final conditions were initiated with denaturation at 94°C for 5 min and 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 1 min. A final extension was carried out at 72°C for 5 min. In the second round, the process was done using conditions similar to those used in the first-round amplification, with the total duration being 25 cycles. In each round, 25 μl of reaction mixture consisted of autoclaved deionized distilled water, Go Taq Flexi buffer, 2 mM MgCl2, 0.2 mM each dNTP, 25 pmol of each primer, and 1.25 U Taq DNA polymerase. In the first-round PCR, 1 μl of DNA template comprised a mix of 0.5 μl (6 pg) M. tuberculosis DNA and 0.5 μl (18.19 pg) P. jirovecii DNA, which was appropriate for the provision of dual bands.
Detection limit of multiplex nested PCR.
The ability of the multiplex nested PCR assay to detect M. tuberculosis and P. jirovecii DNA when the DNA templates were present at various artificially mixed ratios was assessed. Titration experiments were conducted to determine the analytical sensitivity of the PCR for the detection of M. tuberculosis and P. jirovecii DNA.
Sensitivity evaluation of the developed multiplex nested PCR.
All 95 blinded samples from the patients were examined using the multiplex nested PCR developed to test its sensitivity and specificity. The efficiency of the multiplex nested PCR was evaluated using statistical analysis for determination of the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). The results obtained from the multiplex nested PCR and single nested PCR methods were compared with the calculated 95% confidential intervals using the Stata/SE program for Windows (version 9.2; StataCorp LP, College Station, TX).
RESULTS
Detection limit of multiplex nested PCR.
Each of the two pairs of oligonucleotide primers exclusively amplified the target genes of the specific microorganisms. The fragments obtained from the multiplex nested PCR optimization are shown in Fig. 1. The multiplex nested PCR assay could amplify and differentiate the two species M. tuberculosis and P. jirovecii in a single specimen. The expected sizes of the M. tuberculosis and P. jirovecii products were 141 bp and 260 bp, respectively. The limits of detection of M. tuberculosis and P. jirovecii were approximately 0.30 fg and 0.28 fg, respectively.
FIG. 1.
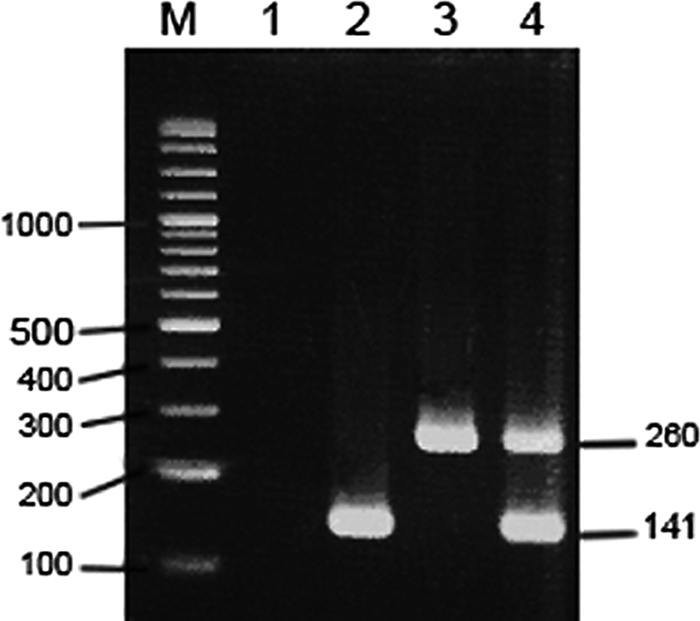
Multiplex nested PCR optimization. Lane M, 100-bp DNA ladder; lane 1, negative control; lane 2, positive M. tuberculosis DNA fragment; lane 3, positive P. jirovecii DNA fragment; lane 4, multiplex nested PCR with the fragments of the two species M. tuberculosis (141 bp) and P. jirovecii DNA (260 bp) together. The numbers on the left and right of the gel are molecular sizes (in base pairs).
Comparison of multiplex nested PCR and single nested PCR results.
The results obtained using the single nested PCR (the “gold standard”) and the multiplex nested PCR are summarized in Table 2. Of the 95 samples, 61 were positive for M. tuberculosis and/or P. jirovecii using the single nested PCR assay. Thirty-eight samples (18 with M. tuberculosis alone and 20 coinfected with M. tuberculosis and P. jirovecii) were positive for M. tuberculosis using IS6110-based detection. Of these samples, 16 were detected in the first- and the second-round amplifications, 19 were detected in the second-round amplification alone, and another 3 were not detected in the second-round amplification, results that are in contrast to the positive results obtained in the first round. A total of 43 samples were positive for P. jirovecii; 23 and 20 of these samples were positive for P. jirovecii alone and coinfection with M. tuberculosis and P. jirovecii, respectively. Of these samples, 20 samples were detected in the first- and the second-round amplifications, while another 23 samples were detected in the second-round amplification alone. The multiplex nested PCR technique could identify 36 cases of TB (19 cases of M. tuberculosis infection alone, 17 cases of coinfection with M. tuberculosis and P. jirovecii) and 35 cases of P. jirovecii pneumonia (18 cases of P. jirovecii infection alone, 17 cases of coinfection with M. tuberculosis and P. jirovecii).
TABLE 2.
Results obtained by single nested PCR and multiplex nested PCR
| Sample no. | No. of samples with the indicated result |
|
|---|---|---|
| Single nested PCR | Multiplex nested PCR | |
| 1 | 18 M. tuberculosis | 16 M. tuberculosis, 2 neg.a |
| 2 | 23 P. jirovecii | 17 P. jirovecii, 1 M. tuberculosis + P. jirovecii, 5 neg. |
| 3 | 20 M. tuberculosis + P. jirovecii | 16 M. tuberculosis + P. jirovecii, 3 M. tuberculosis, 1 P. jirovecii |
| 4 | 34 neg. | 34 neg. |
| Total | 95 | 95 |
neg., negative.
Sensitivity and specificity of multiplex nested PCR.
Using single nested PCR as the gold standard method, the sensitivity, specificity, PPV, and NPV of the multiplex nested PCR in detecting M. tuberculosis and P. jirovecii were determined for each organism, as shown in Tables 3 and 4, respectively. The statistical determinations show the efficiency of the technique developed in the present study for the simultaneous detection of M. tuberculosis and P. jirovecii.
TABLE 3.
Results of multiplex nested PCR compared with those of single nested PCR for M. tuberculosis detection
| Result by multiplex nested PCR | No. (%) of samples with the following single nested PCR resulta: |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 35 (36.84) | 1 (1.05) | 36 (37.89) |
| Negative | 3 (3.16) | 56 (58.95) | 59 (62.11) |
| Total | 38 (40.00) | 57 (60.00) | 95 (100.00) |
By use of gold standard 1 for the single nested PCR. Sensitivity, 92.11% (95% confidence interval, 82.3 to 99.4); specificity, 98.25%; PPV, 97.22%; NPV, 94.92%.
TABLE 4.
Results of multiplex nested PCR compared with those of single nested PCR for P. jirovecii detection
| Result by multiplex nested PCR | No. (%) of samples with the following single nested PCR resulta: |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 35 (36.84) | 0 (0.00) | 35 (36.84) |
| Negative | 8 (8.42) | 52 (54.74) | 60 (63.16) |
| Total | 43 (45.26) | 52 (54.74) | 95 (100.00) |
By use of gold standard 2 for the single nested PCR. Sensitivity, 81.40% (95% confidence interval, 66.6 to 91.6); specificity, 100%; PPV, 100%; NPV, 86.67%.
DISCUSSION
Numerous studies have reported on molecular assays that target M. tuberculosis and P. jirovecii separately. In this report, we describe the successful development of a multiplex nested PCR for the simultaneous detection of M. tuberculosis and P. jirovecii. The assay successfully amplified the positive controls and samples for which nonspecific bands were not observed. The specificity of IS6110 in detecting the M. tuberculosis complex (MTC) using the primer specific sets TB1/TB4 and TB2/TB3 was confirmed by Se Thoe et al. in 1997 (17). These primers did not amplify non-MTC strains (M. fortuitum, M. intracellulare, M. kansasii, M. scrofulaceum, and M. smegmatis). The detection specificity of specific primer set pAZ102-E/pAZ102-H was also confirmed by Wakefield et al. (19). These primers did not amplify DNA from other potential pulmonary pathogens (Candida albicans, other species of Candida, Cryptococcus neoformans, Mycobacterium tuberculosis, Saccharomyces cerevisiae, Aspergillus nidulans) or human DNA.
Clinical sample examination using a single nested PCR revealed 19 additional TB-positive cases and 23 additional P. jirovecii pneumonia-positive cases in the second-round amplification compared with the numbers revealed from the first-round amplification. This confirmed the improvement in sensitivity of the nested PCR over that of the PCR with single-round amplification. A similar contribution of the multiplex nested PCR also occurred. Moreover, the enhanced sensitivity of detection was obtained using multicopy genes IS6110 and mtLSU rRNA. Most reports of studies using IS6110-based detection have claimed sensitivities of over 75% and specificities approaching 100% (14). The sensitivity (92.1%) and specificity (98.2%) of the multiplex nested PCR assay developed in the present study for detecting M. tuberculosis have proved that the assay is reliable enough for diagnostic application. The sensitivity (81.4%) and specificity (100%) of the multiplex nested PCR assay for detecting P. jirovecii also indicated its efficiency for diagnosis. The detection limits determined using DNA solutions diluted prior to amplification were as little as 0.30 fg M. tuberculosis DNA and 0.28 fg P. jirovecii DNA. The amplification products obtained with smaller DNA template amounts were below the limit of quantification. The detection limits of this multiplex nested PCR showed ranges for both M. tuberculosis and P. jirovecii equivalent to those obtained by single nested PCR in previous studies using the same targets (11, 15). These detection limits showed the efficiency of the method for the detection of the two pathogens because they were low. However, the sensitivities acquired from the evaluation were not as high as expected, especially for P. jirovecii detection. The probable reason for this is that amplification of two pathogens at the same time might cause competition for the components of the PCR mixture (i.e., Mg2+ and dNTPs) between the processes of amplification of each gene and inhibition or reduction in the amounts of products from the PCRs in the first-round amplification. This was due to the fact that the assay was developed by modification of two conditions, and the conditions used in detecting M. tuberculosis alone were more sensitive than those used in detecting P. jirovecii.
In conclusion, we report on the first multiplex nested PCR for the simultaneous detection and differentiation of TB and P. jirovecii pneumonia. The rapid, cost-effective development of the multiplex nested PCR will result in an assay more useful than the single nested PCR technique for application to differential diagnosis for a large number of clinical samples.
Acknowledgments
This study was financially supported by Phramongkutklao Hospital's Foundation under Her Royal Highness Princess Maha Chakri Sirindhorn's patronage and the Phramongkutklao College of Medicine.
We thank Chavachol Setthaudom, Immunology Unit, Department of Pathology, Ramathibodi Hospital, Mahidol University.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Abouya, Y. L., A. Beaumel, S. Lucas, A. Dago-Akribi, G. Coulibaly, M. N′Dhatz, J. Konan, A. Yapi, and K. De Cock. 1992. Pneumocystis carinii pneumonia. An uncommon cause of death in African patients with acquired immunodeficiency syndrome. Am. Rev. Respir. Dis. 145:617-620. [DOI] [PubMed] [Google Scholar]
- 2.Aderaye, G., J. Bruchfeld, M. Olsson, and L. Lindquist. 2003. Occurrence of Pneumocystis carinii in HIV-positive patients with suspected pulmonary tuberculosis in Ethiopia. AIDS 17:435-440. [DOI] [PubMed] [Google Scholar]
- 3.Baughman, R. P., M. N. Dohn, and P. T. Frame. 1994. The continuing utility of bronchoalveolar lavage to diagnose opportunistic infection in AIDS patients. Am. J. Med. 97:515-522. [DOI] [PubMed] [Google Scholar]
- 4.Castro, J. G., G. Manzi, L. Espinoza, M. Campos, and C. Boulanger. 2007. Concurrent PCP and TB pneumonia in HIV infected patients. Scand. J. Infect. Dis. 39:1054-1058. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1992. Recommendations for prophylaxis against Pneumocystis carinii pneumonia for adults and adolescents infected with human immunodeficiency virus. MMWR Morb. Mortal. Wkly. Rep. 41:1-11. [PubMed] [Google Scholar]
- 6.Fisk, D. T., S. Meshnick, and P. H. Kazanjian. 2003. Pneumocystis carinii pneumonia in patients in the developing world who have acquired immunodeficiency syndrome. Clin. Infect. Dis. 36:70-78. [DOI] [PubMed] [Google Scholar]
- 7.Huang, L., and K. A. Crothers. 2009. HIV-associated opportunistic pneumonias. Respirology 14:474-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumarasamy, N., S. Solomon, T. P. Flanigan, R. Hemalatha, S. P. Thyagarajan, and K. H Mayer. 2003. Natural history of human immunodeficiency virus disease in southern India. Clin. Infect. Dis. 36:79-85. [DOI] [PubMed] [Google Scholar]
- 9.Lucas, S. B., A. Hounnou, C. Peacock, A. Beaumel, G. Djomand, J. M. N′Gbichi, K. Yeboue, M. Hondé, M. Diomande, C. Giordano, R. Doorly, K. Brattegaard, L. Kestens, R. Smithwick, A. Kadio, N. Ezani, A. Yapi, and K. M. De Cock. 1993. The mortality and pathology of HIV infection in a West African city. AIDS 7:1569-1579. [DOI] [PubMed] [Google Scholar]
- 10.Orlovic, D., R. Kularatne, V. Ferraz, and R. A. J. Smego. 2001. Dual pulmonary infection with Mycobacterium tuberculosis and Pneumocystis carinii in patients infected with human immunodeficiency virus. Clin. Infect. Dis. 32:289-294. [DOI] [PubMed] [Google Scholar]
- 11.Ortona, E., P. Margutti, E. Tamburrini, P. Mencarini, E. Visconti, M. Zolfo, and A. Siracusano. 1997. Detection of Pneumocystis carinii in respiratory specimens by PCR-solution hybridization enzyme-linked immunoassay. J. Clin. Microbiol. 35:1589-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruxrungtham, K., and P. Phanuphak. 2001. Update on HIV/AIDS in Thailand. J. Med. Assoc. Thai. 84(Suppl. 1):S1-S17. [PubMed] [Google Scholar]
- 13.San-Andres, F. J., R. Rubio, J. Castilla, F. Pulido, G. Palao, I. de Pedro, J. R. Costa, and A. del Palacio. 2003. Incidence of acquired immunodeficiency syndrome-associated opportunistic diseases and the effect of treatment on a cohort of 1115 patients infected with human immunodeficiency virus, 1989-1997. Clin. Infect. Dis. 36:1177-1185. [DOI] [PubMed] [Google Scholar]
- 14.Sandhu, G. S., B. C. Kline, L. Stockman, G. D. Roberts, and M. E. Lewis. 24 March 1998. IS6110 based molecular detection of Mycobacterium tuberculosis. U.S. patent 5,731,150.
- 15.Scarpellini, P., S. Racca, P. Cinque, F. Delfanti, N. Gianotti, M. R. Terreni, L. Vago, and A. Lazzarin. 1995. Nested polymerase chain reaction for diagnosis and monitoring treatment response in AIDS patients with tuberculous meningitis. AIDS 9:895-900. [DOI] [PubMed] [Google Scholar]
- 16.Serraino, D., V. Puro, E. Boumis, C. Angeletti, E. Girardi, N. Petrosillo, and G. Ippolito. 2003. Epidemiological aspects of major opportunistic infections of the respiratory tract in persons with AIDS: Europe, 1993-2000. AIDS 17:2109-2116. [DOI] [PubMed] [Google Scholar]
- 17.Se Thoe, S. Y., L. Tay, and E. H. Sng. 1997. Evaluation of Amplicor- and IS6110-PCR for direct detection of Mycobacterium tuberculosis complex in Singapore. Trop. Med. Int. Health 2:1095-1101. [DOI] [PubMed] [Google Scholar]
- 18.Tansuphasawadikul, S., P. N. Amornkul, C. Tanchanpong, K. Limpakarnjanarat, J. Kaewkungwal, S. Likanonsakul, B. Eampokalap, T. Naiwatanakul, D. Kitayaporn, N. L Young, D. J. Hu, and T. D. Mastro. 1999. Clinical presentation of hospitalized adult patients with HIV infection and AIDS in Bangkok, Thailand. J. Acquir. Immune Defic. Syndr. 21:326-332. [DOI] [PubMed] [Google Scholar]
- 19.Wakefield, A. E., F. J. Pixley, S. Banerji, K. Sinclair, R. F. Miller, E. R. Moxon, and J. M. Hopkin. 1990. Amplification of mitochondrial ribosomal RNA sequences from Pneumocystis carinii DNA of rat and human origin. Mol. Biochem. Parasitol. 43:69-76. [DOI] [PubMed] [Google Scholar]
- 20.Wakefield, A. E., F. J. Pixley, S. Banerji, K. Sinclair, R. F. Miller, E. R. Moxon, and J. M. Hopkin. 1990. Detection of Pneumocystis carinii with DNA amplification. Lancet 336:451-453. [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization. 2008. Global tuberculosis control: surveillance, planning, financing. WHO report 2008. World Health Organization. Geneva, Switzerland. http://www.who.int/tb/publications/global_report/2008/pdf/fullreport.pdf.


