Abstract
A pregnant sea lion stranded in the State of Washington was found to have placentitis caused by a unique strain of Coxiella burnetii. This is the first description of coxiellosis in a sea lion and suggests that exposure to sea lions may be a risk factor for contracting Q fever.
CASE REPORT
A pregnant adult female Steller sea lion (Stranding number WDFW2008-058) was found dead on the beach at Westhaven State Park in Westport, WA (lat 46.8981, long 124.1307), on 9 June 2008. The adult female had an estimated weight of 250 to 300 kg and was in fair body condition. Based on the condition of the body and the frequency of beach surveys in the area, the time since death was estimated at approximately 3 days. The animal was in moderate postmortem condition. The abdominal cavity was distended by the enlarged uterus, a near-term fetus, and approximately 2 liters of creamy, brown-pink exudate. Fish bones were dispersed throughout the omentum, and there was pronounced thickening and opacification of the omentum and mesentery. The spleen was moderately enlarged, and there was pronounced injection of the meningeal vasculature, with scattered hemorrhage throughout the surface of the brain. Gross necropsy of the fetus disclosed a near-term male with a weight of 19 kg. The fetus was in good body condition. The liver was friable, with scattered hemorrhage noted randomly through the lobes. The meningeal vessels were congested. Samples of representative organs were fixed in 10% buffered formalin, embedded in wax, sectioned at 5 μm, and stained with hematoxylin and eosin.
The most significant histological findings of the adult female were moderate diffuse lymphoid hyperplasia of the lymph nodes, diffuse pulmonary edema, and mild encephalitis. The fetus showed edema, with aspirated squames through the pulmonary parenchyma. The placenta also showed aggregates of enlarged trophoblasts within the chorioallantoic villi that had vacuolated granular cytoplasm.
Ancillary diagnostic testing was performed at the British Columbia Animal Health Center. Bacterial cultures on blood agar showed light growth of Enterococcus spp. within the fetus lung, brain, spleen, and small intestine and the adult uterus. This was attributed to postmortem overgrowth. Cultures from adult and fetal small intestines in selenite broth followed by plating on XLT4 and Hektoen enteric agar tested negative for Salmonella spp. Pooled tissue (lymph node, lung, spleen, brain, uterus, and thymus) for both the adult and fetus was negative by PCR for Brucella spp. and morbillivirus (3, 5, 8). Brain, heart, and skeletal muscle samples from the adult and the fetus were submitted to the Laboratory of Parasitic Diseases, NIAID, Bethesda, MD, for coccidian PCR testing. Toxoplasma gondii was detected in the adult brain, heart, and skeletal muscle and in the fetal heart by PCR (13). Sarcocystis neurona was also detected in the adult skeletal muscle and in the fetal heart and mediastinal lymph node (12). S. neurona was not detected in the brain of either the mother or the fetus.
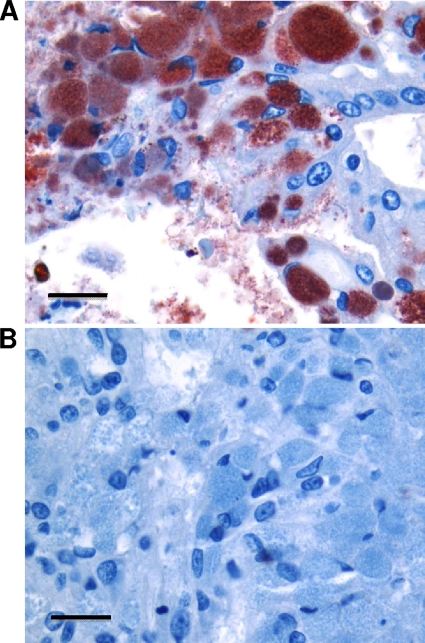
Based on the histopathologic changes in the placenta and on a previous report of placentitis due to Coxiella burnetii in a harbor seal (Phoca vitulina richardsi) (10), formalin-fixed sea lion placental tissue was submitted to the Washington Animal Disease and Diagnostic Laboratory (WADDL), Pullman, WA, for immunohistochemistry analysis (4), which revealed abundant infection of placental trophoblast cells with coccobacilli that were strongly immunoreactive with a polyclonal antibody against C. burnetii (Fig. 1). The immunohistochemistry results were confirmed at the CDC using a polyclonal antibody preparation specific for C. burnetii (data not shown).
FIG. 1.
Immunohistochemical detection of Coxiella burnetii-like bacteria in Steller sea lion placenta. Placenta sections were subjected to immunoperoxidase immunohistochemical staining with polyclonal antiserum to C. burnetii (A) or irrelevant, negative-control antiserum (B). Sections were counterstained with Mayer's hematoxylin. Immunoreactive C. burnetii organisms were located within the cytoplasm and on the apical surfaces of placental trophoblasts (red). This analysis was performed according to a published protocol (4) using a 1:600 dilution of human immune serum against C. burnetii as the primary antibody. Positive-control tissue consisted of placenta from a sheep with confirmed C. burnetii placentitis based upon immunohistochemistry and PCR analysis (data not shown). Bar = 16 μm.
Genomic DNA was purified from the infected sea lion placenta using the QIAamp DNA minikit (Qiagen, Valencia, CA), and PCR was performed to test for multiple C. burnetii genes. The gene targets and primer sequences for PCR are listed in Table 1. Primers targeting the com1, djlA (previously called mucZ), CBU_678, CBU_686, and IS1111A genes from C. burnetii all produced a product. The positive results from these 5 different C. burnetii-specific PCR assays provide molecular evidence for C. burnetii infection in the sea lion placenta.
TABLE 1.
Gene targets and primer sequences for PCR
| Gene target | Gene product | Primers and probe | Oligonucleotide sequence | Detectora | Cycling conditions | Reference |
|---|---|---|---|---|---|---|
| com1 | 27-kDa outer membrane protein | COM1 TaqMan fwd | 5′-AATAAAAACCTCCGCGTTGTCTT-3′ | FAM | 50°C, 2 min | This study |
| COM1 TaqMan rev | 5′-TTGGCAGCGTATTGCGATT-3′ | 95°C, 10 min | ||||
| COM1 probe | 5′-AAAGAACTGCCCATTTTTGGCGGC-3′ | 40 cycles of 95°C, 15 s; 60°C, 60 s | ||||
| com1 | 27-kDa outer membrane protein | Com-1 | 5′-CGTGAAGAACCGTTTGACTG-3′ | Gel | Per reference | 16 |
| Com-4 | 5′-CTTTTCTACCCGGTCGATTTC-3′ | |||||
| djlA | Mucoidy activation protein (MucZ) | Muc-1 | 5′-CGGTGATGAACTGGATTGG-3′ | Gel | Per reference | 16 |
| Muc-4 | 5′-AACCATGCTTCGCACCTTAC-3′ | |||||
| CBU_678 | Putative ADP heptose synthase | PH1SPECF | 5′-AAGCCCTCGATTCATTTTT-3′ | SYBR green | 94°C, 3 min | This study |
| PH1SPECR | 5′-CGCATCACCAGCACCCACAC-3′ | 40 cycles of 94°C, 30 s; 55°C, 30 s; 72°C, 30 s | ||||
| CBU_686 | Pyruvate dehydrogenase | 686F | 5′-TCAGTAGCCATCGAGCACATG-3′ | SYBR green | 94°C, 3 min | This study |
| 686R | 5′-CAGTGGATGCCTTGAGCTTTT-3′ | 40 cycles of 94°C, 30 s; 55°C, 30 s; 72°C, 30 s | ||||
| IS1111a | Multicopy insertion sequence | IS1111F | 5′-CCGATCATTTGGGCGCT-3′ | FAM | 50°C, 2 min | 11 |
| IS1111R | 5′-CGGCGGTGTTTAGGC-3′ | 95°C, 10 min | ||||
| IS1111 probe | 5′-TTAACACGCCAAGAAACGTATCGCTGTG-3′ | 40 cycles of 95°C, 15 s; 60°C, 60 s | ||||
| 16S rRNA gene | rRNA | 16SUPF | 5′-GACGCGTAAAATAGCCATCCAT-3′ | Gel | 95°C, 10 min | This study |
| 16SUPR | 5′-TTAGCCCGAGTTTCCCCAGGTTAT-3′ | 40 cycles of 94°C, 30 s; 56°C, 30 s; 72°C, 60 s | ||||
| 16S rRNA gene | rRNA | CB1F | 5′-ACATGCAAGTCGAACGGCAGCG-3′ | Gel | 95°C, 10 min | This study |
| CB1R | 5′-CATACTCAAGATACCCAGTATCG-3′ | 40 cycles of 94°C, 30 s; 56°C, 30 s; 72°C, 60 s | ||||
| 16S rRNA gene | rRNA | 16SmidF | 5′-TAATCGGAATCACTGGGCGTAAAG-3′ | Gel | 95°C, 10 min | This study |
| 16SmidR | 5′-TTCCGAGGATGTCAAGGGTAGGTA-3′ | 40 cycles of 94°C, 30 s; 56°C, 30 s; 72°C, 60 s | ||||
| 16S rRNA gene | rRNA | 16S3′F | 5-′TGGGGAGCAAACAGGATTAGAGAC-3′ | Gel | 95°C, 10 min | This study |
| 16S3′R | 5′-CATGGTGTGACGGGCGGTGTG-3′ | 40 cycles of 94°C, 30 s; 56°C, 30 s; 72°C, 60 s | ||||
| 16S rRNA gene | rRNA | 16SDNF | 5′-CCGGAGGAAGGTGGGGATGATGT-3′ | Gel | 95°C, 10 min | This study |
| 16SDNR | 5′-CTGAGCTATGGCCCCGAGATGGTG-3′ | 40 cycles of 94°C, 30 s; 56°C, 30 s; 72°C, 60 s |
FAM, 6-carboxyfluorescein; Gel, ethedium bromide-stained agarose gel.
Portions of the com1 and djlA genes and bases 1 to 1482 of the 16S rRNA gene were PCR amplified and sequenced from genomic DNA derived from the infected placenta. For com1, a 624-bp fragment was sequenced (from nucleotides 59 to 682). The sequence of this fragment was 96.6% identical to com1 from the C. burnetii reference strain Nine Mile phase 1 (strain RSA 493), having 21-bp changes. In a published comparison of com1 sequences among 37 isolates of C. burnetii (16), none of the 37 sequences differed from the Nine Mile sequence by more than 3 bp. The sequence of a 724-bp fragment (from nucleotides 42 to 765) of the sea lion Coxiella djlA gene was different from the Nine Mile strain sequence at 20 sites (97.2% identity). Among the 37 isolates mentioned above, none of the djlA gene sequences differed from the Nine Mile sequence at more than 4 bases (16). Thus, the sequence of the com1 and djlA genes reveals a strain of Coxiella that is remarkably distinct from the characterized strains of C. burnetii.
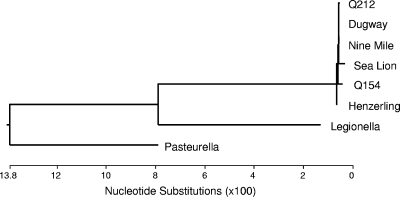
The sequences of 16S rRNA genes among the five fully sequenced strains of C. burnetii differ from the Nine Mile reference strain at only 0 bases, 1 base, or 2 bases. The 16S rRNA gene from the sea lion strain of C. burnetii was sequenced, and it matched the Nine Mile reference sequence at 1,478/1,482 nucleotides (99.7% identical). A phylogenetic tree was constructed using the 16S rRNA sequences of the sea lion strain, 5 sequenced strains of C. burnetii, and the near-neighbor species Legionella pneumophila and Pasteurella multocida (Fig. 2). When the 16S rRNA genes of these other species are compared to known C. burnetii strains and the strain infecting the sea lion, all of the C. burnetii strains, including the sea lion strain, fall into a closely related group. Although it is distinct from other strains of C. burnetii by having 4 nucleotide substitutions instead of 1 or 2, this 16S rRNA sequence is much more like C. burnetii than other near-neighbor species.
FIG. 2.
Phylogenetic tree of 16S rRNA gene sequences from 5 characterized C. burnetii isolates, sea lion Coxiella, and two closely related bacterial species (Legionella pneumophila and Pasteurella multocida). DNA sequencing was performed on purified PCR products. The 16S rRNA gene was amplified using the 5 different primer sets listed in Table 1. Sequences were analyzed and the phylogenetic tree was constructed using the Lasergene 8 suite of nucleic acid analysis software (DNAStar, Madison, WI). Sequences used to construct the tree had the following GenBank accession numbers: for Legionella pneumophila, AE017354; for Pasteurella multocida, AE004439; for Nine Mile, NC 002971; for Henzerling, NC 010117; for Q154, NC 011528; for Dugway, NC 009727; and for Q212, NC 011527. The sequences obtained for this study have the following GenBank accession numbers: for com1, GU797241; for djlA, GU797242; and for 16S rRNA, GU797243.
Coxiella burnetii causes Q fever, a worldwide zoonosis. Acute Q fever is a febrile illness, with hepatitis or pneumonia found in more severe cases (6). Chronic Q fever usually presents as a culture-negative endocarditis. Recent outbreaks of Q fever have been associated with exposure to C. burnetii via inhalation of aerosolized bacteria from infected livestock (cows, sheep, or goats) (1, 14, 15). C. burnetii can replicate to high levels in the placentas of infected animals and is an established endemic abortifacient in goats and sheep (2, 9). A recent case of chronic Q fever in Greenland in a resident whose primary animal exposure was to sled dogs and seals raises the question of whether marine mammals expose humans to C. burnetii (7). The only previous description of C. burnetii infection in a marine mammal was a report of C. burnetii placentitis in a Pacific harbor seal found in California (10). The findings for this Steller sea lion expand the geographic and host range of C. burnetii and show that C. burnetii can replicate to high levels in the placentas of marine mammals, just as it does in terrestrial mammals. This case therefore suggests that persons working with or living near populations of pregnant Steller sea lions could be exposed to concentrated aerosols of C. burnetii and be at risk for Q fever.
Although the precise source of bacterial exposure could not be determined in this case, the uniqueness of this sea lion Coxiella strain makes it possible that this bacterium has a unique and novel marine cycle. Further studies are needed to determine the impact of C. burnetii on the overall health of the marine mammal population. For this sea lion, it is not known if this infection resulted in any clinical symptoms or if the infection contributed to its stranding. Concurrent detection of mild encephalitis and detection of the protozoan pathogens T. gondii and S. neurona may also be significant. Serological studies of marine mammal populations will be needed to determine if C. burnetii infection is widespread among marine mammals.
Acknowledgments
Special thanks go to WDFW/MMI and Cascadia Research Collective staff and stranding interns for their assistance, particularly Josh Oliver, Jessie Huggins, Bethany Diehl, and Lil Luce. We also thank Sherif Zaki and Clifton Drew of the CDC Infectious Diseases Pathology Branch for confirmation of immunohistochemistry.
The NOAA Fisheries John H. Prescott Marine Mammal Rescue Assistance Grant Program, Washington Department of Fish and Wildlife, and Cascadia Research Collective provided funding and support for these research activities.
Footnotes
Published ahead of print on 30 June 2010.
REFERENCES
- 1.Bamberg, W. M., W. J. Pape, J. L. Beebe, C. Nevin-Woods, W. Ray, H. Maguire, J. Nucci, R. F. Massung, and K. Gershman. 2007. Outbreak of Q fever associated with a horse-boarding ranch, Colorado, 2005. Vector Borne Zoonotic Dis. 7:394-402. [DOI] [PubMed] [Google Scholar]
- 2.Berri, M., E. Rousset, J. L. Champion, P. Russo, and A. Rodolakis. 2007. Goats may experience reproductive failures and shed Coxiella burnetii at two successive parturitions after a Q fever infection. Res. Vet. Sci. 83:47-52. [DOI] [PubMed] [Google Scholar]
- 3.Bricker, B. J., D. R. Ewalt, A. P. MacMillan, G. Foster, and S. Brew. 2000. Molecular characterization of Brucella strains isolated from marine mammals. J. Clin. Microbiol. 38:1258-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dilbeck, P. M., and T. F. McElwain. 1994. Immunohistochemical detection of Coxiella burnetii in formalin-fixed placenta. J. Vet. Diagn. Invest. 6:125-127. [DOI] [PubMed] [Google Scholar]
- 5.Hammond, J. A., P. P. Pomeroy, A. J. Hall, and V. J. Smith. 2005. Identification and real-time PCR quantification of Phocine distemper virus from two colonies of Scottish grey seals in 2002. J. Gen. Virol. 86:2563-2567. [DOI] [PubMed] [Google Scholar]
- 6.Hartzell, J. D., R. N. Wood-Morris, L. J. Martinez, and R. F. Trotta. 2008. Q fever: epidemiology, diagnosis, and treatment. Mayo Clinic Proc. 83:574-579. [DOI] [PubMed] [Google Scholar]
- 7.Koch, A., C. B. Svendsen, J. J. Christensen, H. Bundgaard, L. Vindfeld, C. B. Christiansen, M. Kemp, and S. Villumsen. 2010. Q fever in Greenland. Emerg. Infect. Dis. 16:511-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krafft, A., J. H. Lichy, T. P. Lipscomb, B. A. Klaunberg, S. Kennedy, and J. K. Taubenberger. 1995. Postmortem diagnosis of morbillivirus infection in bottlenose dolphins (Tursiops truncatus) in the Atlantic and Gulf of Mexico epizootics by polymerase chain reaction-based assay. J. Wildl. Dis. 31:410-415. [DOI] [PubMed] [Google Scholar]
- 9.Lang, G. H. 1990. Coxiellosis (Q fever) in animals, p. 23-48. In T. J. Marrie (ed.), Q fever, vol. I. The disease. CRC Press, Boca Raton, FL. [Google Scholar]
- 10.Lapointe, J. M., F. M. Gulland, D. M. Haines, B. C. Barr, and P. J. Duignan. 1999. Placentitis due to Coxiella burnetii in a Pacific harbor seal (Phoca vitulina richardsi). J. Vet. Diagn. Invest. 11:541-543. [DOI] [PubMed] [Google Scholar]
- 11.Loftis, A. D., W. K. Reeves, D. E. Szumlas, M. M. Abbassy, I. M. Helmy, J. R. Moriarity, and G. A. Dasch. 2006. Rickettsial agents in Egyptian ticks collected from domestic animals. Exp. Appl. Acarol. 40:67-81. [DOI] [PubMed] [Google Scholar]
- 12.Mansfield, L. S., H. C. Schott II, A. J. Murphy, M. G. Rossano, S. M. Tanhauser, J. S. Patterson, K. Nelson, S. L. Ewart, J. V. Marteniuk, D. D. Bowman, and J. B. Kaneene. 2001. Comparison of Sarcocystis neurona isolates derived from horse neural tissue. Vet. Parasitol. 95:167-178. [DOI] [PubMed] [Google Scholar]
- 13.Owen, M. R., and A. J. Trees. 1998. Vertical transmission of Toxoplasma gondii from chronically infected house (Mus musculus) and field (Apodemus sylvaticus) mice determined by polymerase chain reaction. Parasitology 116:299-304. [DOI] [PubMed] [Google Scholar]
- 14.Porten, K., J. Rissland, A. Tigges, S. Broll, W. Hopp, M. Lunemann, U. van Treeck, P. Kimmig, S. O. Brockmann, C. Wagner-Wiening, W. Hellenbrand, and U. Buchholz. 2006. A super-spreading ewe infects hundreds with Q fever at a farmers' market in Germany. BMC Infect. Dis. 6:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schimmer, B., F. Dijkstra, P. Vellema, P. M. Schneeberger, V. Hackert, R. ter Schegget, C. Wijkmans, Y. van Duynhoven, and W. van der Hoek. 2009. Sustained intensive transmission of Q fever in the south of the Netherlands, 2009. Euro Surveill. 14:19210. [DOI] [PubMed] [Google Scholar]
- 16.Sekeyova, Z., V. Roux, and D. Raoult. 1999. Intraspecies diversity of Coxiella burnetii as revealed by com1 and mucZ sequence comparison. FEMS Microbiol. Lett. 180:61-67. [DOI] [PubMed] [Google Scholar]