Abstract
The emergence of a previously undefined phage type of Salmonella enterica serovar Typhimurium, designated DT191a, occurred in England and Wales in July 2008. The new strain exhibits a number of distinctive phenotypic and genotypic features. This report provides the tools necessary to track S. Typhimurium DT191a globally.
Salmonella enterica serovar Typhimurium is the second most prevalent serovar isolated in Europe, exceeded only by Salmonella enterica serovar Enteritidis (6). Provisional data for 2008 to July 2009 from the Health Protection Agency Salmonella data set show that S. Typhimurium accounted for 2,690 (18.8%) of Salmonella infections from humans in England and Wales. Subtyping of S. Typhimurium by phage typing is well established and not only plays a vital role in detecting outbreaks (11) but also enables the appearance of new phage types or phage types with new characteristics to be monitored.
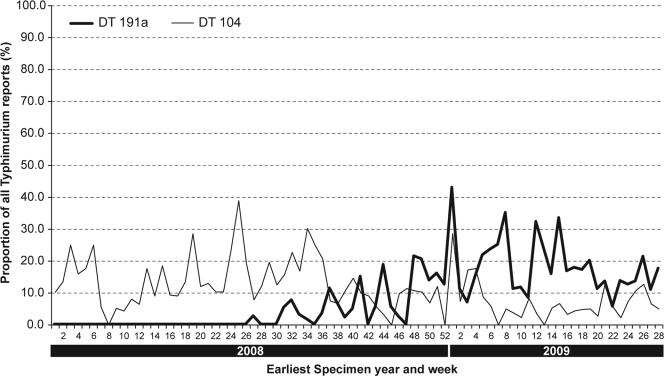
We report here on the emergence, in mid-2008, in England and Wales of a previously undefined phage type of S. Typhimurium and on the further characterization of this phage type by pulsed-field gel electrophoresis (PFGE), variable-number tandem repeat (VNTR) typing, and multilocus sequence typing (MLST). This strain was initially identified in our laboratory, as it did not conform to any of the recognized patterns in the current schemes as described by Callow (4) and Anderson et al. (1). Its unique pattern of phage lysis was subsequently defined as DT191a. The full phage reaction pattern with the panel of 32 typing phages is shown in Table 1, where the only significant difference is in the reaction for phage 18. To date, over 230 cases of DT191a have been typed by the Laboratory of Gastrointestinal Pathogens (LGP), Health Protection Agency (HPA). In England and Wales, DT191a is currently the most common phage type of S. Typhimurium obtained from cases of human infection. The increased isolation rate of this phage type is compared with the level for DT104 in Fig. 1.
TABLE 1.
Phage reactions for the new phage type S. Typhimurium DT191a
| Phage type | Result for indicated phagea |
|||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 32 | 35 | |
| DT191 | SCL | CL | − | SCL | CL | +++ | CL | − | +++ | CL | ++ | +++ | CL | CL | SCL | CL | SCL | CL | ++ | CL | CL | SCL | − | CL | SCL | CL | ± | OL | ++ | OL |
| DT191a | CL | ++ | − | OL | CL | +++ | CL | − | +++ | +++ | −/+ | −/+ | CL | CL | CL | CL | − | ++ | + | OL | OL | ++ | − | CL | SCL | ++ | − | +++ | ++ | OL |
Variable degrees of reaction: −, no reaction; ±, 1 to 20 plaques; +, 21 to 40 plaques; ++, 41 to 80 plaques; +++, 81 to 100 plaques; SCL, semiconfluent lysis; CL, confluent clear lysis; OL, confluent opaque lysis; −/+, variable reaction.
FIG. 1.
Relative proportions of Salmonella enterica serovar Typhimurium DT104 and DT191a typed at the Salmonella Reference Laboratory, United Kingdom, between 2008 and 2009.
In addition to phage typing, all isolates were tested for resistance to a range of antimicrobials by a breakpoint method (8). Of these isolates (n = 231), 226 were resistant only to tetracyclines when the breakpoint of 8 mg/liter was used. Four isolates also carried additional resistance to one or more antimicrobials, and one isolate was fully sensitive. The new strain was therefore defined as resistant to tetracyclines.
Biochemical analysis revealed that all the isolates produced reactions typical of subspecies I, apart from an inability to utilize the sugar dulcitol.
Isolates that react with S. Typhimurium-specific phages are routinely reported as S. Typhimurium without further typing. However, for this study, 17 isolates were fully serotyped according to the Kauffmann-White scheme (9). Two isolates gave reactions typical of serovar Typhimurium, with the antigenic structure 4,5,12:i:1,2. The remainder were monophasic (4,5,12:i:-), as the second-phase flagellar antigen was not detectable by agglutination.
A subset of diphasic and monophasic DT191a isolates were confirmed as S. Typhimurium by MLST, as described by Kidgell et al. (12). When compared with the data in the Salmonella enterica MLST database at the University College Cork (http://mlst.ucc.ie/mlst/dbs/Senterica), all 22 isolates tested by this method were found to be sequence type ST19, regardless of whether they were monophasic or fully serotypeable. ST19 is the central sequence type for S. Typhimurium and is shared by a variety of other S. Typhimurium phage types, including DT104 (13). This result demonstrates that both mono- and diphasic isolates are of the same lineage within S. enterica and supports the suggestion from phage typing that they are typical S. Typhimurium isolates.
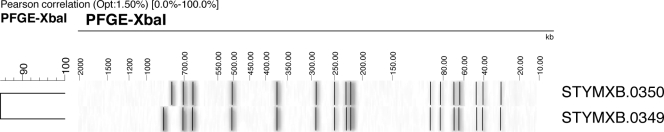
Further subtyping by PFGE using the restriction enzyme XbaI was performed on a selection of isolates (n = 16) as previously described (7, 17). With the exception of a single example, these isolates were shown to have the same PFGE profile, which was designated STYMXB.0350. Pattern STYMXB.0349 (n = 1) differed by the presence of a larger upper band at approximately 800 kb (Fig. 2). Although PFGE has sometimes shown limitations within certain phage types of S. Typhimurium (5, 15), both profiles were new and had not previously been seen in England or Wales or within the PulseNet Europe database.
FIG. 2.
Variation in PFGE profiles noted between isolates of Salmonella serovar Typhimurium DT191a. Opt., optimized.
VNTR typing on a total of 73 isolates was performed using a modified version of the method of Lindstedt et al. (16) as described by Best et al. (2). VNTR profiles were assigned based on the fragment size amplified from each locus (14). For the majority of isolates tested (63/73 isolates), a new single VNTR type, 2-11-5-8-212, was identified (order STTR9-STTR5-STTR6-STTR10-STTR3). However, single-locus variants (SLVs) were observed in 10 isolates on the basis of differences at any of three loci, STTR5, STTR10, and STTR3. SLVs were the result of either the loss or the gain of a single 6-bp repeat at locus STTR5 or locus STTR10 or the gain or loss of up to two 27-bp or 33-bp repeats at locus STTR3 (Table 2). VNTR typing appeared to be more discriminatory than the other methods used here, as seven different VNTR types were observed within the unique PFGE profile STYMXB.0350. Other studies have noted that VNTRs may evolve so rapidly that multiple profiles emerge during an outbreak (10). However, the instability of VNTR loci reported by Call et al. (3) shows that changes are limited to a single locus and allele. This may be reflected by the fact that any changes detected in the loci of the isolates in this study were small. As one VNTR type was predominant (63/73 isolates), it would appear that the VNTR profiles have been relatively stable during the course of this outbreak.
TABLE 2.
Association of VNTR and PFGE profiles
| No. of human cases (n = 73) | VNTR profilea | PFGE designation |
|---|---|---|
| 62 | 2-11-5-8-212 | STYMXB.0350 |
| 1 | 2-11-5-8-212 | STYMXB.0349 |
| 5 | 2-12-5-8-212 | STYMXB.0350 |
| 1 | 2-11-5-8-211 | STYMXB.0350 |
| 1 | 2-10-5-8-212 | STYMXB.0350 |
| 1 | 2-11-5-8-112 | STYMXB.0350 |
| 1 | 2-11-5-8-012 | STYMXB.0350 |
| 1 | 2-11-5-9-212 | STYMXB.0350 |
Differences from the common VNTR profile are highlighted in bold.
Typing of Salmonella isolates responsible for human and animal infections is important for surveillance and outbreak investigations, with many demands being placed on the typing methods used. While a high level of discriminatory power may be required so that unrelated and related isolates can be identified, too much variability complicates the interpretation of the typing data in relation to epidemiologic information (18, 19). All isolates referred to the LGP, HPA, are identified and typed using a variety of phenotypic and molecular methods and are tested for susceptibility to a wide range of antimicrobial drugs. As a result of this, a database of types and subtypes of salmonellas has been developed over a number of decades such that results can be regularly analyzed and reported upon. The type reported here, DT191a, may be an emerging strain of Salmonella with the potential to expand further. Both phenotypically and genotypically, it would appear to be a new variant of S. Typhimurium that is still in circulation within England and Wales. As well as being a new phage type, DT191a exhibits a number of other characteristic features, including resistance to tetracyclines, an inability to utilize the sugar dulcitol, PFGE profile STYMXB.0350, MLST ST19, and typically VNTR type 2-11-5-8-212.
It is currently not known if this type occurs more widely, as not all countries have an organized salmonella surveillance system which reports the resistance patterns, phage types, and molecular subtypes of S. Typhimurium from a variety of sources. Cooperation between veterinary and human public health agencies should enable both rapid detection and control of newly emerging pathogens. S. Typhimurium DT191a may have clinical and biological significance, and the monitoring of its emergence could have important implications for public health control. Whether or not this particular phage type continues to be seen in England and Wales, further characterization and continued surveillance will be important steps for improving our understanding of this strain.
Acknowledgments
We are grateful to the staff at the Salmonella Reference Unit, HPA, and all laboratories in England and Wales that refer isolates of Salmonella for identification and typing.
Satheesh Nair is funded by the MRC, United Kingdom.
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Anderson, E. S., L. R. Ward, M. J. DeSaxe, and J. D. H. De Sa. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. (Lond.) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Best, E. L., B. A. Lindstedt, A. Cook, F. A. Clifton-Hadley, E. J. Threlfall, and E. Liebana. 2007. Multiple-locus variable-number tandem repeat analysis of Salmonella enterica subsp. enterica serovar Typhimurium: comparison of isolates from pigs, poultry and cases of human gastroenteritis. J. Appl. Microbiol. 103:565-572. [DOI] [PubMed] [Google Scholar]
- 3.Call, D. R., L. Orfe, M. A. Davis, S. Lafrentz, and M. S. Kang. 2008. Impact of compounding error on strategies for subtyping pathogenic bacteria. Foodborne Pathog. Dis. 5:505-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callow, B. R. 1959. A new phage-typing scheme for Salmonella typhimurium. J. Hyg. (Lond.) 57:346-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doran, G., D. Morris, C. O'Hare, N. DeLappe, B. Bradshaw, and G. Corbett-Feeney. 2005. Cost-effective application of pulsed-field gel electrophoresis to typing of Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 71:8236-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.European Food Safety Authority. 2009. The community summary report on trends and sources of zoonoses and zoonotic agents in the European Union in 2007. EFSA J. 223:1-215. [Google Scholar]
- 7.Fisher, I. S. T., and E. J. Threlfall. 2005. The Enter-net and Salm-gene databases of food-borne bacterial pathogens causing human infections in Europe and beyond: an international collaboration in the development of intervention strategies. Epidemiol. Infect. 133:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frost, J. A. 1994. Testing for resistance to antibacterial drugs, p. 73-82. In H. Chart (ed.), Methods in practical laboratory bacteriology. CRC Press, New York, NY.
- 9.Grimont, P. A. D., and F. X. Weill. 2007. Antigenic formulas of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France. http://www.pasteur.fr/ip/portal/action/WebdriveActionEvent/oid/01s-000036-089.
- 10.Hopkins, K. L., C. Maguire, E. Best, E. Liebana, and E. J. Threlfall. 2007. Stability of multiple-locus variable-number tandem repeats in Salmonella enterica serovar Typhimurium. J. Clin. Microbiol. 45:3058-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kafatos, G., N. Andrews, I. A. Gillespie, A. Charlett, G. K. Adak, E. De Pinna, and E. J. Threlfall. 2009. Impact of reduced numbers of isolates phage-typed on the detection of Salmonella outbreaks. Epidemiol. Infect. 137:821-827. [DOI] [PubMed] [Google Scholar]
- 12.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 13.Kingsley, R. A., C. L. Msefula, N. R. Thomson, S. Kariuki, K. E. Holt, M. A. Gordon, D. Harris, L. Clarke, S. Whitehead, V. Sangal, K. Marsh, M. Achtman, M. E. Molyneux, M. Cormican, J. Parkhill, C. A. MacLennan, R. S. Heyderman, and G. Dougan. 2009. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19:2279-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsson, J. T., M. Torpdahl, R. F. Petersen, G. Sorensen, B. A. Lindstedt, and E. M. Nielsen. 2009. Development of a new nomenclature for Salmonella Typhimurium multilocus variable number of tandem repeats analysis (MLVA). Euro Surveill. 14:pii=19174. [PubMed] [Google Scholar]
- 15.Lawson, A. J., M. Desai, S. J. O'Brien, R. H. Davies, L. R. Ward, and E. J. Threlfall,2004. Molecular characterisation of an outbreak strain of multiresistant Salmonella enterica serovar Typhimurium DT104 in the UK. Clin. Microbiol. Infect. 10:143-147. [DOI] [PubMed] [Google Scholar]
- 16.Lindstedt, B. A., T. Vardund, L. Aas, and G. Kapperud. 2004. Multiple-locus variable-number tandem repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59:163-172. [DOI] [PubMed] [Google Scholar]
- 17.Peters, T. M. 2009. Pulsed-field gel electrophoresis for molecular epidemiology of food pathogens, p. 59-70. In D. A. Caugant (ed.), Molecular epidemiology of microorganisms: methods in molecular biology, vol. 551. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 18.Torpdahl, M., G. Sørensen, B. A. Lindstedt, and E. M. Nielsen. 2007. Tandem repeat analysis for surveillance of human Salmonella Typhimurium infections. Emerg. Infect. Dis. 13:388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Belkum, A. 1999. The role of short sequence repeats in epidemiologic typing. Curr. Opin. Microbiol. 2:306-311. [DOI] [PubMed] [Google Scholar]