Abstract
A multicenter clinical study was conducted to evaluate the performance characteristics of the Abbott RealTime CT/NG assay, a multiplex real-time PCR assay, for simultaneous detection of Chlamydia trachomatis and Neisseria gonorrhoeae. The specimens were collected from a total of 3,832 male and female subjects at 16 geographically diverse sites. Specimens included male and female urine samples, male urethral swabs, female endocervical swabs, and self-collected and clinician-collected vaginal swabs. Specimens were tested with the automated Abbott RealTime CT/NG assay, Aptima Combo 2 assay (Gen-Probe), ProbeTec ET CT/GC assay (Becton Dickinson), and culture for N. gonorrhoeae. The Aptima Combo 2 assay, the ProbeTec assay, and the N. gonorrhoeae culture were used as the reference assays. For each subject, a patient infected status (PIS) was determined based on the combined results from the reference assays. The overall prevalence in female subjects was 8.9% for C. trachomatis and 3.8% for N. gonorrhoeae. The overall male prevalence was 18.2% for C. trachomatis and 16.7% for N. gonorrhoeae. The overall sensitivity and specificity of the Abbott RealTime CT/NG assay were 92.4% and 99.2% for C. trachomatis and 96.9% and 99.7% for N. gonorrhoeae, respectively. In comparison, the sensitivity and specificity, respectively, for the Aptima Combo 2 assay were 94.5% and 99.0% for C. trachomatis and 96.1% and 99.5% for N. gonorrhoeae, and those for the ProbeTec ET assay were 90.3% and 99.5% for C. trachomatis and 92.0% and 97.3% for N. gonorrhoeae in this study. The Abbott RealTime CT/NG assay offers C. trachomatis and N. gonorrhoeae dual detection with high sensitivity and specificity. The automated assay provides a useful alternative nucleic acid amplification assay for clinical laboratories and clinicians.
Chlamydia trachomatis and Neisseria gonorrhoeae are the two most prevalent bacterial sexually transmitted infections (STIs) reported to the Centers for Disease Control and Prevention (CDC) (2). Together these two infections accounted for over 1.5 million cases of STIs in 2008. The CDC estimates that sexually transmitted diseases (STDs) cost the health care system $1.5 billion annually. Since these infections, especially chlamydia, are most often asymptomatic, the CDC recommends yearly screening for chlamydia in all sexually active women ages 16 to 25 years. Since coinfections are common, many diagnostic test platforms assay for both organisms. Although there are several commercial assays available for performing nucleic acid amplification tests (NAATs), which are now recommended by the CDC as the test of choice, new assays and new platforms are needed by both private and public health programs.
The Abbott RealTime CT/NG assay is a new real-time PCR assay for the direct, qualitative detection of the plasmid DNA of Chlamydia trachomatis and the genomic DNA of Neisseria gonorrhoeae in female endocervical or vaginal swab specimens, in male urethral swab specimens, and in male and female urine specimens from symptomatic and asymptomatic individuals. There were several sets of objectives of this study: (i) to determine the performance characteristics of the Abbott RealTime CT/NG assay compared to those of other amplified assays using specimens obtained from symptomatic and asymptomatic male and female subjects from clinical sites with high or low prevalence of Chlamydia trachomatis and/or Neisseria gonorrhoeae; (ii) to demonstrate the precision of the Abbott RealTime CT/NG assay using a precision panel consisting of combinations of high and low concentrations of Chlamydia trachomatis and Neisseria gonorrhoeae specimens; (iii) to evaluate the clinical sensitivity, clinical specificity, positive predictive value (PPV), and negative predictive value (NPV) of the Abbott RealTime CT/NG assay in symptomatic and asymptomatic individuals from high- and low-prevalence sites; (iv) to compare the performance of the new Abbott assay with that of the Gen-Probe Aptima Combo 2 assay for female endocervical and vaginal swabs, male urethral swabs, and male and female urine specimens; (v) to compare the performance of the Abbott assay with those of the Becton Dickinson ProbeTec ET Chlamydia trachomatis and Neisseria gonorrhoeae amplified DNA assays (ProbeTec ET) for female endocervical swabs and male and female urine specimens; and (vi) to evaluate the procedure for self-collection of a vaginal swab specimen using the Abbott multi-Collect specimen collection kit.
MATERIALS AND METHODS
Overview.
This study was designed as a multicenter evaluation of the Abbott multi-Collect specimen collection kit, the performance (i.e., precision, clinical sensitivity, clinical specificity, PPV, and NPV) of the Abbott RealTime CT/NG assay, and its comparison to two FDA-cleared nucleic acid amplification tests (NAATs) for Chlamydia trachomatis and Neisseria gonorrhoeae. The NAAT comparator methods used were the Gen-Probe Aptima Combo 2 assay (3, 5) and the Becton Dickinson ProbeTec ET (9, 17). Testing was performed by ICON Laboratories according to the comparator assay package inserts. The gonococcal culture (GC) methods used modified Thayer Martin medium for isolation, and identification was confirmed by the bioMerieux API NH assay and the EY Laboratories Gonocheck II system; the Pharmacia Diagnostics Phadebact Monoclonal GC test was used for confirmation. Results from the NAATs and GC cultures were used to construct a patient infected status (PIS) algorithm (10).
Patient population: subjects and clinical specimens.
Subjects meeting the following criteria were eligible for the study.
(i) Inclusion criteria.
An individual was eligible for the study if he or she met all of the following criteria. Prior to the performance of any study-specific procedure, the individual or his/her legal representative signed and dated an institutional review board (IRB)-approved consent form, was 18 years of age or older, had been sexually active within the past 6 months, and met one or more of the following criteria: seeking screening for STI and/or a medical checkup, seeking treatment for STI-related symptoms, had sexual contact with an infected partner, had been previously treated for an STI and/or had a history of STIs, or had a new sexual partner(s), multiple sexual partners, or reported inconsistent condom use.
(ii) Exclusion criteria.
An individual was ineligible for the study if he or she met any of the following criteria: reported using or having completed antimicrobial therapy within 21 days of enrollment in the study, being unable or unwilling to provide the appropriate specimens, having voided within 1 h of specimen collection, or having insufficient medical and/or sexual history to document the information required for the study. The study involved a single visit.
Specimen collection.
The following specimens were collected from each subject.
(i) Female subjects.
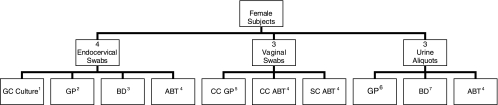
A total of 10 specimens were collected from each female subject: one self-collected vaginal swab, one urine sample divided into three aliquots, four endocervical swabs, and two clinician-collected vaginal swabs (Fig. 1). Female subjects were instructed to self-collect a vaginal swab specimen using an Abbott multi-Collect specimen collection kit. Study staff provided the Abbott self-collected vaginal swab instructions to female subjects prior to self-collection. Self-collected vaginal swab and urine specimens were collected first from each female subject. Clinicians then obtained a GC swab from the endocervix. The order of the remaining endocervical and clinician-collected vaginal swabs was randomized in order to minimize collection bias.
FIG. 1.
Sample collection protocol for female participants. Superscript numbers indicate procedures: 1, gonococcal culture; 2, Gen-Probe Aptima unisex swab specimen collection kit for endocervical swab specimens; 3, Becton Dickinson ProbeTec ET Chlamydia trachomatis/Neisseria gonorrhoeae (CT/GC) amplified DNA assay collection kit for endocervical specimens; 4, Abbott multi-Collect specimen collection kit; 5, Gen-Probe Aptima vaginal swab specimen collection kit for clinician-collected swab specimens; 6, Gen-Probe Aptima urine specimen collection kit for urine specimens; 7, Becton Dickinson ProbeTec ET urine processing kit for urine specimens.
(ii) Male subjects.
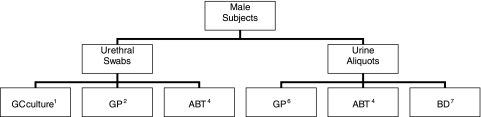
A total of six specimens were collected from each male subject: three urethral swabs and one urine sample divided into three aliquots (Fig. 2). The first urethral swab was used for GC. The order of collection for the other two urethral swabs was randomized in order to minimize collection bias. The male subjects then self-collected a urine specimen.
FIG. 2.
Sample collection protocol for male participants. Superscript numbers indicate procedures: 1, gonococcal culture; 2, Gen-Probe Aptima unisex swab specimen collection kit for male urethral swab specimens; 4, Abbott multi-Collect specimen collection kit; 6, Gen-Probe Aptima urine specimen collection kit for urine specimens; 7, Becton Dickinson ProbeTec ET urine processing kit for urine specimens.
Abbott RealTime CT/NG assay.
The target for C. trachomatis is a cryptic plasmid, and that for N. gonorrhoeae is the Opa gene. The C. trachomatis PCR primers target a 122-bp target within the plasmid. This short sequence is highly conserved in all C. trachomatis serovars but is not found in other species. A new variant strain of C. trachomatis (nvCT) has been characterized and contains a 377-bp deletion in the cryptic plasmid within the region targeted by the first primer set. A second set of PCR primers targets 140 bp located outside this deleted region. The 122-bp target of the N. gonorrhoeae OPA gene is conserved in all GC strains but is not found in non-N. gonorrhoeae strains, such as a respiratory Neisseria sp. The internal-control PCR primers target a 136-bp sequence not found in chlamydiae or neisseriae. During each round of PCR amplification, the fluorescent probes anneal to the amplified target DNA, if present. The probes are labeled with different fluorescent molecules allowing C. trachomatis, N. gonorrhoeae, and the internal control to be distinguished from each other. The probes are single-stranded, linear DNA oligonucleotides modified with a fluorescent moiety covalently linked to one end of the probe and a quenching moiety to the other end. In the absence of target sequences, the probes adopt a conformation that brings the quencher close enough to the excited fluorophore to absorb its energy before it can be fluorescently emitted. When the probe binds to its complementary sequence in the target, the fluorophore and the quencher are held apart, allowing fluorescent emission and detection. Since this fluorescence occurs during every cycle, the PCR can be read in real time.
The Abbott RealTime CT/NG assay was performed on the Abbott m2000 system by the four testing sites in accordance with the Abbott RealTime CT/NG clinical testing protocol, clinical brochure, specimen collection and transport, and self-collected vaginal swab instructions for use with the Abbott multi-Collect specimen collection kit. Turnaround time was approximately 6 h, 20 min. Hands-on time was approximately 1 h, 15 min. The specimen collection sites were geographically diverse. They included a primary care physician's office, an obstetrics-gynecology practice, a gynecology practice, a private-practice STD clinic, five public health department STI clinics, a university hospital adult emergency department, a county hospital sexually transmitted disease/infection clinic, a university infectious disease clinic, and four clinics serving the gay, lesbian, bisexual, and transgendered community. Specimens were collected from subjects in accordance with the Abbott RealTime CT/NG specimen collection protocol. The Abbott RealTime CT/NG results were not used for medical management of subjects from whom specimens were collected for this study.
Specimen collection, handling, and storage were performed in accordance with the Abbott RealTime CT/NG clinical brochure and comparator method package inserts. After the specimens were collected, aliquoted, labeled, and prepared for shipment, the specimens for NAAT comparator method testing were sent to the central laboratory (ICON Laboratories Inc., Farmingdale, NY) for testing. The NAAT comparator method specimens were tested in the central laboratory. The GC specimens were tested in the ICON central laboratory or local public health laboratories.
Abbott RealTime CT/NG testing was performed at three external clinical testing sites and one internal site (precision testing only) that represented the intended user of the Abbott RealTime CT/NG assay.
The assay was performed in accordance with the Abbott RealTime CT/NG testing protocol and the Abbott RealTime CT/NG clinical brochure. The Abbott assay was performed on the Abbott m2000 system, consisting of the m2000sp instrument for sample preparation and the m2000rt instrument for amplification and detection. Each of the testing sites was provided one Abbott m2000 system. The site technologists were trained to operate the Abbott m2000 system and to perform the Abbott RealTime CT/NG assay prior to initiation of the study. There were no testing protocol deviations. Daily, weekly, monthly, and as-needed maintenance were performed on the Abbott m2000sp and the Abbott m2000rt according to the procedures outlined in the service and maintenance section of the Abbott m2000sp and the Abbott m2000rt operation manuals.
The laboratory surfaces and equipment used for the Abbott RealTime CT/NG assay were monitored on a monthly basis for contamination by screening for amplification product. The monitoring procedure was performed using the procedure outlined in the Abbott clinical brochure.
Precision.
Abbott RealTime CT/NG assay precision was evaluated by testing a coded 45-member precision panel that consisted of nine unique members repeated five times within the panel (Table 1). The panel was commercially prepared from dilutions of heat-treated, cultured, whole bacteria as the source of the Chlamydia and Neisseria. The source material for Chlamydia trachomatis was Vero/LGV-II, strain 434. The source material for Neisseria gonorrhoeae was ATCC isolate 27628. The targeted concentrations for C. trachomatis and N. gonorrhoeae for the nine panel members are listed in Table 1. Both source materials for C. trachomatis and N. gonorrhoeae were diluted in Abbott specimen transport buffer, and aliquots were dispensed into Abbott specimen transport tubes. The design of the precision study was based upon the National Committee for Clinical Laboratory Standards (NCCLS) EP10-A2 guideline (12a). A total of three reagent lots were used. Each of the three sites, two external and one internal site, tested two of the three amplification lots for 5 days for a total of 10 runs per site, 30 runs total.
TABLE 1.
Numbers of organisms tested for the precision panel
| Panel member | C. trachomatis IFU/assaya | N. gonorrhoeae CFU/assay |
|---|---|---|
| 1 | 4,500 | 2,000 |
| 2 | 4,500 | 20 |
| 3 | 0.75 | 20 |
| 4 | 45 | 0 |
| 5 | 0 | 20 |
| 6 | 4,500 | 0 |
| 7 | 0 | 2,000 |
| 8 | 0 | 0 |
| 9 | 0.2 | 0.1 |
IFU, inclusion-forming units.
Neisseria gonorrhoeae culture.
Three clinical laboratories conducted N. gonorrhoeae culture testing using MTM II agar: ICON Central Laboratories, BDC Laboratory, and Miami-Dade County Health Department. The Biolog Jembec system was used to transport specimens for GC to ICON Central Laboratories for testing with the bioMerieux API NH assay.
Gen-Probe Aptima Combo 2 assay.
The Gen-Probe Aptima Combo 2 results are qualitative and were reported as positive for C. trachomatis rRNA, presumed negative for C. trachomatis rRNA, or indeterminate and as positive for N. gonorrhoeae rRNA, presumed negative for N. gonorrhoeae rRNA, or indeterminate. Repeatedly equivocal results were reported as indeterminate.
Becton Dickenson ProbeTec ET assay.
The Becton Dickinson ProbeTec ET Chlamydia trachomatis and Neisseria gonorrhoeae results are qualitative and were reported as positive, negative, or indeterminate. Repeatedly inhibitory specimens were reported as indeterminate.
Data analysis: interpretation of assay results.
The Abbott RealTime CT/NG assay is a qualitative assay. Results for N. gonorrhoeae were reported as positive or negative. Results for C. trachomatis were reported as positive, negative, or equivocal. A specimen with an initial interpretation of equivocal for C. trachomatis was retested before a final interpretation was made. One replicate of the negative control, designated by the software as the “control,” and two replicates of the cutoff control, designated by the software as the “calibrator,” were required with each run. For each assayed analyte (C. trachomatis or N. gonorrhoeae), the software calculated the mean target cycle number (CN) of the cutoff controls and then added a predetermined number of cycles to this average to generate the decision cycle (assay cutoff [assay CO]). If the tested sample generated a cycle number less than or equal to the assay CO, a “positive” interpretation and a numerical result greater than or equal to zero were reported. The numerical result (delta cycle [DC]) corresponds to the difference in cycle number between the CO and the sample CN. A higher number in DC was indicative of a greater amount of analyte present in the assayed sample. No DC calculation was performed for samples with a cycle number greater than the assay CO or for samples with no cycle number determination.
C. trachomatis samples with a cycle number less than or equal to the assay CO were interpreted as “positive.” C. trachomatis samples with a cycle number beyond the assay CO were interpreted as “equivocal.” C. trachomatis samples with no evidence of amplification were interpreted as “negative.” Samples with an initial interpretation of “equivocal” for C. trachomatis were retested. For these samples, retest results were used to determine the final result interpretation. If the C. trachomatis retest result was positive, the sample was interpreted as “positive.” If the C. trachomatis retest result was negative, the sample was interpreted as “negative.” If the C. trachomatis retest was equivocal, the sample was reported as indeterminate. (The C. trachomatis “indeterminate” designation is specific to this U.S. study.) Indeterminate C. trachomatis samples were not included in the final evaluation of sensitivity and specificity. For N. gonorrhoeae, no equivocal range was established. Only the initial N. gonorrhoeae results were used for sample result interpretation.
All data analyses were performed by SAS version 6.12 or higher on a UNIX operating system. All assay results were included in the data analyses if they were considered valid according to the assay procedure and in compliance with the applicable protocol requirements. The primary data analyses for clinical sensitivity and clinical specificity of C. trachomatis and N. gonorrhoeae were calculated on the basis of infection status for symptomatic and asymptomatic subjects, by specimen type, by gender, by site, and for all sites combined.
Patient infected status for Chlamydia trachomatis.
The patient infection status was defined as follows.
(i) Infection present.
A female subject was defined as infected with C. trachomatis if a minimum of one positive result was reported by each of the reference NAATs (Gen-Probe, Becton Dickinson) (two comparator platforms). A male subject was defined as infected with C. trachomatis if a minimum of two positive results were reported by the reference NAAT. The PIS was different for men in order to spare men from being subjected to four urethral swabs; we did not collect urethral swabs for the BD assay. Therefore, the PIS was adjusted.
(ii) Infection absent.
A female subject was defined as not infected with C. trachomatis if at least one of the reference NAATs reported negative results for all sample types. A male subject was defined as not infected with C. trachomatis if two or more reference NAAT results were negative.
Patient infection status for Neisseria gonorrhoeae.
A female subject was defined as infected with N. gonorrhoeae if a minimum of one positive result was reported by each of the reference NAATs (Gen-Probe, Becton Dickinson). A male subject was defined as infected with N. gonorrhoeae if a minimum of two positive results were reported by the reference NAAT.
If the N. gonorrhoeae culture result was positive, the subject was defined as infected regardless of NAAT results.
RESULTS
A total of 3,832 male and female, asymptomatic and symptomatic subjects were enrolled at 16 sites by licensed clinicians. Summaries describing the study population and documenting study eligibility are presented in Tables 2 to 4. A total of 2,014 female subjects were enrolled, of which 1,132 (56.2%) were asymptomatic and 882 (48.8%) were symptomatic. A total of 1,818 male subjects were enrolled, of which 909 (50.0%) were asymptomatic and 909 (50.0%) were symptomatic. Male and female subjects were classified as asymptomatic if the subject reported no symptoms. Male and female subjects were classified as symptomatic if the subject reported STD-related symptoms. Counting replicate samples for all the different assays, a total of 31,048 specimens were estimated to have been collected: 20,140 from female subjects and 10,908 from male subjects.
TABLE 2.
Subjects' reasons for visit
| Visit reason and status | No. of subjects | % of subjectsa |
|---|---|---|
| Reason | ||
| STD-related symptomsb | 1,370 | 35.75 |
| Sexual contact | 313 | 8.17 |
| STD screenc | 1,342 | 35.02 |
| Medical checkup | 807 | 21.06 |
| Status | ||
| First visit | 2,171 | 56.65 |
| Repeat visit | 1,661 | 43.35 |
Total = 3,832 subjects.
STD-related symptoms/sexual contact included in total number.
STD screen/medical checkup and sexual contact/STD screen included in total number.
TABLE 4.
Demographics of study participants
| Criterion | No. of subjects | % of subjectsa |
|---|---|---|
| Gender | ||
| Male | 1,818 | 47.44 |
| Female | 2,014 | 52.56 |
| Age | ||
| <18b | 3 | 0.08 |
| 18-23 | 1,064 | 27.77 |
| 24-29 | 880 | 22.96 |
| 30-35 | 580 | 15.14 |
| 36-41 | 499 | 13.02 |
| ≥42 | 806 | 21.03 |
| Race | ||
| African-American | 2,572 | 67.12 |
| American Indian (Alaska Native) | 11 | 0.29 |
| American Indian (Alaska Native)/African-American | 4 | 0.10 |
| Asian | 28 | 0.73 |
| Caucasian | 1,198 | 31.26 |
| Caucasian/Asian | 1 | 0.03 |
| Unknown | 18 | 0.47 |
| Ethnicity | ||
| Hispanic/Latino | 577 | 15.06 |
| Non-Hispanic/Latino | 3,255 | 84.94 |
| Marital status | ||
| Single | 2,790 | 72.81 |
| Married | 673 | 17.56 |
| Separated | 107 | 2.79 |
| Divorced | 219 | 5.72 |
| Widowed | 43 | 1.12 |
| Education level | ||
| Primary | 205 | 5.35 |
| Secondary | 1,923 | 50.18 |
| College | 1,522 | 39.72 |
| Postgraduate | 180 | 4.70 |
| No response | 2 | 0.05 |
Total = 3,832 subjects.
Subjects did not meet study inclusion criteria.
Precision testing.
Overall precision testing of the panel was excellent, indicating that for C. trachomatis, 45 IFU/assay could be detected 100% of the time and 0.75 IFU/assay could be detected 95.3% of the time (Table 5). Similarly for N. gonorrhoeae, 20 CFU/assay could be detected 100% of the time (Table 6). Combinations of high-concentration and low-concentration mixtures of C. trachomatis and N. gonorrhoeae did not affect the ability of either organism to be detected (Tables 5 and 6).
TABLE 5.
Results of precision testing for C. trachomatis
| Panel membera | No. testedb | No. positive | Mean delta cycle | SDc |
||||
|---|---|---|---|---|---|---|---|---|
| Within-run component | Between-run component | Between-lot component | Between-site component | Total | ||||
| 1 | 150 | 150 | 14.78 | 0.300 | 0.194 | 0.066 | 0.137 | 0.388 |
| 2 | 149 | 149 | 15.15 | 0.385 | 0.139 | 0.285 | 0.000 | 0.499 |
| 3 | 149 | 149 | 3.12 | 0.591 | 0.241 | 0.000 | 0.047 | 0.640 |
| 4 | 150 | 150 | 8.89 | 0.385 | 0.156 | 0.169 | 0.162 | 0.477 |
| 5 | 148 | 0 | ||||||
| 6 | 148 | 148 | 16.88 | 0.167 | 0.207 | 0.149 | 0.215 | 0.373 |
| 7 | 150 | 0 | ||||||
| 8 | 149 | 1 | 0.67 | |||||
| 9 | 148 | 103 | 1.09 | 0.637 | 0.000 | 0.192 | 0.000 | 0.665 |
C. trachomatis concentrations were targeted approximately to 4,500 IFU/assay in members 1, 2, and 6 and to 45 IFU/assay in member 4. The concentration for member 3 was targeted approximately to 0.75 IFU/assay and that for member 9 to 0.2 IFU/assay, both below the claimed assay limit of detection. Members 5, 7, and 8 did not contain any C. trachomatis organisms.
Invalid replicates were excluded from the analysis.
The SD is based on positive replicates only. For member 9, analysis of all replicates with a cycle number (n = 133), including those beyond the assay cutoff, resulted in a total SD of 0.966. The total variability contains within-run, between-run, between-lot, and between-site variability.
TABLE 6.
Results of precision testing for N. gonorrhoeae
| Panel membera | No. testedb | No. positive | Mean delta cycle | SDc |
||||
|---|---|---|---|---|---|---|---|---|
| Within-run component | Between-run component | Between-lot component | Between-site component | Total | ||||
| 1 | 150 | 150 | 13.43 | 0.382 | 0.172 | 0.000 | 0.147 | 0.444 |
| 2 | 149 | 149 | 7.89 | 0.430 | 0.064 | 0.097 | 0.166 | 0.475 |
| 3 | 149 | 149 | 8.24 | 0.270 | 0.149 | 0.057 | 0.060 | 0.319 |
| 4 | 150 | 0 | ||||||
| 5 | 148 | 148 | 7.80 | 0.231 | 0.198 | 0.040 | 0.185 | 0.358 |
| 6 | 147 | 0 | ||||||
| 7 | 150 | 150 | 13.59 | 0.539 | 0.191 | 0.000 | 0.205 | 0.608 |
| 8 | 149 | 0 | ||||||
| 9 | 148 | 56 | 0.58 | 0.386 | 0.000 | 0.000 | 0.120 | 0.404 |
N. gonorrhoeae concentrations were targeted approximately to 2,000 CFU/assay in members 1 and 7 and to 20 CFU/assay in members 2, 3, and 5. Member 9 was targeted to 0.1 CFU/assay, below the claimed assay limit of detection. Members 4, 6, and 8 did not contain any N. gonorrhoeae organisms.
Invalid replicates were excluded from the analysis.
The SD is based on positive replicates only. For member 9, analysis of all replicates with a cycle number (n = 148), including those beyond the assay cutoff, resulted in a total SD of 0.978. The total variability contains within-run, between-run, between-lot, and between-site variability.
Clinical specimen testing.
A total of 8,712 female endocervical and vaginal, male urethral, and male and female urine specimens were evaluated for C. trachomatis using the Abbott RealTime CT/NG assay. A total of 8,743 female endocervical and vaginal, male urethral, and male and female urine specimens were evaluated for N. gonorrhoeae using the Abbott RealTime CT/NG assay. Overall prevalence in females for C. trachomatis was 8.9%, and that for N. gonorrhoeae was 3.8%. Overall prevalence for males for C. trachomatis was 18.2%, and that for N. gonorrhoeae was 16.7%.
C. trachomatis.
The sensitivity and specificity of the Abbott RealTime CT/NG assay for C. trachomatis were 92.4% and 99.2% (Table 7), while the sensitivity and specificity for the Aptima Combo 2 assay for C. trachomatis were 94.5% and 99.0%, and those for the ProbeTec ET assay were 90.3% and 99.5% for C. trachomatis in this study (Table 7).
TABLE 7.
Clinical sensitivities and specificities of three assaysa for C. trachomatis
| Specimen typeb and symptom status | % sensitivity (range) |
% specificity (range) |
||||
|---|---|---|---|---|---|---|
| ABT m2000 | GP AC2 | BD ProbeTec | ABT m2000 | GP AC2 | BD ProbeTec | |
| Endocervical | ||||||
| Symptomatic | 87.7 (78.5-93.9) | 91.4 (83.0-96.5) | 88.8 (79.7-94.7) | 99.7 (98.9-100.0) | 99.4 (98.5-99.8) | 99.1 (98.0-99.7) |
| Asymptomatic | 80.9 (66.7-90.9) | 78.7 (64.3-89.3) | 78.3 (63.6-89.1) | 99.4 (98.4-99.8) | 98.6 (97.4-99.4) | 99.8 (99.1-100.0) |
| CC vaginal | ||||||
| Symptomatic | 92.5 (84.4-97.2) | 92.6 (84.6-97.2) | NAc | 98.8 (97.6-99.5) | 98.7 (97.5-99.4) | NA |
| Asymptomatic | 87.2 (74.3-95.2) | 85.1 (71.7-93.8) | NA | 99.1 (98.0-99.7) | 98.2 (96.8-99.1) | NA |
| SC vaginal | ||||||
| Symptomatic | 94.7 (86.9-98.5) | NA | NA | 99.0 (97.9-99.6) | NA | NA |
| Asymptomatic | 84.8 (71.1-73.7) | NA | NA | 98.9 (97.7-99.6) | NA | NA |
| Female urine | ||||||
| Symptomatic | 92.6 (84.6-97.2) | 93.8 (86.2-98.0) | 90.9 (82.2-96.3) | 99.5 (98.7-99.9) | 99.4 (98.5-99.8) | 99.7 (98.8-100.0) |
| Asymptomatic | 95.7 (85.2-99.5) | 93.5 (82.1-98.6) | 91.3 (79.2-97.6) | 99.2 (98.2-99.7) | 99.2 (98.2-99.8) | 99.7 (98.8-100.0) |
| Urethral | ||||||
| Symptomatic | 93.3 (86.6-96.5) | 98.4 (95.3-99.7) | NA | 98.3 (97.0-99.1) | 98.5 (97.2-99.3) | NA |
| Asymptomatic | 88.6 (80.1-94.4) | 91.2 (83.4-96.1) | NA | 99.1 (97.9-99.7) | 99.1 (98.0-99.7) | NA |
| Male urine | ||||||
| Symptomatic | 97.3 (93.7-99.1) | 99.5 (97.0-100.0) | 91.0 (85.7-94.7) | 99.7 (98.9-100.0) | 99.4 (98.4-99.8) | 99.0 (97.9-99.6) |
| Asymptomatic | 97.8 (92.3-99.7) | 98.9 (94.0-100.0) | 95.5 (88.9-98.8) | 99.6 (98.7-100.0) | 99.5 (98.5-99.9) | 99.4 (98.4-99.9) |
| All types | ||||||
| Symptomatic | 93.7 (91.6-95.4) | 96.4 (94.5-97.7) | 90.4 (86.7-93.4) | 99.2 (98.8-99.4) | 99.1 (98.7-99.4) | 99.3 (98.8-99.6) |
| Asymptomatic | 90.1 (86.6-93.0) | 91.0 (87.3-93.9) | 90.1 (84.7-94.0) | 99.2 (98.9-99.5) | 98.9 (98.5-99.2) | 99.7 (99.3-99.9) |
| Overall | 92.4 (90.7-94.0) | 94.5 (92.9-95.9) | 90.3 (87.4-92.7) | 99.2 (99.0-99.4) | 99.0 (98.7-99.2) | 99.5 (99.2-99.7) |
GP AC2, GenProbe Aptima Combo 2; BD ProbeTec, Becton Dickinson ProbeTec; ABT m2000, Abbott m2000.
CC, clinician collected; SC, self-collected.
NA, not applicable.
N. gonorrhoeae.
The sensitivity and specificity of the Abbott RealTime CT/NG assay for N. gonorrhoeae were 96.9% and 99.7%, respectively (Table 8). In comparison, the sensitivity and specificity for the Aptima Combo 2 assay for N. gonorrhoeae were 96.1% and 99.5%, and those for the ProbeTec assay were 92.0% and 97.3% for N. gonorrhoeae in this study (Table 8).
TABLE 8.
Clinical sensitivities and specificities of three assaysa for N. gonorrhoeae
| Specimen typeb and symptom status | % sensitivity (range) |
% specificity (range) |
||||
|---|---|---|---|---|---|---|
| ABT m2000 | GP AC2 | BD ProbeTec | ABT m2000 | GP AC2 | BD ProbeTec | |
| Endocervical | ||||||
| Symptomatic | 87.1 (70.2-96.4) | 90.6 (75.0-98.0) | 87.5 (71.0-96.5) | 99.7 (99.0-100.0) | 99.4 (98.6-99.8) | 99.6 (98.8-99.9) |
| Asymptomatic | 91.3 (72.0-98.9) | 90.9 (70.8-98.9) | 91.3 (72.0-98.9) | 100.0 (99.5-100.0) | 99.7 (99.0-100.0) | 98.9 (97.8-99.6) |
| CC vaginal | ||||||
| Symptomatic | 96.8 (83.3-99.9) | 93.8 (79.2-99.2) | NAc | 99.9 (99.2-100.0) | 99.3 (98.4-99.8) | NA |
| Asymptomatic | 95.7 (78.1-99.9) | 95.7 (78.1-99.9) | NA | 99.4 (98.5-99.8) | 99.7 (98.9-100.0) | NA |
| SC vaginal | ||||||
| Symptomatic | 96.7 (82.8-99.9) | NA | NA | 99.7 (98.9-100.0) | NA | NA |
| Asymptomatic | 95.7 (78.1-99.9) | NA | NA | 100.0 (99.4-100.0) | NA | NA |
| Female urine | ||||||
| Symptomatic | 93.8 (79.2-99.2) | 84.4 (67.2-94.7) | 76.7 (57.7-90.1) | 99.7 (99.0-100.0) | 99.6 (98.8-99.9) | 95.6 (93.7-97.0) |
| Asymptomatic | 87.0 (66.4-97.2) | 82.6 (61.2-95.0) | 85.7 (63.7-97.0) | 99.6 (98.7-99.9) | 99.4 (98.5-99.8) | 96.9 (95.3-98.1) |
| Urethral | ||||||
| Symptomatic | 99.2 (97.0-99.9) | 99.2 (97.0-99.9) | NA | 99.3 (98.3-99.8) | 99.2 (98.1-99.7) | NA |
| Asymptomatic | 81.8 (48.2-99.7) | 81.8 (48.2-97.7) | NA | 99.8 (99.1-100.0) | 99.7 (98.9-100.0) | NA |
| Male urine | ||||||
| Symptomatic | 98.8 (96.4-99.7) | 97.9 (95.2-99.3) | 94.9 (91.3-97.3) | 99.5 (98.5-99.9) | 99.7 (98.8-100.0) | 97.0 (95.2-98.2) |
| Asymptomatic | 100.0 (71.5-100.0) | 100.0 (71.5-100.0) | 100.0 (69.2-100.0) | 100.0 (99.4-100.0) | 99.5 (98.7-99.9) | 95.7 (93.8-97.2) |
| All types | ||||||
| Symptomatic | 97.8 (96.3-98.8) | 97.1 (95.3-98.3) | 92.3 (88.6-95.0) | 99.6 (99.4-99.8) | 99.4 (99.1-99.7) | 97.5 (96.7-98.1) |
| Asymptomatic | 92.1 (85.5-96.3) | 90.0 (81.9-95.3) | 90.7 (79.7-96.9) | 99.8 (99.6-99.9) | 99.6 (99.3-99.8) | 97.2 (96.4-97.9) |
| Overall | 96.9 (95.4-98.1) | 96.1 (94.3-97.4) | 92.0 (88.7-94.6) | 99.7 (99.6-99.8) | 99.5 (99.3-99.7) | 97.3 (96.8-97.8) |
GP AC2, GenProbe Aptima Combo 2; BD ProbeTec, Becton Dickinson ProbeTec; ABT m2000, Abbott m2000.
CC, clinician collected; SC, self-collected.
NA, not applicable.
Positive and negative predictive values.
The hypothetical positive predictive values (PPVs) and negative predictive values (NPVs) were calculated for this assay at prevalence rates ranging from 0.5 to 30% for C. trachomatis and N. gonorrhoeae and are shown in Table 9.
TABLE 9.
Hypothetical positive and negative predictive values for C. trachomatis and N. gonorrhoeae at various prevalence levels for the Abbott RealTime CT/NG assay
| Prevalence rate (%) | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|
| C. trachomatis | ||||
| 0.5 | 92.4 | 99.2 | 36.7 | 100.0 |
| 1.0 | 92.4 | 99.2 | 53.8 | 99.9 |
| 2.0 | 92.4 | 99.2 | 70.2 | 99.8 |
| 5.0 | 92.4 | 99.2 | 85.9 | 99.6 |
| 10.0 | 92.4 | 99.2 | 92.8 | 99.2 |
| 15.0 | 92.4 | 99.2 | 95.3 | 98.7 |
| 20.0 | 92.4 | 99.2 | 96.7 | 98.1 |
| 25.0 | 92.4 | 99.2 | 97.5 | 97.5 |
| 30.0 | 92.4 | 99.2 | 98.0 | 96.8 |
| N. gonorrhoeae | ||||
| 0.5 | 96.9 | 99.7 | 61.9 | 100.0 |
| 1.0 | 96.9 | 99.7 | 76.5 | 100.0 |
| 2.0 | 96.9 | 99.7 | 86.8 | 99.9 |
| 5.0 | 96.9 | 99.7 | 94.4 | 99.8 |
| 10.0 | 96.9 | 99.7 | 97.3 | 99.7 |
| 15.0 | 96.9 | 99.7 | 98.3 | 99.5 |
| 20.0 | 96.9 | 99.7 | 98.8 | 99.2 |
| 25.0 | 96.9 | 99.7 | 99.1 | 99.0 |
| 30.0 | 96.9 | 99.7 | 99.3 | 98.7 |
Self-collected vaginal swab survey results.
Upon completing the entire specimen collection process, female subjects were asked to complete a survey evaluating the vaginal swab self-collection process. This evaluation included the Abbott self-collected vaginal swab instructions and the ease of use of the Abbott multi-Collect specimen collection kit. Surveys were completed by 99.75% (2,009 of 2,014) of the female subjects. Survey results were not available for five subjects. Not all subjects provided responses to each question. The written instructions were understood by 99.25% (1,994 of 2,009) of respondents, and 96.27% (1,934 of 2,009) found the diagrams helpful. The self-collection process was successfully completed by 98.26% (1,974 of 2,009) of the respondents. The majority of respondents found the transport tube and swab contained within the kit easy to use. The self-collected vaginal swab was most preferred by respondents (30.51%), followed by the urine specimen (26.18%). Respondents least preferred the clinician-collected vaginal swab (13.89%). No preference in specimen collection was expressed by 29.87% (600 of 2,009) of respondents. Abbott RealTime CT/NG results for the self-collected vaginal swab were considered invalid and excluded from the analyses for respondents who failed to perform the self-collection in accordance with the instructions and who did not request another kit to perform a new collection.
DISCUSSION
In this multicenter trial, a new NAAT platform, the Abbott RealTime CT/NG assay, for the real-time, simultaneous detection of chlamydia and gonorrhea was evaluated. Based on two comparator assays, its clinical sensitivity and specificity were excellent for all specimen types, including a self-administered vaginal swab. Overall sensitivities and specificties of 92.4% and 99.2%, respectively, for C. trachomatis and 96.9% and 99.7%, respectively, for N. gonorrhoeae provided exceptional performance. The assay performed well for both asymptomatic and symptomatic patients. Of note is the superior sensitivity of male urine over male urethral swabs for asymptomatic men, confirming urine as the recommended sample of choice for testing men (1, 4). The very high sensitivity and specificity of vaginal swabs, whether clinician or self-collected, compared to the lower sensitivity of female urine also supported the vaginal swab as the recommended specimen of choice in screening women for both chlamydia and gonorrhea (1). The lack of any statistical difference between the sensitivity and specificity between clinician-collected and self-collected vagina swabs also indicated that self-collected vaginal swabs are excellent screening specimens.
Using a patient infected status (PIS), defined by having positive test results from two different commercially available platforms, the GenProbe Aptima Combo 2 and the Becton Dickinson ProbeTec, and/or two different test types in the case of gonorrhea, was useful in demonstrating the overall performance of this real-time PCR assay. The definition of the PIS has been an evolving estimate for a number of years as assay platforms and test types have evolved for molecular testing (10). This conservative definition of a PIS based on two different comparator platforms has the possible effect of providing slightly higher sensitivity estimates and slightly lower estimates of specificity than if only two tests from any one (or more) assay type are used (10), as has been done for earlier diagnostic trials for chlamydia and gonorrhea (5, 17, 18).
The somewhat lower sensitivity for female urine samples compared to that of vaginal samples has been noted in other studies of NAATs as well (5-7, 17). What was interesting was that there were a number of participants that were positive by only the urine sample for chlamydia. This apparent “urethral dysuria syndrome” has been described in several reports where there are specimens that are positive only from the urine and not from the cervix (8, 11-15).
The assay was extremely reproducible in the precision study and could accurately detect organisms from high to very low organism loads as well as in mixtures of high- and low-concentration combinations of C. trachomatis and N. gonorrhoeae. This is particularly important in detecting mixed infections.
The ease of use of the automated sample preparation and extraction process and the rapid work flow are also conducive to efficient laboratory handling, as the hands-on time is approximately 1 h, 15 min, and low compared to those of other NAAT platforms, and the time to results is approximately 6 h (16).
In summary, the Abbott RealTime CT/NG assay performed on the m2000 platform was highly accurate, reproducible, sensitive, and specific. Nucleic acid amplification tests are the most sensitive assays available to date for detecting chlamydia and gonorrhea in clinical specimens, and the RealTime CT/NG assay adds to the group of commercially available assays that are available to laboratories as choices for superior diagnostic performance.
TABLE 3.
Subjects' medical history
| Criterion | No. of subjects | % of subjectsa |
|---|---|---|
| Antibiotic use in the past 21 days | ||
| Yes | 12b | 0.31 |
| No | 3,820 | 99.69 |
| Currently menstruating | ||
| Yes | 104 | 2.71 |
| No | 1,910 | 49.84 |
| Currently pregnant | ||
| Yes | 221 | 5.77 |
| No | 1,793 | 46.79 |
Total = 3,832 subjects.
Subjects did not meet study inclusion criteria.
Acknowledgments
We thank all the participants in the trial, as well as the investigators and clinicians at the participating collection sites: Julie King at Medford Women's Clinic, Medford, OR; Jeffery Rosen at Clinical Research of South Florida, Coral Gables, FL; Catherine Dean, Medex Healthcare Research Inc., St. Louis, MO; Druid Clinic, Baltimore, MD; Eastern Clinic, Baltimore, MD; Johns Hopkins Adult Emergency Department, Baltimore, MD; Leandro Mena, Crossroads Clinic, Jackson, MS; Wayne A. Duffus, Richland County Department of Health STD Clinic, Columbia, SC; Jose G. Castro, Miami-Dade County Health Department STD Clinic, Miami, FL; William McCormack, SUNY Healthcare Service Center, Kings County Hospital STD Clinic, Brooklyn, NY; Tyrone Malloy, Soapstone Center for Clinical Research, Decatur, GA; Gary Simon, George Washington University Medical Faculty Associates, Washington, DC; Julie Ann Buenting, Howard Brown Clinic, Chicago, IL; Melanie Thompson, AIDS Research Consortium of Atlanta (ARCA), Atlanta, GA; Richard Elion, Whitman-Walker Clinic, Washington, DC; and Clifford Kinder, Kinder Medical Group, Miami, FL. We also thank Hong Yu for her assistance in the manuscript preparation.
This study was funded by Abbott Molecular, Des Plaines, IL.
Footnotes
Published ahead of print on 28 July 2010.
REFERENCES
- 1.Association of Public Health Laboratories. 2009. Guidelines for the laboratory testing of STDs. APHL/CDC panel summary reports: laboratory diagnostic testing for Chlamydia trachomatis and Neisseria gonorrhoeae and laboratory diagnostic testing for Treponema pallidum. APHL, Atlanta, GA. http://www.aphl.org/aphlprograms/infectious/std/Pages/stdtestingguidelines.aspx.
- 2.Centers for Disease Control and Prevention. 2010. Sexually transmitted disease surveillance, 2008. Centers for Disease Control and Prevention, Atlanta, GA.
- 3.Chernesky, M. A., D. H. Martin, E. W. Hook III, D. Willis, J. Jordan, S. Wang, J. R. Lane, D. Fuller, and J. Schachter. 2005. Ability of new APTIMA CT and APTIMA GC assays to detect Chlamydia trachomatis and Neisseria gonorrhoeae in male urine and urethral swabs. J. Clin. Microbiol. 43:127-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaydos, C. A., D. V. Ferrero, and J. Papp. 2008. Laboratory aspects of screening men for Chlamydia trachomatis in the new millennium. Sex. Transm. Dis. 35(Suppl.):S45-S50. [DOI] [PubMed] [Google Scholar]
- 5.Gaydos, C. A., T. C. Quinn, D. Willis, A. Weissfeld, E. W. Hook, D. H. Martin, D. V. Ferrero, and J. Schachter. 2003. Performance of the APTIMA Combo 2 assay for the multiplex detection of Chlamydia trachomatis and Neisseria gonorrheae in female urine and endocervical swab specimens. J. Clin. Microbiol. 41:304-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hook, E. W., III, S. F. Ching, J. Stephens, K. F. Hardy, K. R. Smith, and H. H. Lee. 1997. Diagnosis of Neisseria gonorrhoeae infections in women by using the ligase chain reaction on patient-obtained vaginal swabs. J. Clin. Microbiol. 35:2129-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hook, E. W., III, K. Smith, C. Mullen, J. Stephens, L. Rinehart, M. S. Pate, and H. H. Lee. 1997. Diagnosis of genitourinary Chlamydia trachomatis infections by using the ligase chain reaction on patient-obtained vaginal swabs. J. Clin. Microbiol. 35:2133-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marrazzo, J. M., R. E. Johnson, T. A. Greem, W. E. Stamm, J. Schachter, G. Bolan, E. W. Hook III, R. B. Jones, D. H. Martin, M. E. St. Louis, and C. M. Black. 2005. Impact of patient characteristics on performance of nucleic acid amplification tests and DNA probe for detection of Chlamydia trachomatis in women with genital infections. J. Clin. Microbiol. 43:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin, D. H., C. Cammarata, B. Van Der Pol, R. B. Jones, T. C. G. C. A. Quinn, K. Crotchfelt, J. Schachte, J. Moncada, D. Jungkind, B. Turner, and C. Peyton. 2000. Multicenter evaluation of AMPLICOR and automated COBAS AMPLICOR CT/NG tests for Neisseria gonorrhoeae. J. Clin. Microbiol. 38:3544-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin, D. H., M. Nsuami, J. Schachter, E. W. Hook III, D. Ferrero, T. C. Quinn, and C. Gaydos. 2004. Use of multiple nucleic acid amplification tests to define the infected patient “gold standard” in clinical trials of new diagnostic tests for Chlamydia trachomatis infections. J. Clin. Microbiol. 42:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moncada, J., J. Schachter, E. W. Hook III, D. Ferrero, C. Gaydos, T. C. Quinn, D. Willis, A. Weissfeld, and D. H. Martin. 2004. The effect of urine testing in evaluations of the sensitivity of the Gen-Probe APTIMA Combo 2 assay on endocervical swabs for Chlamydia trachomatis and Neisseria gonorrhoeae: the infected patient standard reduces sensitivity of single site evaluation. Sex. Transm. Dis. 31:273-277. [DOI] [PubMed] [Google Scholar]
- 12.Morris, R. E., J. Lagault, and C. Baker. 1993. Prevalence of isolated urethral asymptomatic Chlamydia trachomatis infection in the absence of cervical infection in incarcerated adolescent girls. Sex. Transm. Dis. 20:198-200. [DOI] [PubMed] [Google Scholar]
- 12a.NCCLS. 2002. EPI0-A2 guideline. NCCLS, Wayne, PA.
- 13.Paavonen, J. 1979. Chlamydia trachomatis-induced urethritis in female partners of men with nongonococcal urethritis. Sex. Transm. Dis. 6:69-71. [DOI] [PubMed] [Google Scholar]
- 14.Pasternack, R., P. Vuorinen, T. Pitkajarvi, M. Koskela, and A. Miettinen. 1997. Comparison of manual Amplicor PRC, Cobas Amplicor PCR, and LcX assays for detection of Chlamydia trachomatis infection in women by using urine specimens. J. Clin. Microbiol. 35:402-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shafer, M. A., J. Moncada, C. B. Boyer, K. Betsinger, S. D. Flinn, and J. Schachter. 2003. Comparing first-void urine specimens, self-collected vaginal swabs, and endocervical specimens to detect Chlamydia trachomatis and Neisseria gonorrhoeae by a nucleic acid amplification test. J. Clin. Microbiol. 41:4395-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Der Pol, B. 2009. Beyond the result: enhancing CT/GC amplification testing, p. 21. Assoc. Mol. Pathol. 19 to 22 November 2009, Kissimmee, FL.
- 17.Van Der Pol, B., D. Ferrero, L. Buck-Barrington, E. Hook III, C. Lenderman, T. C. Quinn, C. A. Gaydos, J. Moncada, G. Hall, M. J. Tuohy, and B. R. Jones. 2001. Multicenter evaluation of the BDProbeTec ET system for the detection of Chalmydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J. Clin. Microbiol. 39:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Der Pol, B., T. C. Quinn, C. A. Gaydos, K. Crotchfelt, J. Schachte, J. Moncada, D. Jungkind, D. H. Martin, B. Turner, C. Peyton, and R. B. Jones. 2000. Evaluation of the AMPLICOR and automated COBAS AMPLICOR CT/NG tests for the detection of Chlamydia trachomatis. J. Clin. Microbiol. 38:1105-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]