Abstract
Pulsed-field gel electrophoresis (PFGE) is a standard typing method for isolates from Salmonella outbreaks and epidemiological investigations. Eight hundred sixty-six Salmonella enterica isolates from eight serotypes, including Heidelberg (n = 323), Javiana (n = 200), Typhimurium (n = 163), Newport (n = 93), Enteritidis (n = 45), Dublin (n = 25), Pullorum (n = 9), and Choleraesuis (n = 8), were subjected to PFGE, and their profiles were analyzed by random forest classification and compared to conventional hierarchical cluster analysis to determine potential predictive relationships between PFGE banding patterns and particular serotypes. Cluster analysis displayed only the underlying similarities and relationships of the isolates from the eight serotypes. However, for serotype prediction of a nonserotyped Salmonella isolate from its PFGE pattern, random forest classification provided better accuracy than conventional cluster analysis. Discriminatory DNA band class markers were identified for distinguishing Salmonella serotype Heidelberg, Javiana, Typhimurium, and Newport isolates.
Salmonellosis is an important public health issue (18); according to the Centers for Disease Control and Prevention (CDC), 800,000 to 4,000,000 cases of nontyphoidal Salmonella-related illnesses occur annually in the United States (20). Salmonellosis is most often attributed to the consumption of contaminated foods, such as poultry, beef, pork, eggs, and fresh produce. Knowledge of how Salmonella is disseminated through the food chain is important in understanding how food animals and/or food processing procedures contribute to product contamination and to subsequent human infection by this pathogen.
Traditionally, phenotypic methods such as serotyping have been used for identification of Salmonella isolates in outbreak investigations. However, phenotypic methods have limited utility for epidemiologic analysis of Salmonella transmission because of their poor discriminatory ability for closely related isolates (8, 17). Standard serotyping methods, which rely on the detection of somatic (O) and flagellar (H) antigens present on the cell surface of Salmonella, are tedious and time-consuming (10, 12). Genotyping methods have been developed for genetic discrimination of Salmonella isolates in outbreaks. Pulsed-field gel electrophoresis (PFGE) is a standard typing method used in Salmonella outbreak investigations (6, 13, 17). While it is also labor-intensive, many laboratories have used PFGE to determine strain relatedness and to confirm outbreaks of a bacterial disease. Thus, the ability to deduce the serotype of a Salmonella isolate based on its PFGE profile would be highly attractive, in that it would limit the need for both PFGE and traditional serotyping. Liebana et al. (13) analyzed several methods for molecular typing of five selected serovars of Salmonella and indicated that serotypes of isolates could be deduced based on PFGE patterns. Gaul et al. (7) presented an analysis of 674 isolates from 12 Salmonella serotypes that separated into 66 different XbaI PFGE subtypes. The 66 subtypes could be separated into groups of specific serotypes by cluster analysis. Thus, PFGE fingerprint profiling can potentially provide an alternative method for screening and identifying Salmonella serotypes.
Hierarchical cluster analysis is commonly used to group bacterial isolates with similar PFGE patterns to understand their similarities and differences and to find or characterize the relationships among isolates. The hierarchical clustering algorithms form clusters in a hierarchical fashion, resulting in a tree-like dendrogram. Cluster analysis of PFGE patterns is typically performed by using software such as BioNumerics (Applied Maths, Inc., Austin, TX). Hierarchical clustering is an unsupervised clustering algorithm which does not use serotype information in the analysis. Classification is a supervised analysis (4) and can be used to distinguish the serotypes of samples based on their PFGE profiles. Supervised classification analysis is considered more appropriate and efficient than unsupervised analysis (hierarchical clustering tree) for prediction/classification purposes (19). A classification algorithm is developed from the available sample data set, with the ultimate goal to accurately predict the serotypes of future samples, such as those encountered during an outbreak. Typically, the available data are partitioned into a training set and a test set. The classification algorithm is built from the training samples, and then its prediction rule is applied to the test samples. Development of a classification (prediction) model involves two phases: (i) building a classification model, including determining the classification algorithm, identifying the most relevant PFGE features (bands), and fitting the prediction model to training data; and (ii) assessing the performance of the prediction model.
A classification model is a mathematical function constructed based on the training samples from the selected classification algorithm and the selected features that can discriminate the serotypes. The objective is to search for a prediction function and feature subset that minimizes the probability of misclassification error. In the development of a prediction model, the most important issue is the ability of the model to predict the type of a future sample. To ensure an unbiased assessment of accuracy, the prediction model is developed with one (training) data set; the model is then applied to another (test) data set to estimate the predictive accuracy. Cross validation is typically used to evaluate the performance of a prediction model. Cross validation involves repeatedly splitting the data, creating a training set containing most of the samples, and applying the prediction rule to the test set, made up of the remaining samples.
Supervised classification is the most widely used method for analyzing pharmacogenomic data for safety assessment, disease diagnostics and prognostics, and prediction of responses for patient assignment (4). Random forest classification has recently been applied widely in genomic research (1). The random forest method has been shown to be superior to classical classification algorithms, such as the κ-nearest-neighbor method and linear discriminate analysis (11), which require preselection of the potential predictors to optimize the performance. In this study, a classification algorithm for use in predicting Salmonella serotypes was developed by random forest supervised analysis by directly modeling the relationships between the serotypes and PFGE banding patterns for eight Salmonella serotypes. Since both PFGE and serotyping are labor-intensive and serotyping requires reagents that are often cost prohibitive, the purpose of this study is to present an approach, as a complement to cluster analysis, for rapidly predicting the serotypes of unknown Salmonella isolates based on PFGE fingerprinting analysis.
MATERIALS AND METHODS
Bacterial isolates.
A total of 866 Salmonella enterica isolates from eight serotypes were collected from food-producing animals, production facilities, and clinical diagnostic samples and genotyped by PFGE during previous studies (5, 9, 14-16). Of the 866 isolates, the first 784 isolates were used to develop the classification model for comparison with cluster analysis. An additional 82 isolates were then added only for validation of the classification model. The 784 isolates consisted of eight Salmonella serotypes: Heidelberg (n = 322), Javiana (n = 150), Typhimurium (n = 135), Newport (n = 91), Enteritidis (n = 44), Dublin (n = 25), Pullorum (n = 9), and Choleraesuis (n = 8). The additional 82 isolates consisted of five Salmonella serotypes: Heidelberg (n = 1), Javiana (n = 50), Typhimurium (n = 28), Newport (n = 2), and Enteritidis (n = 1). When isolates were used as validation, their serotype information was hidden to avoid any potential bias.
Cluster analysis.
The bacterial isolates were fingerprinted by the XbaI-PFGE method, using the PulseNet protocol developed by the CDC (22). The gel images were processed and analyzed by BioNumerics software. The images were normalized by use of standard molecular markers, and banding patterns were compared. Similarity analysis was performed using Dice coefficients, with a 1.0% band position tolerance and 1.56% optimization, and isolates were separated into similarity clusters by the unweighted-pair group method using average linkages.
Classification.
The random forest classification algorithm was used to distinguish the serotypes of samples based on their PFGE profiles (3). The PFGE profiles of the 784 isolates, with 71 band classes of various sizes, were generated using BioNumerics software. The 71 band classes were coded as 1 and 0, representing the presence and absence of a band, respectively. In the classification analysis, the 784 PFGE profiles were partitioned into a training set and a separate test set. The model development involved two phases: (i) building of a classification model, including determination of the classification algorithm, identification of the most relevant PFGE features (band classes), and fitting of the prediction model to training data; and (ii) assessment of the performance of the prediction model. The leave-one-out cross-validation (LOOCV) approach was used in the analysis and to evaluate the performance of the prediction model. This approach used a single observation from the original sample as a test datum and the remaining observations as the training data. This was repeated such that each observation in the sample was used once as the test datum. The predictive error was calculated as the proportion of misclassification.
The classification model, developed based on 784 isolates, was further validated using 110 Salmonella isolates as test samples. This set contained 28 isolates from S. Heidelberg (n = 17) and S. Enteritidis (n = 11), randomly selected from the isolates of these serovars present in the training set, and 82 isolates belonging to five trained serotypes but not initially included in the training set. For the testing, all 110 isolates were provided in a serotype-blinded format to minimize the potential for bias.
Identification of discriminatory markers for four serotypes.
Additional classification analysis was performed to identify those PFGE features associated with the Salmonella serotypes Heidelberg, Javiana, Typhimurium, and Newport. Only the four largest serotype groups were analyzed. Classification analysis of S. Heidelberg isolates versus non-S. Heidelberg isolates was performed to identify S. Heidelberg-specific markers. In LOOCV, the ranking of each band class was recorded when the left-out sample was classified accurately. The five highest-ranked band classes were the S. Heidelberg-specific markers. The S. Javiana-specific, S. Typhimurium-specific, and S. Newport-specific markers were identified similarly.
For each band class identified, a measure of association between the band class and the serotype was computed by comparing the observed number of bands (O) to the expected number (E) under the (independent) model that the band class was not associated with any serotypes. Specifically, if the band class was not associated with any serotypes, then the number of bands observed for the serotype would be proportional to its sample size. The proportions of the sample sizes for the four serotypes are as follows: Heidelberg, P = 322/784 = 0.411; Javiana, P = 150/784 = 0.191; Typhimurium, P = 135/784 = 0.172; and Newport, P = 91/784 = 0.116. If T is the total number of bands observed in a band class, then the expected numbers of bands are as follows: Heidelberg, E = 0.411 × T; Javiana, E = 0.191 × T; Typhimurium, E = 0.172 × T; and Newport, E = 0.116 × T. The ratio of the observed number of bands over the expected number was computed as a measure of the association between serotype and band class. An O/E ratio of 1 implies no association, while a ratio of >1 indicates overrepresentation and one of <1 indicates underrepresentation. For example, for the 188-kb band class, the total number of isolates observed was 518. The observed number of bands of serotype Heidelberg was 319, and the expected number was 518 × 0.411 = 212.8; the O/E ratio was 1.50, indicating overrepresentation. The O/E ratios of the five top-ranked band classes were computed for each of the four serotypes.
RESULTS
Cluster analysis.
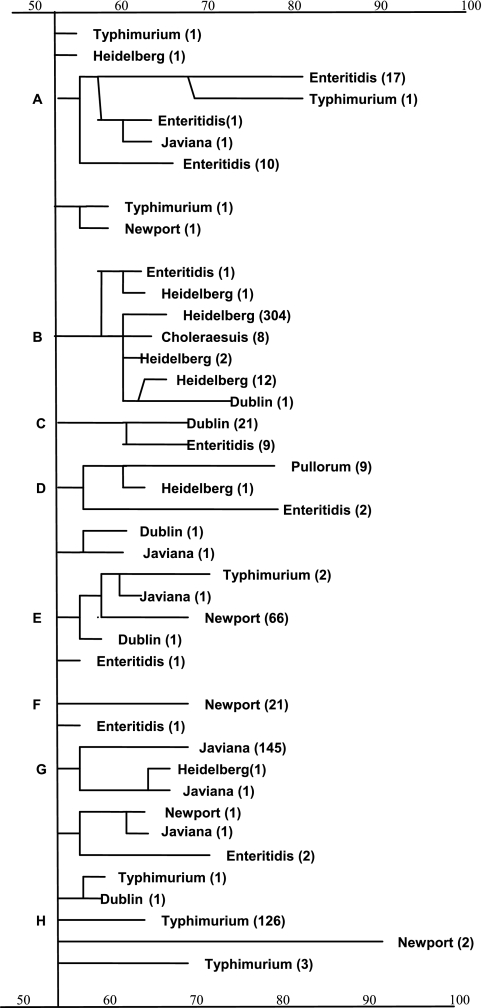
Eight major clusters were identified at a similarity level of 54%. Figure 1 shows a simplified dendrogram obtained with the results of cluster analysis for the 784 isolates. The eight clusters, A, B, C, D, E, F, G, and H, comprised mainly the isolates of Salmonella serotypes Enteritidis, Heidelberg, Dublin, Pullorum, Newport, Newport, Javiana, and Typhimurium, respectively. Both clusters E and F were predominantly S. Newport isolates. Many minor tree nodes with 1 to 3 isolates could not be grouped within a serotype cluster, and also, many isolates were grouped incorrectly into a different serotype cluster. To summarize, 32 isolates did not fall into one of the predominant serotype clusters, including isolates of S. Heidelberg (n = 3), S. Javiana (n = 5), S. Typhimurium (n = 9), S. Newport (n = 4), S. Enteritidis (n = 7), and S. Dublin (n = 4). In addition, 8 isolates of S. Choleraesuis were grouped in S. Heidelberg cluster B, 9 isolates of S. Enteritidis were grouped in S. Dublin cluster C, and 87 of 91 isolates of S. Newport were separated into clusters E and F.
FIG. 1.
Cluster analysis of XbaI-PFGE fragment patterns of 784 Salmonella isolates at a 54% similarity level. The dendrogram shows a simplified tree structure for 784 isolates from eight serotypes. The numbers in parentheses indicate the numbers of isolates in branches or leaves. There are eight major clusters: S. Enteritidis (A), S. Heidelberg (B), S. Dublin (C), S. Pullorum (D), S. Newport (E), S. Newport (F), S. Javiana (G), and S. Typhimurium (H).
Classification analysis.
Table 1 shows the results of the classification analysis using the random forest algorithm. The numbers on the diagonal show the correct classification for the eight serotypes. The overall accuracy rate was 96.3%. The number of misclassified isolates for the eight serotypes was 29, including isolates of Salmonella serotypes Heidelberg (n = 2), Typhimurium (n = 5), Newport (n = 5), Enteritidis (n = 9), Dublin (n = 5), Pullorum (n = 1), and Choleraesuis (n = 2). The supervised classification approach was able to distinguish six of the eight isolates of S. Choleraesuis from those of S. Heidelberg, while the hierarchical cluster analysis grouped these two serotypes together (cluster B) (Fig. 1). Classification by the random forest approach correctly identified 86 of 91 isolates of S. Newport, with most of them grouped in clusters E and F by hierarchical clustering.
TABLE 1.
Classification analysis of XbaI-PFGE fragment patterns of 784 Salmonella isolates by the random forest method, based on LOOCVa
| Predicted serotype | True Serotype |
|||||||
|---|---|---|---|---|---|---|---|---|
| Heidelberg | Javiana | Typhimurium | Newport | Enteritidis | Dublin | Pullorum | Choleraesuis | |
| Heidelberg | 320 | 0 | 0 | 1 | 2 | 1 | 0 | 2 |
| Javiana | 1 | 150 | 1 | 1 | 1 | 0 | 0 | 0 |
| Typhimurium | 1 | 0 | 130 | 3 | 3 | 3 | 1 | 0 |
| Newport | 0 | 0 | 1 | 86 | 3 | 1 | 0 | 0 |
| Enteritidis | 0 | 0 | 3 | 0 | 35 | 0 | 0 | 0 |
| Dublin | 0 | 0 | 0 | 0 | 0 | 20 | 0 | 0 |
| Pullorum | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 |
| Choleraesuis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| Total (n) | 322 | 150 | 135 | 91 | 44 | 25 | 9 | 8 |
LOOCV, leave-one-out cross-validation.
The classification model developed with the 784 isolates was applied to 110 Salmonella isolates for further validation. Of these, the 28 resampled serotype-hidden isolates of S. Heidelberg and S. Enteritidis from the original 784 isolates were classified accurately, as expected (see Table SA in the supplemental material). For the 82 additional isolates whose serotypes were hidden, the prediction accuracy was 93.9% (77/82 isolates) (see Table SB in the supplemental material), which is slightly lower than the 96.3% accuracy rate estimated from the LOOCV analysis (details are given in Tables SB and SC in the supplemental material).
Identification of discriminatory markers for four serotypes.
Table 2 lists the five top-ranked discriminatory PFGE band classes identified for each of four serotypes. The prediction accuracies were 99.5%, 99.0%, 98.6%, and 98.7% for S. Heidelberg, S. Javiana, S. Typhimurium, and S. Newport, respectively. The O/E column shows the association measure of overrepresentation or underrepresentation between the band class and its corresponding serotype, with an O/E ratio of >1 indicating overrepresentation of the bands for the serotype and a ratio of <1 indicating underrepresentation. Because of interdependent classes among the bands, the most discriminatory band class does not necessarily have the largest (or smallest) O/E ratio (see Discussion). All five top-ranked markers were overrepresented for S. Heidelberg. For S. Typhimurium, S. Newport, and S. Javiana, the five top-ranked markers contained both overrepresented and underrepresented band classes. The 188-kb and 181-kb marker bands were overrepresented markers for S. Heidelberg and underrepresented markers for S. Typhimurium. In general, if a band class is a marker for two serotypes, it will be an overrepresented marker in one serotype and an underrepresented marker in another serotype.
TABLE 2.
Five top-ranked band classes for S. Heidelberg, S. Javiana, S. Typhimurium, and S. Newport
| Serotype (n) | Band size (kbp) | Total no. of bands | O | E | O/Ea |
|---|---|---|---|---|---|
| S. Heidelberg (322) | 188 | 518 | 319 | 212.8 | 1.5 |
| 621 | 360 | 291 | 147.9 | 2.0 | |
| 206 | 285 | 264 | 117.1 | 2.3 | |
| 181 | 466 | 313 | 191.4 | 1.6 | |
| 78 | 546 | 318 | 224.3 | 1.4 | |
| S. Javiana (150) | 477 | 201 | 93 | 38.5 | 2.4 |
| 196 | 120 | 101 | 23.0 | 4.4 | |
| 94 | 161 | 1 | 30.8 | 0.0 | |
| 281 | 194 | 118 | 37.1 | 3.2 | |
| 167 | 114 | 83 | 21.8 | 3.8 | |
| S. Typhimurium (135) | 188 | 518 | 11 | 89.2 | 0.1 |
| 43 | 172 | 103 | 29.6 | 3.5 | |
| 746 | 107 | 83 | 18.4 | 4.5 | |
| 181 | 466 | 7 | 80.2 | 0.1 | |
| 373 | 195 | 88 | 33.6 | 2.6 | |
| S. Newport (91) | 104 | 123 | 71 | 14.3 | 5.0 |
| 305 | 185 | 70 | 21.5 | 3.3 | |
| 999 | 45 | 33 | 5.2 | 6.3 | |
| 36 | 150 | 59 | 17.4 | 3.4 | |
| 266 | 415 | 2 | 48.2 | 0.0 |
An O/E ratio of 1 implies no association, a ratio of >1 indicates overrepresentation, and a ratio of <1 indicates underrepresentation.
DISCUSSION
Traditional Salmonella serotyping is time-consuming and requires specialized skills and reagents. Wise et al. (21) developed a laboratory method utilizing repetitive sequence-based PCR to predict Salmonella enterica serotypes, while several others have reported a good correlation between PFGE patterns and Salmonella serotypes (2, 7, 13). Gaul et al. (7) suggested using hierarchical cluster analysis of PFGE with XbaI restriction as a possible alternative method for screening and identifying Salmonella serotypes. The current work shows that supervised random forest classification analysis provides a more efficient alternative method for determining Salmonella serotypes than conventional hierarchical cluster analysis.
Hierarchical cluster analysis is generally considered to be unsupervised in the sense that the isolates are grouped based only on the pairwise similarities among their PFGE profiles, without using serotype information. The dendrogram itself does not define a specific set of disjointed clusters that can be correlated with serotype; typically, the cutting of the dendrogram to form clusters (Fig. 1) is based on subjective visual analysis. Cluster analysis generally does not attempt to study the correlation between PFGE profiles and serotypes. It only arranges the isolates into subsets with similar PFGE profiles to distinguish their underlying phylogenetic structures or to discover new subtypes. Figure 1 suggests that the isolates of S. Newport consist of two subserotypes and that the isolates of S. Choleraesuis have similar profiles to those of some isolates of S. Heidelberg.
To predict serotypes from PFGE profiles, the supervised classification approach uses serotype information to optimize predictive accuracy. In developing the prediction model, a classification algorithm identified a subset of discriminatory bands of various sizes (second column in Table 2). A discriminatory marker set consists of those bands that are unique for characterization of one serotype. Therefore, in cases where two variable bands contribute equally to the same serotype, only one would normally be selected if the inclusion of another band does not increase the accuracy. The discriminatory ability of a selected band marker for its corresponding serotype relies not only on the percentage of this band found in all isolates with the same serotype but also on the percentage of this band not found in other serotype isolates. As described in Results, a band with an O/E ratio of >1 indicates that this band is overrepresented for the serotype, which means that this band appeared more often in the PFGE band profiles of this serotype than in those of others, and vice versa. A discriminatory band class usually has an O/E ratio of much more than 1 or much less than 1 (last columns in Table 2). The discriminative ability and O/E ratio are related but not equivalent. For example, as shown in Table 2, the band at 188 kb is predicted to be a marker for S. Heidelberg, but its O/E ratio of 1.5 is smaller than that of the second-ranked band class, 621 kb, with an O/E ratio of 2.0. This is probably because 518 of 784 (66%) isolates from the eight serotypes had this band, while 99% (319 of 322 isolates) of S. Heidelberg isolates shared this band. Conversely, only 90% (291 of 322 isolates) of S. Heidelberg isolates were found to have the band of 621 kb in their PFGE profiles (Table 2). This work shows the results of marker prediction only for Salmonella serotypes Heidelberg, Javiana, Typhimurium, and Newport. The predictions for the remaining four serotypes could not be validated because of the small sample sizes. Even for the four marker sets presented, the underrepresented markers identified may change when more samples are analyzed. Therefore, while the results in the current study are promising, future prediction models will require more data from additional Salmonella serotypes that are associated with human infections.
This study involved 866 isolates from eight serotypes. The classification model was developed from and the results are applicable only to isolates from these eight serotypes, since the model was trained only on them. Isolates which are not from one of the eight serotypes will be misclassified. The random forest algorithm can be expanded to include more serotypes in the training model, for example, isolates from the top 10 or top 20 most frequently reported Salmonella serotypes. However, considering that there are more than 2,500 serotypes of Salmonella in the world, it is impossible to cover all serotypes due to data limitation. One remedy is to create an “unknown” group for isolates from minor serotypes. An additional 28 new isolates, from Salmonella serotypes Agona, Albany, Brandenburg, Bredeney, Derby, Infantis, London, Mbandaka, Montevideo, Muenchen, Ohio, Worthington, and Anatum, with 1 to 3 isolates per serotype, were further evaluated. The isolates that were not from one of the serotypes in the training set (Table 1) were classified into a new, “unknown” group. Eleven of the 28 isolates with unknown serotypes were predicted correctly (see Table SC in the supplemental material). The percentage of misprediction of nontrained serotypes will decrease with increasing data size for each serotype in the training group.
The purpose of this paper was to propose a classification approach for more efficient and accurate prediction of serotype and identification of discriminatory markers. Although the traditional serotyping method is still the fundamental solution for serotype identification, the application of random forest classification to PFGE data should provide a good tool for predicting serotype. The classification approach should be considered a complement to cluster analysis, which maintains wide applicability for bacterial phylogenetics. Additionally, based on the study results, unsupervised cluster analysis can provide clues to serotype identity and supervised classification analysis then complements these results to provide further serotype confirmation. These conclusions concur with those of Gaul et al. (7), who suggested that “when unable to serotype by conventional methods, PFGE would be a possible alternative in serotype determination or may be used to screen isolates for possible serotypes before actual serotyping.” While the results presented in this study include representatives of the top 5 Salmonella serovars associated with human disease in the United States (http://www.cdc.gov/ncidod/dbmd/phlisdata/salmtab/2006/SalmonellaTable1_2006.pdf), further predictive models that may be developed based on a larger data set, including more isolates representing various serotypes involved in food-borne disease and outbreaks, should lead to further refinement in both model development and validation. The method potentially also has the ability to classify isolates that are “untypeable” based on conventional serotyping. Since random forest classification is based on the genotype rather than on expression of surface antigens, the isolates could be differentiated into unique groups for classification.
Supplementary Material
Acknowledgments
We are grateful to John Sutherland and Ching-Wei Chang for critical readings of the manuscript.
Wei-Jiun Lin acknowledges support of a fellowship from the Oak Ridge Institute for Science and Education, administered through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
The views presented in this paper are those of the authors and do not necessarily represent those of the U.S. Food and Drug Administration.
Footnotes
Published ahead of print on 14 July 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Ahn, H., H. Moon, M. J. Fazzari, N. Lim, J. J. Chen, and R. L. Kodell. 2007. Classification by ensembles from random partitions of high-dimensional data. Comput. Stat. Data Anal. 51:6166-6179. [Google Scholar]
- 2.Avery, S. M., E. Liebana, C. A. Reid, M. J. Woodward, and S. Buncic. 2002. Combined use of two genetic fingerprinting methods, pulsed-field gel electrophoresis and ribotyping, for characterization of Escherichia coli O157 isolates from food animals, retail meats, and cases of human disease. J. Clin. Microbiol. 40:2806-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiman, L. 2001. Random forests. Mach. Learn. 45:5-32. [Google Scholar]
- 4.Chen, J. J. 2007. Key aspects of analyzing microarray gene-expression data. Pharmacogenomics 8:473-482. [DOI] [PubMed] [Google Scholar]
- 5.Foley, S. L., D. G. White, P. F. McDermott, R. D. Walker, B. Rhodes, P. J. Fedorka-Cray, S. Simjee, and S. Zhao. 2006. Comparison of subtyping methods for differentiating Salmonella enterica serovar Typhimurium isolates obtained from food animal sources. J. Clin. Microbiol. 44:3569-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garaizar, J., N. Lopez-Molina, I. Laconcha, D. Lau Baggesen, A. Rementeria, A. Vivanco, A. Audicana, and I. Perales. 2000. Suitability of PCR fingerprinting, infrequent-restriction-site PCR, and pulsed-field gel electrophoresis, combined with computerized gel analysis, in library typing of Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 66:5273-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaul, S. B., S. Wedel, M. M. Erdman, D. L. Harris, I. T. Harris, K. E. Ferris, and L. Hoffman. 2007. Use of pulsed-field gel electrophoresis of conserved XbaI fragments for identification of swine Salmonella serotypes. J. Clin. Microbiol. 45:472-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, J. R., C. Clabots, M. Azar, D. J. Boxrud, J. M. Besser, and J. R. Thurn. 2001. Molecular analysis of a hospital cafeteria-associated salmonellosis outbreak using modified repetitive element PCR fingerprinting. J. Clin. Microbiol. 39:3452-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaldhone, P., R. Nayak, A. M. Lynne, D. E. David, P. F. McDermott, C. M. Logue, and S. L. Foley. 2008. Characterization of Salmonella enterica serovar Heidelberg from turkey-associated sources. Appl. Environ. Microbiol. 74:5038-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, J. W., J. B. Lee, M. Park, and S. H. Song. 2005. An extensive comparison of recent classification tools applied to microarray data. Comput. Stat. Data Anal. 48:869-885. [Google Scholar]
- 12.Li, J., K. Nelson, A. C. McWhorter, T. S. Whittam, and R. K. Selander. 1994. Recombinational basis of serovar diversity in Salmonella enterica. Proc. Natl. Acad. Sci. U. S. A. 91:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liebana, E., D. Guns, L. Garcia-Migura, M. J. Woodward, F. A. Clifton-Hadley, and R. H. Davies. 2001. Molecular typing of Salmonella serotypes prevalent in animals in England: assessment of methodology. J. Clin. Microbiol. 39:3609-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynne, A. M., L. L. Dorsey, D. E. David, and S. L. Foley. 2009. Characterisation of antibiotic resistance in host-adapted Salmonella enterica. Int. J. Antimicrob. Agents 34:169-172. [DOI] [PubMed] [Google Scholar]
- 15.Lynne, A. M., P. Kaldhone, D. David, D. G. White, and S. L. Foley. 2009. Characterization of antimicrobial resistance in Salmonella enterica serotype Heidelberg isolated from food animals. Foodborne Pathog. Dis. 6:207-215. [DOI] [PubMed] [Google Scholar]
- 16.Lynne, A. M., B. S. Rhodes-Clark, K. Bliven, S. Zhao, and S. L. Foley. 2008. Antimicrobial resistance genes associated with Salmonella enterica serovar Newport isolates from food animals. Antimicrob. Agents Chemother. 52:353-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen, J. E., M. N. Skov, E. J. Threlfall, and D. J. Brown. 1994. Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J. Med. Microbiol. 40:15-22. [DOI] [PubMed] [Google Scholar]
- 18.Scaria, J., R. U. Palaniappan, D. Chiu, J. A. Phan, L. Ponnala, P. McDonough, Y. T. Grohn, S. Porwollik, M. McClelland, C. S. Chiou, C. Chu, and Y. F. Chang. 2008. Microarray for molecular typing of Salmonella enterica serovars. Mol. Cell. Probes 22:238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon, R. 2003. Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data. Br. J. Cancer 89:1599-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voetsch, A. C., T. J. Van Gilder, F. J. Angulo, M. M. Farley, S. Shallow, R. Marcus, P. R. Cieslak, V. C. Deneen, and R. V. Tauxe. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl. 3):S127-S134. [DOI] [PubMed] [Google Scholar]
- 21.Wise, M. G., G. R. Siragusa, J. Plumblee, M. Healy, P. J. Cray, and B. S. Seal. 2009. Predicting Salmonella enterica serotypes by repetitive sequence-based PCR. J. Microbiol. Methods 76:18-24. [DOI] [PubMed] [Google Scholar]
- 22.Wonderling, L., R. Pearce, F. M. Wallace, J. E. Call, I. Feder, M. Tamplin, and J. B. Luchansky. 2003. Use of pulsed-field gel electrophoresis to characterize the heterogeneity and clonality of Salmonella isolates obtained from the carcasses and feces of swine at slaughter. Appl. Environ. Microbiol. 69:4177-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.