Abstract
In the context of hematopoietic stem cell transplantation, adenovirus infections are associated with relevant mortality and morbidity. Detection of adenovirus DNA by quantitative PCR is the “gold standard” for these patients. A total of 150 samples, namely, 78 whole-blood, 22 cerebrospinal fluid, 24 digestive biopsy, and 26 stool samples, from 29 patients, including 24 hematopoietic stem cell transplant recipients, were tested for the detection of adenovirus using an in-house real-time quantitative PCR assay (A. Heim, C. Ebnet, G. Harste, and P. Pring-Akerblom, J. Med. Virol. 70:228-239, 2003) and the commercially available Adenovirus R-Gene kit. Adenovirus DNA was automatically isolated from whole-blood samples (Magna Pure LC system; Roche) or was manually extracted from other specimens (QIAamp; Qiagen) using the appropriate kit. The intra- and interassay reproducibilities and sensitivities were evaluated with cell culture supernatant dilutions. Of the 150 samples tested, 86 were found to be positive and 55 were found to be negative using both techniques. Nine (6%) discordant results were obtained. In most cases, discrepant results concerned samples with low viral loads. Quantitative results for all concordant positive samples were analyzed using the Spearman correlation test. A good correlation between the results of the in-house assay and those of the kit assay was obtained (r = 0.95; P < 0.001). Regarding the threshold cycle value for internal control spiked samples, none of the 150 samples tested contained a PCR inhibitor. In conclusion, a relevant correlation of results between the in-house assay and the kit assay, as well as the high-quality reproducibility and sensitivity of the kit assay, warranted its use for follow-up of hematopoietic stem cell transplantation recipients.
Human adenoviruses are nonenveloped double-stranded DNA viruses. To date, 55 different serotypes have been identified and grouped into seven different species (Human Adenovirus A through Human Adenovirus G) (13, 14, 30, 31). In immunocompetent patients, adenovirus infections lead to benign enteric or respiratory symptoms; and direct diagnosis of local infection by cell culture, indirect immunofluorescence, or immunochromatography is adequate (25). Adenoviruses may persist in a latent form after infection, and adenovirus reactivation may cause fatal infections in immunosuppressed patients (18).
In the context of hematopoietic stem cell transplantation (HSCT) between matched unrelated donors, adenovirus infections are associated with relevant mortality and morbidity in correlation with a delayed adenovirus-specific cellular immune response. The pathogenesis of adenovirus disease is associated with an early stage of viral dissemination in whole blood (WB) from the respiratory or gastrointestinal tract and allows the detection of adenovirus in the peripheral blood compartment (6).
Clinical management of adenovirus infection in HSCT recipients is based on the early diagnosis of adenovirus infection and preemptive treatment with antiviral drugs (i.e., cidofovir) (2) or adoptive immunotherapy strategies (19). In this case, detection of adenovirus DNA by quantitative PCR (qPCR) is the “gold standard” for these patients. On the one hand, the sensitivity of molecular methods allows early detection of adenovirus disseminated infection, defined by the presence of adenovirus DNA in whole blood (9, 23), and helps prevent severe clinical virological complications in HSCT recipients (11). On the other hand, precise quantification of adenovirus in whole blood is a useful indicator of adenovirus disease evolution after HSCT (7). In addition, adenovirus DNA detection in stool samples by PCR could be associated with adenovirus DNA detection in whole blood. Combining these two parameters provides an optimal tool for managing the risk of adenovirus-associated morbidity and mortality (22). Adenovirus is also detected in cerebrospinal fluid (CSF) and digestive biopsy specimens, since adenovirus can be implicated in central nervous system infections in immunocompromised patients (32) and since the most frequently involved site of adenovirus infection is the gastrointestinal tract (5).
Several in-house PCRs have been described to detect all or some of the 55 adenovirus serotypes (4, 10, 12, 28) and are of clinical interest for adenovirus detection in immunosuppressed patients, but no kit for quantitative detection of adenovirus was commercially available, until recently.
A total of 150 samples, including CSF, digestive biopsy, whole-blood, and stool specimens from 29 patients were tested for the detection and quantification of the adenovirus load using an in-house real-time qPCR assay (12) and the Adenovirus R-Gene kit, in order to validate the use of this recent commercially available kit for clinical application.
MATERIALS AND METHODS
Samples.
This study included a total of 150 samples, namely, 78 whole-blood, 24 digestive biopsy, 22 CSF, and 26 stool samples. All samples were obtained from 29 patients during routine virological detection over 2 years (2008 and 2009). Most of the patients (n = 24) had undergone HSCT. The DNA extracted from the samples was stored at −80°C until quantification by either the in-house assay or the kit assay.
DNA purification.
The DNA extraction procedure differed according to sample type.
DNA was extracted from 200 μl of CSF samples using the blood and body spin fluid protocol and a QIAamp DNA blood minikit (Qiagen, Courtaboeuf, France), as recommended by the manufacturer. Total DNA was eluted in a final volume of 50 μl.
DNA was automatically isolated from 200 μl of whole blood using a MagNa Pure LC system (Roche, Meylan, France) and the DNA blood high-performance protocol, according to the manufacturer's instructions. DNA was eluted in 100 μl.
Stools from HSCT recipients were stored at −20°C until retrospective DNA quantification. DNA was extracted from a known quantity of stool sample comprised between 180 and 220 mg using a QIAamp DNA stool minikit (Qiagen), according to the manufacturer's instructions. DNA eluted in 200 μl was stored at −80°C. In addition, four stool samples were extracted using an adapted protocol in the MagNa Pure LC system instrument (Roche) and the DNA blood high-performance protocol: a weighed sample of stools (200 ± 10 mg) was diluted in 2 ml of distilled water and centrifuged at 3,000 × g for 20 min, and the supernatant was then centrifuged at 20,000 × g for 10 min.
The digestive biopsy specimens were ground in 500 μl of distilled water. A mixture of 15 μl of proteinase K (20 μg/μl) and 100 μl of ATL tissue lysis buffer (Qiagen) was added to 200 μl of the ground biopsy specimen, and the whole blend was incubated for 3 h at 56°C. The DNA extraction was then performed using the QIAamp DNA blood minikit (Qiagen), as described above for CSF samples.
Adenovirus real-time qPCR assay.
The two adenovirus quantification assays were carried out on the same DNA extract using the same instrument, the ABI Prism 7000 instrument (Applied Biosystems, Courtaboeuf, France), according to the manufacturer's instructions. Highly positive samples were diluted before quantification.
In-house assay.
The real-time assay was routinely performed as described by Heim et al. (12) on an ABI Prism 7000 sequence detection system (Applied Biosystems), using 2× TaqMan mix (Applied Biosystems). qPCR amplified a sequence of a conserved region of the adenovirus hexon gene using the forward primer 5′-GCC ACG GTG GGG TTT CTA AAC TT-3′ and the reverse primer 5′-GCC CCA GTG GTC TTA CAT GCA CAT C-3′. The TaqMan probe sequence was 5′-6-carboxyfluorescein (FAM)-TGC ACC AGA CCC GGG CTC AGG TAC TCC GA-6-carboxytetramethylrhodamine-3′. The quantification threshold was 30 copies of adenovirus genome per 10 μl of DNA extract. A plasmid DNA containing a partial human adenovirus serotype 2 hexon sequence, kindly provided by A. Heim, was serially diluted and used as a quantification standard for the real-time qPCR. Appropriate negative and positive controls were included in all the experiments. There was no internal control included in this assay.
Commercial assay.
A kit containing the new commercially available real-time PCR test, the Adenovirus R-Gene kit (Argene Inc., NY), was retrospectively used with previously extracted DNA. This kit was developed to be used for analysis of whole-blood, plasma, bronchoalveolar lavage, nasopharyngeal secretion, and stool specimens. The kit assay is a 5′-nuclease real-time PCR targeting the hexon gene of adenovirus. The adenovirus ready-to-use premix contains the primers, probe, polymerase, and buffer needed for specific amplification. Four quantification standards with 50, 500, 5,000, and 50,000 copies/reaction mixture, respectively, are included in the kit, as are sensitivity-control (10 copies/reaction mixture), negative-control, and internal-control PCR assays.
The kit assay is validated for detection of serotypes 1 to 52. Discrepant results were systematically controlled using the in-house assay.
Appropriate negative and positive controls were included in all the experiments.
Results expression.
Results were expressed as the numbers of copies per milliliter of whole blood and the numbers of copies per gram of stool specimen. The results obtained with CSF and digestive biopsy samples were qualitative because they are likely to be influenced by the sampling procedure and the sample size.
Intra- and interassay reproducibilities and sensitivity threshold of the Adenovirus R-Gene kit.
The sensitivity and the precision of the kit assay were evaluated using a 10-fold dilution, in molecular biology-grade water, of DNA extracted from the adenovirus serotype 2 cell culture supernatant. Intra-assay variability was determined by amplifying a serial dilution (undiluted to a 1:106 dilution) of DNA extract in five replicate assays, whereas interassay variability was determined by amplifying the six dilutions in six different experiments.
Statistical analysis.
The Spearman correlation test was performed using Statview software for Windows (version 5; Abacus Concepts). P values below 0.05 were considered significant in all analyses.
RESULTS
Detection of PCR inhibitors.
All 150 biological samples (CSF, biopsy, whole-blood, and stool samples) were tested for PCR inhibitors using the PCR included in the kit assay. PCR inhibition was defined by spiking the samples with an internal control that resulted in a threshold cycle (CT) that was considered to be validated if it was equal to the CT expected value ± 2 cycles. None of the 150 samples tested contained a PCR inhibitor.
Correlation between in-house qPCR assay and the Adenovirus R-Gene kit. (i) Qualitative results.
Detection of adenovirus DNA in digestive biopsy specimens and CSF was analyzed only in a qualitative way in order that the results could be compared to the routinely acquired results. Among the 22 CSF samples, 1 sample with a negative result using the in-house assay presented a positive result using the kit assay. In addition to this, 2 digestive biopsy samples of the 24 tested presented discrepant results between the two assays; they were negative using the kit assay and positive using the in-house assay (Table 1). Those three samples with discordant results presented very low viral loads and had CT values greater than 35 cycles (range, 35.43 to 42.92 cycles).
TABLE 1.
Comparison of qualitative detection of adenovirus DNA in different samples using an in-house real-time qPCR assay and the Adenovirus R-Gene kita
| Sample type | No. of samples giving: |
|||||
|---|---|---|---|---|---|---|
| Concordant results |
Discordant results |
|||||
| Detectable | Nondetectable | Total | Detectable using in-house assay | Detectable using kit assay | Total | |
| Cerebrospinal fluid (n = 22) | 1 | 20 | 21 | 1 | 0 | 1 |
| Digestive biopsy specimen (n = 24) | 11 | 11 | 22 | 2 | 0 | 2 |
| Whole blood (n = 78) | 55 | 19 | 74 | 2 | 2 | 4 |
| Stools (n = 26) | 19 | 5 | 24 | 0 | 2 | 2 |
| All samples (n = 150) | 86 | 55 | 141 | 5b | 4c | 9 |
The in-house real-time qPCR assay is that of Heim et al. (12).
The CT values for discrepant samples were 35.43, 35.53, 38.25, 40.63, and 33.62.
The CT values for discrepant samples were 42.92, 37.66, 40.68, and 38.96.
The qualitative results obtained from whole-blood and stool samples (manually or automatically extracted) were discrepant for four whole-blood specimens (two positive using the kit assay and two positive using the in-house assay) and two stool specimens (Table 1). The CT values were also very high (range, 33.62 to 40.68 cycles). Overall, nine samples (6%) presented discrepant results, whatever type of sample was used. All samples with discrepant results obtained using both assays were controlled by repeated performance of the in-house assay and the kit assay, and only samples with repeated discrepant results with both assays were considered discrepant.
(ii) Quantitative results.
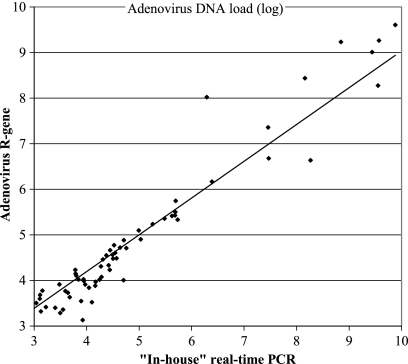
Adenovirus DNA was quantified using 104 stool samples and whole-blood specimens. The adenovirus loads in the tested samples ranged from 1.35 to 9.88 log10 units and were expressed as the numbers of copies/ml in whole-blood samples and as the numbers of copies/g in stool samples, with the stool samples being weighed before the extraction step. Samples with viral loads higher than 50,000 copies/reaction mixture were diluted before quantification. A good correlation between the adenovirus DNA loads obtained using the kit assay and the in-house assay described by Heim et al. (12) was found (Fig. 1). The Spearman correlation coefficient was found to be 0.83 (P < 0.0001).
FIG. 1.
Comparison of quantitative results for adenovirus loads in blood and stool samples using an in-house real-time qPCR assay (12) and the Adenovirus R-Gene kit.
The median difference between the viral loads obtained using the commercial and the in-house assays was 0.19 log10 copy/ml (range, 0.02 to 0.7 log10 copy/ml) for whole-blood samples and 0.31 log10 copy/g (range, 0.2 to 1.73 log10 copy/g) for stool samples. Whatever the type of sample, the median difference between the two results was 0.2 log10 copy, considering that a difference of <0.5 log10 copy is nonsignificant for clinical diagnosis.
Performance characteristics of commercial kit.
The sensitivity of the kit assay was determined using serial dilutions of adenovirus DNA purified from a strain of adenovirus serotype 2 isolated from an HSCT recipient (the undiluted DNA was quantified to be 7.55 log10 copies/ml). The coefficient of variation of the CT values ranged from 0.73 to 2.24% for intra-assay repeatability (n = 5 repetitions in the same series) and from 1.09 to 2.40% for interassay reproducibility (n = 6 repetitions in six different PCRs) (Table 2).
TABLE 2.
Intra- and interassay reproducibilities of the Adenovirus R-Gene kit using serial dilutions of an adenovirus serotype 2 strain isolated from a hematopoietic stem cell recipient
| Dilution | Intra-assay results (n = 5) |
Interassay results (n = 6) |
||
|---|---|---|---|---|
| Range of CT values | Coefficient of variation (%) | Range of CT values | Coefficient of variation (%) | |
| Undiluted (3.52 × 107) | 18.49-19.12 | 1.38 | 18.72-19.19 | 1.09 |
| 1:10 (3.52 × 106) | 22.09-22.67 | 0.96 | 22.34-23.03 | 1.15 |
| 1:102 (3.52 × 105) | 25.84-26.27 | 0.73 | 26.21-27.79 | 2.11 |
| 1:103 (3.52 × 104) | 29.00-29.57 | 0.89 | 29.57-30.74 | 1.71 |
| 1:104 (3.52 × 103) | 32.31-33.90 | 1.89 | 32.77-35.16 | 2.40 |
| 1:105 (3.52 × 102) | 35.07-37.10 | 2.24 | NRa | |
| 1:106 (3.52 × 101) | NDb | NR | ||
| Negative control | ND | ND | ||
NR, not realized.
ND, nondetectable.
The threshold of detection was found to be 2 log10 copies/ml, corresponding to 8.8 copies/reaction mixture (volume of reaction mixture, 25 μl).
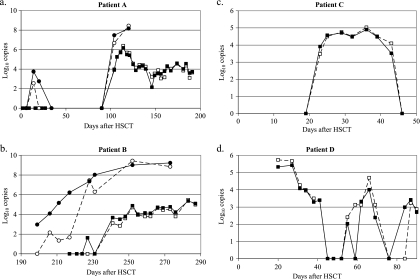
Kinetics of adenovirus load in hematopoietic stem cell recipients.
Adenovirus DNA was quantified in successive samples from four HSCT recipients, identified as patients A to D, using the kit assay and the in-house assay (12).
Whichever test was used, the kinetics of the adenovirus loads in stool and whole-blood samples were parallel. Patient A presented low and then very high viral loads (about 8 log10 copies/g) in stool samples; consistent values were obtained when the results of both qPCR assays with stool as well as whole-blood samples were compared (Fig. 2 a). Patient B presented an increasing viral load in stool samples preceding an increase in the viral load in whole blood. The viral loads in whole blood obtained using both techniques were stackable. In stool specimens, the viral loads quantified using the kit assay were more sensitive and consistent with adenovirus infection evolution (Fig. 2b).
FIG. 2.
Monitoring of adenovirus load in stool (•) and/or blood (▪) samples from four HSCT recipients identified as patients A to D using an in-house real-time qPCR assay (12) (dashed line) or the Adenovirus R-Gene kit (solid line).
For patients C and D, whole-blood samples were tested. Patient C presented an increasing adenovirus load from days 19 to 36 post-HSCT and then a decreasing load up to day 46 post-HSCT, whereas patient D presented decreasing viral loads, fluctuating from 5 log10 copies/ml to undetectable (Fig. 2c and d). The results and kinetics of the viral load were concordant between the two assays.
Adenovirus DNA quantification in whole-blood and stool samples from HSCT recipients using the kit assay allowed diagnosis and follow-up of the adenovirus infections. The increase in the viral load in stool samples preceded detection of the viral load in whole blood.
DISCUSSION
Several in-house methods for adenovirus quantification in different samples have been described (4, 10, 12, 28), and their use has been validated in different clinical situations (7, 11, 15, 16, 20, 21, 24, 27). Previously, some PCR kits validated for qualitative detection of adenovirus were commercially available (i.e., Adenovirus Real-Time PCR [Diagenode sa, Liège, Belgium], Adenovirus PCR kit [Eurobio, Courtaboeuf, France], Adenovirus Real-Time PCR kit [Shanghai ZJ Bio-Tech Co., Ltd., Shanghai, China]). The recently commercialized Adenovirus R-Gene kit allows quantitative detection of adenovirus loads. It had to be validated for clinical use for HSCT recipients. With that objective, 150 CSF, digestive biopsy, stool, or whole-blood samples were selected for adenovirus DNA load detection or quantification using the kit assay and the in-house assay described by Heim et al. (12), already in use for clinical diagnosis. qPCR assays were performed using the ABI Prism 7000 system, but the kit assay has also been validated for use with the most commonly used real-time PCR systems, including the LightCycler 530 and instruments 560 (Roche), the SmartCycler instruments (Cepheid), the ABI FAM and VIC instruments (Applied Biosystems), and the Rotor-gene Green and Yellow instruments (Corbett Research).
Before quantification, adenovirus DNA was extracted using specific automated or manual techniques frequently used in virological laboratories. The automated method was chosen only for DNA extraction from whole blood. For other specimens, specific manual kits were selected for optimal DNA extraction, and a pretreatment step consisting of heating in the presence of proteinase K was added for digestive biopsy samples (data not shown). In addition, in order to permit quantification of adenovirus DNA from stools, stool samples were accurately weighed before DNA purification. The adenovirus load in stools, expressed as the numbers of copies/g of stool, could thus be compared from one technique to another; the kinetics of the adenovirus load in stool samples could also be established.
An internal control, included in the kit assay, allowed PCR-inhibitory agents to be excluded from the samples tested. DNA extraction quality control was also available in the kit, but it could not be used because the PCR was carried out using previously purified, stored DNA. The evaluation of the analytical performance of the kit assay was performed using DNA extracted from an adenovirus serotype 2 strain isolated from an HSCT recipient. The coefficient of variation was less than 5%, regardless of the viral load, for both the intra- and interassay reproducibilities. The sensitivity, evaluated to be 2 log10 copies/ml, is better than the sensitivities obtained using the in-house assay and the plasmid dilution method. The comparison of the results obtained with both methods tested displayed a good correlation for the qualitative and quantitative detection of adenovirus DNA. The rate of agreement by comparison of the results of the two assays was 96% (9 discrepant results/150 samples), whatever specimen was tested (CSF, biopsy, WB, or stool specimens). The type of sample did not influence the rate of agreement between the techniques (Table 1). Since adenovirus DNA detection using the kit assay was carried out retrospectively, discrepant results were systematically controlled using the in-house assay to eliminate false-negative results due to DNA degradation. All the confirmed discrepant results concerned samples with low viral loads. Furthermore, quantitative results obtained using the two different assays were stackable for a large range of values. Thus, both techniques are comparable and reliable for clinical testing of HSCT recipients' samples.
For many years, molecular diagnosis of adenovirus infection by qPCR has been the gold standard for HSCT recipient follow-up. The former method of reference, cell culture, is now used only for determining the serotype of the adenoviruses isolated in epidemiological studies. The diagnosis of adenovirus infection in peripheral sites (i.e., in stool samples or nasal washings) could be performed using immunochromatographic methods; these techniques are quite simple and quick to perform but are insufficiently sensitive to be used in establishing preemptive treatment. Moreover, diagnosis of adenovirus disease, defined by the detection of adenovirus at a sterile site (i.e., CSF or whole blood), could be achieved only using PCR methods. The quantification of the adenovirus DNA load is useful for detecting adenovirus reactivation and infection, which are life-threatening for immunocompromised patients, notably, HSCT recipients. Among HSCT recipients, adults receiving unrelated cord blood transplantation and children are particularly at risk of severe adenovirus reactivation (23, 26, 29). The detection of adenovirus DNA both in whole blood and in stools has been described to be beneficial for the establishment of preemptive antiviral treatment in such patients (8). The results of a study, currently in progress in our center, will increase interest in the use of qPCR with stool samples for the better management of HSCT recipients (unpublished data).
At present, the treatment of adenovirus infection is based on the use of antiviral drugs, notably, cidofovir (3); but a residual high mortality rate may be observed even after patients receive cidofovir (26). The role of adenovirus-specific cellular immune recovery is preponderant in controlling adenovirus infection (22). The current development of adoptive immunotherapy strategies based on iatrogenic reconstitution of expanded virus-specific cytotoxic T lymphocytes provides hope that it will be a means of treatment of HSCT recipients with adenovirus infections (17). The generation of clinical-grade adenovirus-specific cytotoxic T lymphocytes for adoptive T-cell transfer is a delicate process that takes almost 2 weeks (1). The early and precise diagnosis of adenovirus infection using a standardized qPCR will be part of adoptive immunotherapy strategies by predicting adenoviremia in order to initiate the preparation of adenovirus-specific T cells. In addition, the monitoring of antiviral treatment or adoptive immunotherapy is also based on the quantification of the adenovirus load in whole blood (21).
Finally, the results obtained using this commercially available kit are equivalent to those obtained using the widely used in-house assay described by Heim et al. (12). The benefit of this ready-to-use kit is the possibility of standardizing results between clinical laboratories, leading to the possible homogenization of treatment protocols between hospitals. The presence of an internal control in the kit assay is also an advantage, particularly for the determination of the adenovirus load in stools. The kit characteristics and results obtained here warrant the use of the kit assay for HSCT recipient follow-up.
Acknowledgments
We are particularly grateful to A. Heim (Germany) for access to the in-house real-time qPCR protocol and for providing the plasmid DNA. We also thank Martine Didier and Segolene Pernet for excellent technical assistance.
None of us has any financial interest in a business or commercial entity. We have no conflicts of interest to declare.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Aissi-Rothe, L., V. Decot, V. Venard, H. Jeulin, A. Salmon, L. Clement, A. Kennel, C. Mathieu, J. H. Dalle, G. Rauser, C. Cambouris, M. de Carvalho, J. F. Stoltz, P. Bordigoni, and D. Bensoussan. 2010. Rapid generation of full clinical-grade human antiadenovirus cytotoxic T cells for adoptive immunotherapy. J. Immunother. 33:414-424. [DOI] [PubMed] [Google Scholar]
- 2.Bordigoni, P., A. S. Carret, V. Venard, F. Witz, and A. Le Faou. 2001. Treatment of adenovirus infections in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 32:1290-1297. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti, S., K. E. Collingham, R. H. Stevens, D. Pillay, C. D. Fegan, and D. W. Milligan. 2000. Isolation of viruses from stools in stem cell transplant recipients: a prospective surveillance study. Bone Marrow Transplant. 25:277-282. [DOI] [PubMed] [Google Scholar]
- 4.Claas, E. C., M. W. Schilham, C. S. de Brouwer, P. Hubacek, M. Echavarria, A. C. Lankester, M. J. van Tol, and A. C. Kroes. 2005. Internally controlled real-time PCR monitoring of adenovirus DNA load in serum or plasma of transplant recipients. J. Clin. Microbiol. 43:1738-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Mezerville, M. H., R. Tellier, S. Richardson, D. Hebert, J. Doyle, and U. Allen. 2006. Adenoviral infections in pediatric transplant recipients: a hospital-based study. Pediatr. Infect. Dis. J. 25:815-818. [DOI] [PubMed] [Google Scholar]
- 6.Echavarria, M., M. Forman, M. J. van Tol, J. M. Vossen, P. Charache, and A. C. Kroes. 2001. Prediction of severe disseminated adenovirus infection by serum PCR. Lancet 358:384-385. [DOI] [PubMed] [Google Scholar]
- 7.Erard, V., M. L. Huang, J. Ferrenberg, L. Nguy, T. L. Stevens-Ayers, R. C. Hackman, L. Corey, and M. Boeckh. 2007. Quantitative real-time polymerase chain reaction for detection of adenovirus after T cell-replete hematopoietic cell transplantation: viral load as a marker for invasive disease. Clin. Infect. Dis. 45:958-965. [DOI] [PubMed] [Google Scholar]
- 8.Feuchtinger, T., P. Lang, and R. Handgretinger. 2007. Adenovirus infection after allogeneic stem cell transplantation. Leuk. Lymphoma 48:244-255. [DOI] [PubMed] [Google Scholar]
- 9.Flomenberg, P., V. Piaskowski, R. L. Truitt, and J. T. Casper. 1995. Characterization of human proliferative T cell responses to adenovirus. J. Infect. Dis. 171:1090-1096. [DOI] [PubMed] [Google Scholar]
- 10.Gu, Z., S. W. Belzer, C. S. Gibson, M. J. Bankowski, and R. T. Hayden. 2003. Multiplexed, real-time PCR for quantitative detection of human adenovirus. J. Clin. Microbiol. 41:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustafson, I., A. Lindblom, Z. Yun, H. Omar, L. Engstrom, I. Lewensohn-Fuchs, P. Ljungman, and K. Broliden. 2008. Quantification of adenovirus DNA in unrelated donor hematopoietic stem cell transplant recipients. J. Clin. Virol. 43:79-85. [DOI] [PubMed] [Google Scholar]
- 12.Heim, A., C. Ebnet, G. Harste, and P. Pring-Akerblom. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228-239. [DOI] [PubMed] [Google Scholar]
- 13.Ishiko, H., and K. Aoki. 2009. Spread of epidemic keratoconjunctivitis due to a novel serotype of human adenovirus in Japan. J. Clin. Microbiol. 47:2678-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, M. S., B. Harrach, R. D. Ganac, M. M. Gozum, W. P. Dela Cruz, B. Riedel, C. Pan, E. L. Delwart, and D. P. Schnurr. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 81:5978-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalpoe, J. S., P. L. van Der Heiden, R. M. Barge, S. Houtzager, A. C. Lankester, M. J. van Tol, and A. C. Kroes. 2007. Assessment of disseminated adenovirus infection using plasma PCR in adult allogeneic stem cell transplant recipients receiving reduced intensity or myeloablative conditioning. Eur. J. Haematol. 78:314-321. [DOI] [PubMed] [Google Scholar]
- 16.Lankester, A. C., M. J. van Tol, E. C. Claas, J. M. Vossen, and A. C. Kroes. 2002. Quantification of adenovirus DNA in plasma for management of infection in stem cell graft recipients. Clin. Infect. Dis. 34:864-867. [DOI] [PubMed] [Google Scholar]
- 17.Leen, A. M., G. D. Myers, C. M. Bollard, M. H. Huls, U. Sili, A. P. Gee, H. E. Heslop, and C. M. Rooney. 2005. T-cell immunotherapy for adenoviral infections of stem-cell transplant recipients. Ann. N. Y. Acad. Sci. 1062:104-115. [DOI] [PubMed] [Google Scholar]
- 18.Leen, A. M., and C. M. Rooney. 2004. Adenovirus as an emerging pathogen in immunocompromised patients. Br. J. Haematol. 128:135-144. [DOI] [PubMed] [Google Scholar]
- 19.Lenaerts, L., E. De Clercq, and L. Naesens. 2008. Clinical features and treatment of adenovirus infections. Rev. Med. Virol. 18:357-374. [DOI] [PubMed] [Google Scholar]
- 20.Leruez-Ville, M., V. Minard, F. Lacaille, A. Buzyn, E. Abachin, S. Blanche, F. Freymuth, and C. Rouzioux. 2004. Real-time blood plasma polymerase chain reaction for management of disseminated adenovirus infection. Clin. Infect. Dis. 38:45-52. [DOI] [PubMed] [Google Scholar]
- 21.Lion, T., R. Baumgartinger, F. Watzinger, S. Matthes-Martin, M. Suda, S. Preuner, B. Futterknecht, A. Lawitschka, C. Peters, U. Potschger, and H. Gadner. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114-1120. [DOI] [PubMed] [Google Scholar]
- 22.Myers, G. D., C. M. Bollard, M. F. Wu, H. Weiss, C. M. Rooney, H. E. Heslop, and A. M. Leen. 2007. Reconstitution of adenovirus-specific cell-mediated immunity in pediatric patients after hematopoietic stem cell transplantation. Bone Marrow Transplant. 39:677-686. [DOI] [PubMed] [Google Scholar]
- 23.Robin, M., S. Marque-Juillet, C. Scieux, R. Peffault de Latour, C. Ferry, V. Rocha, J. M. Molina, A. Bergeron, A. Devergie, E. Gluckman, P. Ribaud, and G. Socie. 2007. Disseminated adenovirus infections after allogeneic hematopoietic stem cell transplantation: incidence, risk factors and outcome. Haematologica 92:1254-1257. [DOI] [PubMed] [Google Scholar]
- 24.Schilham, M. W., E. C. Claas, W. van Zaane, B. Heemskerk, J. M. Vossen, A. C. Lankester, R. E. Toes, M. Echavarria, A. C. Kroes, and M. J. van Tol. 2002. High levels of adenovirus DNA in serum correlate with fatal outcome of adenovirus infection in children after allogeneic stem-cell transplantation. Clin. Infect. Dis. 35:526-532. [DOI] [PubMed] [Google Scholar]
- 25.Storch, G. A. 2001. Diagnostic virology, p. 493-531. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 26.Symeonidis, N., A. Jakubowski, S. Pierre-Louis, D. Jaffe, E. Pamer, K. Sepkowitz, R. J. O'Reilly, and G. A. Papanicolaou. 2007. Invasive adenoviral infections in T-cell-depleted allogeneic hematopoietic stem cell transplantation: high mortality in the era of cidofovir. Transpl. Infect. Dis. 9:108-113. [DOI] [PubMed] [Google Scholar]
- 27.Teramura, T., M. Naya, T. Yoshihara, G. Kanoh, A. Morimoto, and S. Imashuku. 2004. Adenoviral infection in hematopoietic stem cell transplantation: early diagnosis with quantitative detection of the viral genome in serum and urine. Bone Marrow Transplant. 33:87-92. [DOI] [PubMed] [Google Scholar]
- 28.Vabret, A., S. Gouarin, M. Joannes, C. Barranger, J. Petitjean, S. Corbet, J. Brouard, F. Lafay, J. F. Duhamel, B. Guillois, and F. Freymuth. 2004. Development of a PCR-and hybridization-based assay (PCR Adenovirus Consensus) for the detection and the species identification of adenoviruses in respiratory specimens. J. Clin. Virol. 31:116-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Tol, M. J., A. C. Kroes, J. Schinkel, W. Dinkelaar, E. C. Claas, C. M. Jol-van der Zijde, and J. M. Vossen. 2005. Adenovirus infection in paediatric stem cell transplant recipients: increased risk in young children with a delayed immune recovery. Bone Marrow Transplant. 36:39-50. [DOI] [PubMed] [Google Scholar]
- 30.Walsh, M. P., A. Chintakuntlawar, C. M. Robinson, I. Madisch, B. Harrach, N. R. Hudson, D. Schnurr, A. Heim, J. Chodosh, D. Seto, and M. S. Jones. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4:e5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh, M. P., J. Seto, M. S. Jones, J. Chodosh, W. Xu, and D. Seto. 2010. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J. Clin. Microbiol. 48:991-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waruiru, C., M. A. Slatter, C. Taylor, V. Ramesh, T. J. Flood, M. Abinun, A. J. Cant, and A. R. Gennery. 2007. Outcome of hematopoietic stem cell transplantation in severe combined immune deficiency with central nervous system viral infection. Pediatr. Infect. Dis. J. 26:129-133. [DOI] [PubMed] [Google Scholar]