Abstract
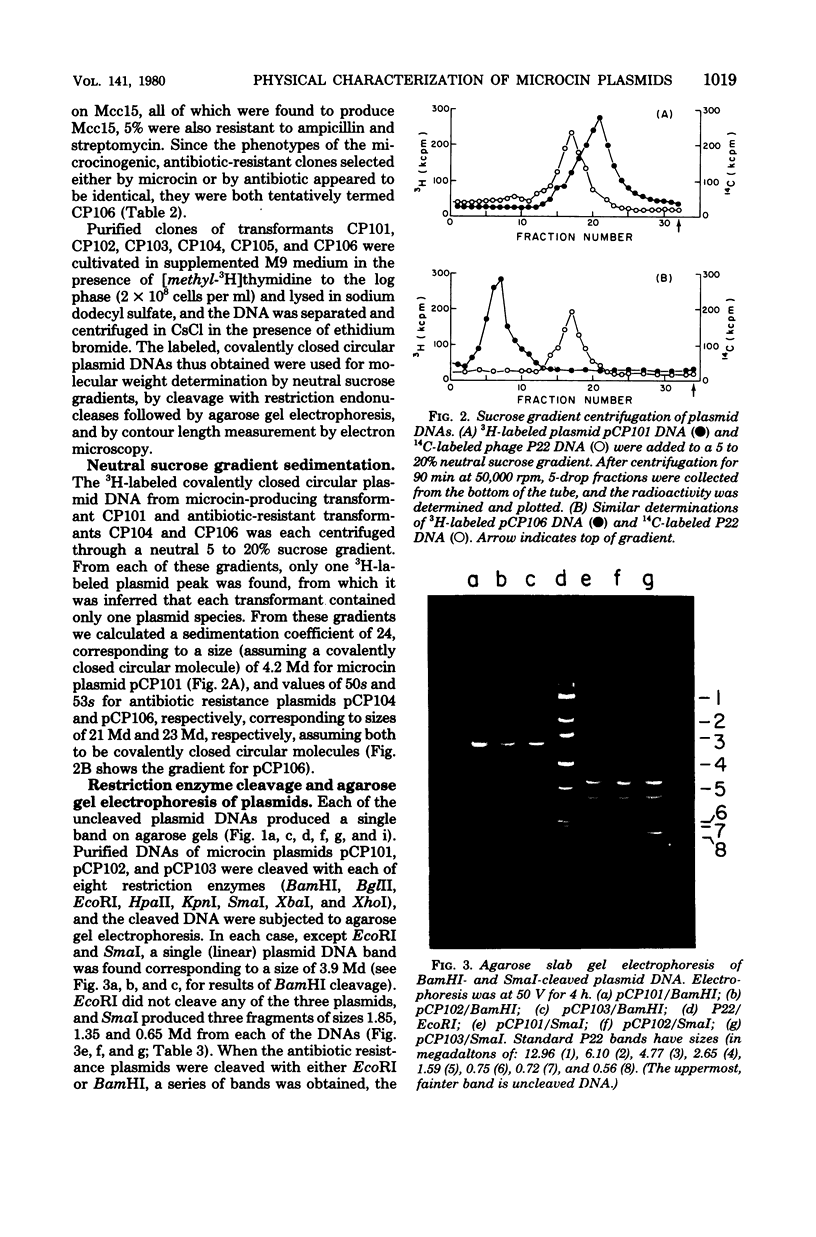
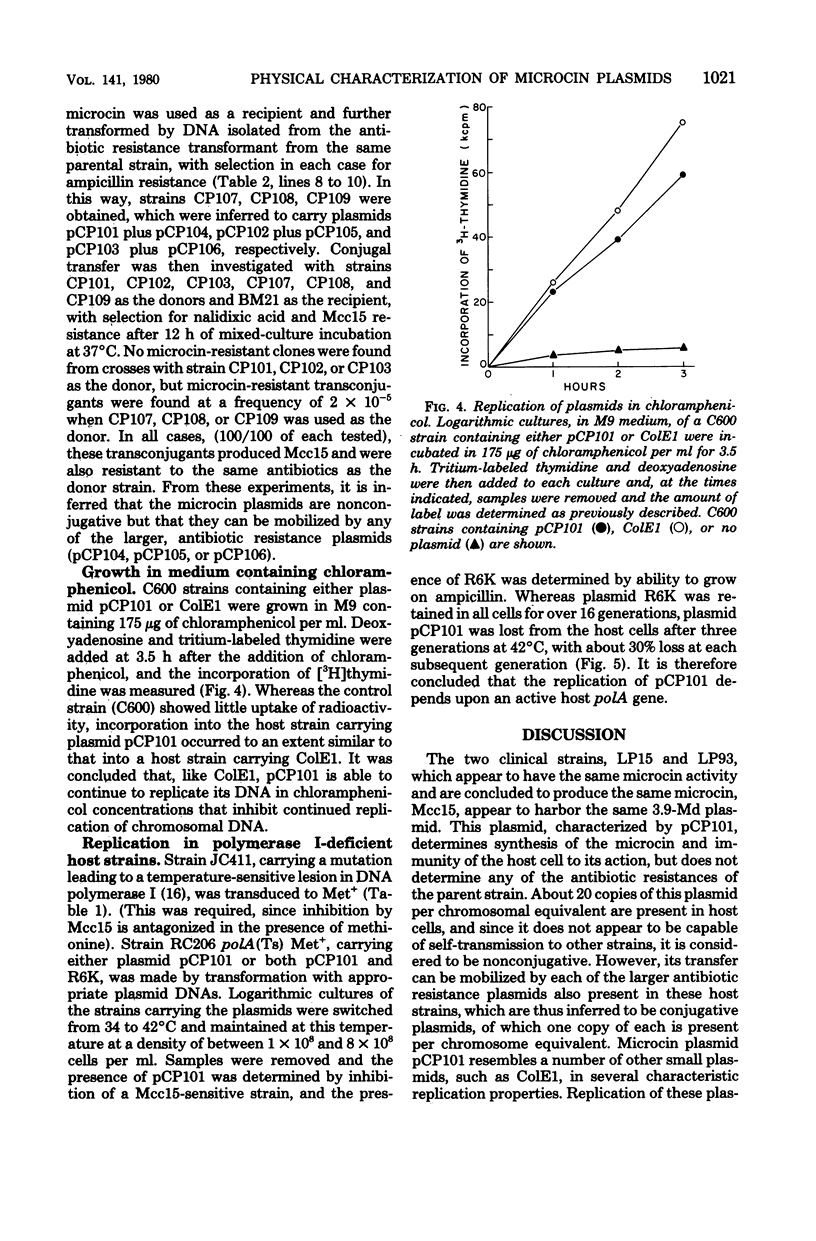
Plasmid deoxyribonucleic acid (DNA) isolated from each of three antibiotic-resistant clinical strains of Escherichia coli producing the same microcin showed multiple bands upon agarose gel electrophoresis. Transformants selected either for microcin resistance or ampicillin resistance yielded plasmid DNA corresponding in size to only one of the multiple bands. Plasmids, isolated from all three hosts, which determined microcin resistance and microcin production measured about 4 megadaltons by sucrose density, restriction enzyme, and contour length analyses; cleavage of the DNAs by each of eight restriction enzymes showed the same response, and DNA-DNA hybridization indicated complete homology. The antibiotic resistance plasmids of the three host strains were uniformly larger, were of different sizes, and showed different restriction enzyme cleavage patterns. One of these R plasmids (pCP106) also determined the synthesis of the same microcin, and DNA-DNA hybridization studies indicated an approximate 2.4-megadalton homology with the 4-megadalton microcin plasmid pCP101. The microcin plasmids were present at approximately 20 copies per genome equivalent and were nonconjugative, whereas the R plasmids had a copy number of about 1, were conjugative, and could mobilize the microcin plasmid. Microcin plasmid pCP101 showed replication properties similar to those of a number of small multicopy plasmids such as ColE1.
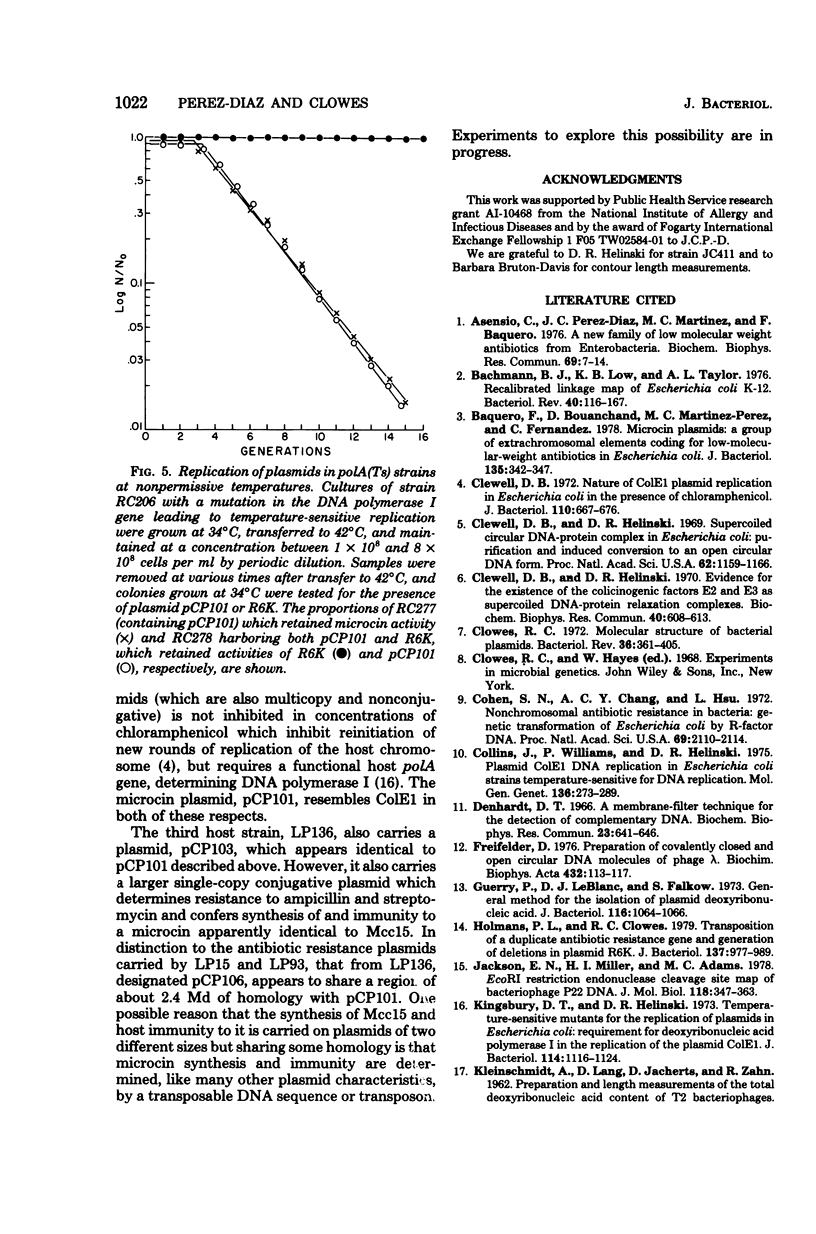
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asensio C., Pérez-Díaz J. C. A new family of low molecular weight antibiotics from enterobacteria. Biochem Biophys Res Commun. 1976 Mar 8;69(1):7–14. doi: 10.1016/s0006-291x(76)80264-1. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F., Bouanchaud D., Martinez-Perez M. C., Fernandez C. Microcin plasmids: a group of extrachromosomal elements coding for low-molecular-weight antibiotics in Escherichia coli. J Bacteriol. 1978 Aug;135(2):342–347. doi: 10.1128/jb.135.2.342-347.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Evidence for the existence of the colicinogenic factors E2 and E3 as supercoiled circular DNA-protein relaxation complexes. Biochem Biophys Res Commun. 1970 Aug 11;40(3):608–613. doi: 10.1016/0006-291x(70)90947-2. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J., Williams P., Helinski D. R. Plasmid ColE1 DNA replication in Escherichia coli strains temperature-sensitive for DNA replication. Mol Gen Genet. 1975;136(4):273–289. doi: 10.1007/BF00341713. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Preparation of covalently closed and open circular DNA molecules of phage lambda. Biochim Biophys Acta. 1976 Apr 15;432(1):113–117. doi: 10.1016/0005-2787(76)90047-2. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P. L., Clowes R. C. Transposition of a duplicate antibiotic resistance gene and generation of deletions in plasmid R6K. J Bacteriol. 1979 Feb;137(2):977–989. doi: 10.1128/jb.137.2.977-989.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E. N., Miller H. I., Adams M. L. EcoRI restriction endonuclease cleavage site map of bacteriophage P22DNA. J Mol Biol. 1978 Jan 25;118(3):347–363. doi: 10.1016/0022-2836(78)90233-4. [DOI] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. Temperature-sensitive mutants for the replication of plasmids in Escherichia coli: requirement for deoxyribonucleic acid polymerase I in the replication of the plasmid ColE 1 . J Bacteriol. 1973 Jun;114(3):1116–1124. doi: 10.1128/jb.114.3.1116-1124.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LURIA S. E., HUMAN M. L. A nonhereditary, host-induced variation of bacterial viruses. J Bacteriol. 1952 Oct;64(4):557–569. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D., Mitani M. Simplified quantitative electron microscopy of biopolymers. Biopolymers. 1970;9(3):373–379. doi: 10.1002/bip.1970.360090310. [DOI] [PubMed] [Google Scholar]
- Lang D. Molecular weights of coliphages and coliphage DNA. 3. Contour length and molecular weight of DNA from bacteriophages T4, T5 and T7, and from bovine papilloma virus. J Mol Biol. 1970 Dec 28;54(3):557–565. doi: 10.1016/0022-2836(70)90126-9. [DOI] [PubMed] [Google Scholar]
- McGilvray D., Umbarger H. E. Regulation of transaminase C synthesis in Escherichia coli: conditional leucine auxotrophy. J Bacteriol. 1974 Nov;120(2):715–723. doi: 10.1128/jb.120.2.715-723.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisioka T., Mitani M., Clowes R. C. Molecular recombination between R-factor deoxyribonucleic acid molecules in Escherichia coli host cells. J Bacteriol. 1970 Jul;103(1):166–177. doi: 10.1128/jb.103.1.166-177.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Clowes R. C., Cohen S. N., Curtiss R., 3rd, Datta N., Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976 Mar;40(1):168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]