Abstract
As the pandemic (H1N1) 2009 influenza virus continues to infect human populations globally, reports on epidemiologically linked animal infections are also on the rise. Since December 2009, pandemic (H1N1) 2009-like viruses have been isolated in pigs from different swine farms of South Korea. Genetic and phylogenetic analyses of viral segments demonstrated several events of human-to-swine transmission with no apparent signs of reassortment. These events were also supported by serological surveillance in pig sera collected from April to December, suggesting that reverse transmission probably started between June and July with a drastic increase in prevalence the following months. Although molecular characterization indicates that the swine isolates are generally stable, some viruses are genetically evolving, most notably in their surface proteins. Animal studies (ferrets and mice) reveal that swine pandemic isolates epitomize biological properties attributed to the currently circulating human pandemic viruses, including replication kinetics and efficient transmission, indicating their potential to return to circulation among humans. Overall, these results indicate widespread human-to-animal transmission of pandemic (H1N1) 2009 influenza viruses in South Korea. With the significant role of pigs in the ecology of influenza viruses, these transmission events should be closely monitored and minimized to prevent the risk of generating viruses with greater human health concerns.
In June 2009, a global pandemic was declared by the World Health Organization (WHO) for the emergence and rapid spread of a novel influenza A (H1N1) virus (6, 7). The causative virus strain, termed as the pandemic (H1N1) 2009 influenza virus, is highly transmissible among humans and contains a unique reassortment of gene segments derived from viruses of the triple reassortant swine North American lineage and the avian-like swine Eurasian lineage (12, 39). At present, the mortality rate due to infection with the pandemic virus is relatively low among humans, where the majority of laboratory-confirmed infections result in self-limiting, uncomplicated influenza (44). Fatal cases are largely often associated with preexisting medical conditions (40). Experts have already demonstrated that the virus is pathogenic in mammalian hosts like mice, ferrets, and nonhuman primates (18, 24, 26). Furthermore, pigs have been shown to be susceptible and can transmit the virus (3, 18, 20, 30).
Accordingly, natural cases of reverse zoonosis into turkeys and primarily pigs have been increasing considerably in different continents since the first detection of the virus among pigs in a Canadian swine farm (16, 41), as reflected in reports through the weekly disease information of the Paris-based World Organization for Animal Health Information Database (28). Due to dual susceptibility to both human and animal influenza viruses, pigs are considered important intermediate hosts, acting as “mixing vessels” for genetic reassortment (4, 17, 23, 33). Such events may consequently lead to generation of novel reassortant influenza viruses which can cause human pandemics or, as in the current influenza pandemic, a reassortant virus with potentially enhanced pathogenicity and lethality.
Here we report the detection and isolation of the pandemic (H1N1) 2009 influenza viruses isolated from various swine farms in South Korea. Virus isolates were genetically characterized to determine whether these swine viruses have undergone any evolutions that would significantly alter their overall phenotype. Subsequently, pathogenicity and transmissibility in ferrets were tested and compared with local Korean human pandemic viruses and a recent Korean swine H1N1 virus.
MATERIALS AND METHODS
Viruses and virus isolation.
A/Swine/Korea/CAN01/2004 (Sw/Korea/CAN01/04) is a recent swine influenza H1N1 virus strain isolated from a Korean swine farm in 2004 (29). The 50% tissue culture infective doses (TCID50) of viruses were determined by infection in Madin-Darby canine kidney cells (MDCK).
Pandemic (H1N1) 2009 influenza viruses were isolated using homogenized lung tissue samples of commercially slaughtered pigs, which came in from different swine farms, by infection into MDCK cells. Briefly, the samples were processed and inoculated onto monolayers of MDCK cells and then incubated for 1 h at 37°C to allow for viral adsorption to the cells. The cells were rinsed two times each with 1× cold phosphate-buffered solution (PBS), before and after sample inoculation, after which appropriate culture growth medium containing a final concentration of 1 μg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin solution (Sigma-Aldrich) was added. Viral growth was determined by observing morphological changes in the cells (cytopathic effects) and by hemagglutinin (HA) assay. Subtyping was done by using two multiplex reverse transcription-PCR (RT-PCR) assays and sequencing as previously described (5).
Genomic sequencing and phylogenetic analysis.
Viral RNA was extracted from cell culture isolates using a QIAamp viral RNA mini kit (Qiagen, Valencia, CA). RT-PCR was carried out under standard conditions using influenza-specific primers (14). Nucleotide sequencing of the amplified products was carried out using a DNA sequencer (model 377; Applied Biosystems, PerkinElmer, Foster City, CA) and a Taq dye deoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, CA). The sequences were resolved using the ABI Prism collection program (PerkinElmer, Foster City, CA). The DNA sequences were compiled and edited using the Lasergene sequence analysis software package (Lasergene, version 5.07; DNAStar, Inc., Madison, WI). Multiple sequence alignments were made using Clustal_X (1, 38). The rooted phylograms were prepared using the neighbor-joining (NJ) algorithm and then plotted using the program NJ plot (31).
Serologic assays.
Hemagglutination inhibition (HI) assays were performed as previously described (21) to determine the seroprevalence of pandemic (H1N1) 2009 viruses since April 2009. All collected swine sera were heat inactivated at 56°C for 30 min and pretreated with receptor-destroying enzyme (RDE) from Vibrio cholerae (Denka Seiken, Tokyo, Japan) to remove nonspecific serum inhibitors. The sera were then tested for antibody against the swine pandemic virus Sw/Korea/SCJ01/09 by the HI technique with 0.5% turkey red blood cells (RBC). The HI titer was determined by the reciprocal of the last dilution that contained turkey RBC with no agglutination. A neutralization test was done on selected HI-positive sera to confirm results (21). Virus-neutralizing titers were determined by infection of MDCK cells and expressed as the reciprocal of the highest dilution of serum that gave 50% neutralization of 100 TCID50 of virus after incubation at 37°C for 72 h (19). Hemagglutination assays were performed according to WHO/World Organization for Animal Health (OIE) recommendations.
Replication and transmission studies.
Groups of four 15- to 18-week-old specific-pathogen-free (SPF) ferrets (Marshall Bio Resources, New York, NY) were instillated intranasally (i.n.) with 105 TCID50 of Korea/CJ01/09, Sw/Korea/SCJ01/09, or Sw/Korea/CAN01/04 virus in 1 ml of sterile PBS (divided between two plastic syringes for separate inoculation of each nostril) under isoflurane anesthesia. To monitor virus transmission, two seronegative animals were cohoused at 1 day postinfection (dpi) in adjacent transmission cages fitted in the same isolator separated by a screen mesh (5 cm apart) preventing direct or indirect animal contact but allowing the spread of influenza virus through the respiratory droplets from experimentally infected animals. Clinical signs among tested ferrets were noted while body temperatures were recorded daily, beginning 1 day before experimental inoculation. Groups of 12 5-week-old BALB/c mice (Samtaco, Seoul, Korea) were i.n. inoculated with 30 μl 105 TCID50/ml of Korea/CJ01/09, Sw/Korea/SCJ01/09, or Sw/Korea/CAN01/04. All viruses and animal experiments including serologic testing were handled in an enhanced biosafety level 3 (BSL-3+) containment facility approved by the Korea Centers for Disease Control and Prevention.
Experimental sampling and virus titration.
Nasal washes and rectal swabs were collected daily from all ferrets for 12 days. Different tissue samples (tonsils, trachea, lungs, liver, spleen, kidney cecum, and rectum) were harvested on 3 and 5 dpi from infected ferrets to examine tissue distribution of test viruses. Lung tissue samples of mice were collected at 2, 4, 6, 8, and 10 dpi. To prevent cross-contamination, different sterile instruments were used for collecting each tissue. Swabs, nasal washes, and collected tissue samples were homogenized in 1× PBS containing antibiotics. Tissue homogenates and supernatants were clarified by centrifugation at 12,000 × g, and supernatants were transferred to new tubes. All samples were immediately serially diluted 10-fold and then inoculated into 11-day-old embryonated chicken eggs for virus titration as computed by the Reed and Muench method with results expressed as log10 50% egg infective dose per milliliter or gram of tissue collected (EID50/ml or EID50/g) (32). The limit of virus detection was set to 0.7 log10 EID50/ml. Standard deviations for virus titers and body temperatures are indicated appropriately. Student's t test was used for statistical analysis.
Nucleotide sequence accession numbers.
Complete sequences of the eight viral gene segments of all viruses collected from humans and swine were submitted in GenBank. Accession numbers assigned to the sequences determined in this study are HM189430 to HM189669.
RESULTS
Virus isolation from field samples.
A total of 42 (eight on 4 December 2009, two on 10 December 2009, seven on 24 December 2009, and 15 on 2 January 2010) swine influenza viruses were isolated from 456 swine lung tissue samples (9.21%) collected in swine slaughterhouses within Chungbuk province, South Korea, since November 2009. The virus isolates were subtyped as H1N1 using multiplex RT-PCR and subsequent sequencing. Nucleotide sequence analysis of the amplicons indicated that each of the eight RNA segments of all the isolates have high nucleotide homology to pandemic (H1N1) 2009 influenza viruses currently circulating among humans as determined through BLAST homologies (∼99% sequence identities). Positive detection and isolation of the pandemic virus among local pigs were promptly reported to corresponding authorities. No other subtypes of swine influenza virus were detected or isolated during the span of the sampling period.
Phylogenetic analyses.
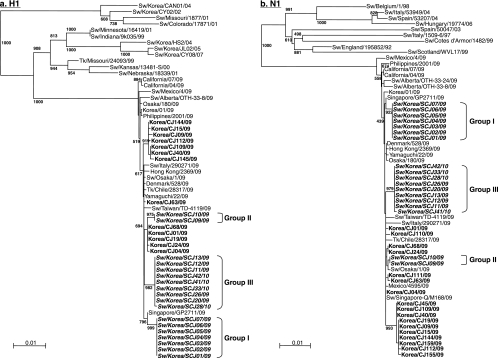
The pandemic (H1N1) 2009 isolates from swine were aligned with sequences from human and other animal influenza viruses available in GenBank which bore the highest homology (∼99% identity on all gene segments) with the viruses in the study. Local human pandemic (H1N1) 2009 influenza viruses isolated from patients suffering from respiratory diseases in Chungbuk National University Hospital were also included. Pairwise sequence analysis of the H1 gene segments showed that the pandemic viruses from swine form at least three groups: group I (4 December 2009 isolates, Sw/Korea/SCJ01/09 to Sw/Korea/SCJ08/09) isolates are more closely related to a human pandemic (H1N1) 2009 influenza virus from Singapore (Singapore/GP2711/09; ≥99.7% sequence homology), while group II (10 December 2009 isolates, Sw/Korea/SCJ09/09 and Sw/Korea/SCJ10/09) and group III (24 December 2009 and 2 January 2010 isolates, Sw/Korea/SCJ11/09 to Sw/Korea/SCJ42/10) isolates clustered separately with local Korean human isolates (99.4 to 99.8%) (Fig. 1a). Phylogeny of the neuraminidase (NA) genes also reflected the same clustering observed in HA genes (Fig. 1b). Average homology of viruses between the three groups is about 99.2 and 99.5% (HA and NA, respectively), whereas isolates within each group of isolation are highly identical to each other, sharing as high as 100% nucleotide sequence identities. Therefore, phylogenetic alignment of the surface genes (HA and NA) suggest that these viruses may have been derived from different human pandemic viruses.
FIG. 1.
(a and b) Phylogenetic analysis of the HA and NA genes of pandemic (H1N1) 2009 influenza virus isolates from South Korea. Phylogenetic trees of the nucleotide sequences for the H1 (a) and N1 (b) genes of pandemic viruses isolated from pigs and humans in this study compared with nucleotide sequences from selected swine, human, and avian influenza virus strains available in GenBank are shown. The nucleotide sequences were aligned using Clustal_X (1, 38), and the phylograms were generated by the neighbor-joining method using the tree-drawing program NJ plot (31). The scale represents the number of substitutions per nucleotide. Branch labels record the stability of the branches over 1,000 bootstrap replicates. Only bootstrap values ≥400% are shown in each tree. Sw, swine; Tk, turkey.
Phylogenies of all the internal genes revealed that all are genetically related to human pandemic (H1N1) 2009 influenza viruses currently circulating worldwide. Specifically, the PB2, PB1, PA, NP, and NS genes were placed into the lineage of triple reassortant swine viruses predominantly found in North American pigs, while the M genes also fell under the Eurasian avian-like swine H1N1 viruses like their virus precursors (7, 12; data not shown). Interestingly, the group I isolates appear to cluster consistently with Sw/Singapore-Q/M168/09, the first reported cases of swine pandemic (H1N1) influenza virus in Singapore. In contrast, results from group II and III isolates indicated that they are more phylogenetically related to human pandemic viruses isolated from the same region. Overall, phylogenetic analyses presumably suggest that there were at least three individual events of human-to-swine transmissions.
Molecular characterization of viral genes.
To determine whether the viruses have undergone significant molecular modifications during the course of natural transmission in pigs, the deduced amino acid sequences of the viral genes, generated and aligned using the MegAlign program of the Lasergene sequence analysis software package (Lasergene, version 5.07; DNAStar, Inc., Madison, WI), were examined. Synonymous to all isolates, the cleavage site contains a single basic amino acid residue (Arg), and residues at positions 187 and 222 (H1 numbering) retained human-type cell receptor specificities (Table 1) (27, 36). However, three consensus substitutions in group I isolates were found at sites (Ser84Asn, Asp86Gly, Asp94Glu) near the second stabilizing shell of the protein (Cys90 and Pro92; H1 numbering) and a conserved component of the H1 receptor binding pocket (Tyr91) (Table 2) (43). Isolates within group II and group III also bear mutations in their HA1 regions at positions 74 (Ser to Asn) and 116 (Iso to Met), respectively. In the N1 protein, at least two consensus mutations could be observed in group II and III isolates (Table 2). All potential glycosylation sites in the surface proteins of group I and II isolates appear to have been coherently conserved with reference to the CA/04/09 virus, while group III isolates had gained an additional N-linked glycosylation (Asn35) in their NA. Furthermore, all Korean pandemic (H1N1) 2009 influenza isolates (human and swine) have consensus mutations in their HA1 (Pro83Ser, Ser203Thr) and N1 (Val106Iso) proteins compared to virus strain CA/04/09, indicating uniform genetic substitution at these positions (Table 2).
TABLE 1.
Molecular analysis, residue positions, and comparison of amino acid sequences of different gene products of swine pandemic (H1N1) 2009 influenza virus isolates with selected H1N1 virusesa
| Strain | HA gene |
NA gene |
PB2 |
PB1-F2e | M 31 | NS 92 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cleavage site | RBS |
||||||||||||
| 187 (190) | 222 (225) | 223 (226) | Stalk deletion | 275 | 590 | 591 | 627 | 701 | |||||
| BM/1/18 | SIQSR/GLF | D | D | Q | No | H | G | Q | K | D | Yes | S | D |
| Sw/KR/CAN01/04 | SIQSR/GLF | D | G | Q | No | H | S | R | E | D | Yes | S | D |
| CA/04/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| SG/GP2711/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| Sw/AB/OTH33-8/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| Sw/SG-Q/M168/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| Tk/CL/6504-3/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| KR/CJ01/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| KR/CJ19/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| KR/CJ24/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| KR/CJ63/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| KR/CJ112/09 | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| Sw/KR/SCJ01/09b | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| Sw/KR/SCJ07/09b | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| Sw/KR/SCJ09/09c | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| Sw/KR/SCJ10/09c | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| Sw/KR/SCJ24/09d | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
| Sw/KR/SCJ33/10d | SIQSR/GLF | D | D | Q | No | H | S | R | E | D | No | N | D |
Molecular marker(s) previously identified to impact replication, pathogenesis, or drug resistance in respective segments are indicated. Numbers are position of residue, with H3 numbering in parentheses. AB, Alberta; BM, Brevig Mission; CA, California; CL, Chile; KR, Korea; SG, Singapore; RBS, receptor binding site; Sw, swine; Tk, turkey.
Group I swine isolates.
Group II swine isolates.
Group III swine isolates.
Expression of the PB1-F2 protein.
TABLE 2.
Substitutions in HA and NA proteins of pandemic (H1N1) 2009 influenza viruses isolated from human and swine in South Koreaa
| Strain | Mutation at indicated amino acid position |
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HA1 region |
HA2 region |
NA protein |
|||||||||||||||||||||||||||||
| 74 | 83 | 84 | 86 | 94 | 116 | 129 | 184 | 197 | 198 | 203 | 249 | 271 | 321 | 334 | 352 | 354 | 411 | 460 | 476 | 29 | 35 | 95 | 96 | 100 | 123 | 248 | 305 | 334 | 416 | 424 | |
| CA/04/09 | S | P | S | D | D | I | S | T | T | Y | S | V | P | I | A | H | Q | V | I | M | I | S | S | G | V | S | N | S | S | D | V |
| Sw/AL/OTH33-8/09 | S | A | N | V | V | G | E | L | C | ||||||||||||||||||||||
| Sw/SP/M168/09 | S | N | A | T | V | I | D | ||||||||||||||||||||||||
| Korea/CJ01/09 | S | A | T | V | I | D | |||||||||||||||||||||||||
| Korea/CJ09/09 | S | T | V | E | I | I | D | T | I | ||||||||||||||||||||||
| Korea/CJ15/09 | S | T | V | L | I | I | D | T | I | ||||||||||||||||||||||
| Korea/CJ24/09 | S | A | T | L | V | I | D | ||||||||||||||||||||||||
| Korea/CJ63/09 | S | P | A | T | V | I | D | ||||||||||||||||||||||||
| Korea/CJ112/09 | S | T | S | V | I | D | T | I | |||||||||||||||||||||||
| Sw/Korea/SCJ01/09b | S | N | G | E | A | T | V | I | I | ||||||||||||||||||||||
| Sw/Korea/SCJ07/09b | S | N | G | E | A | T | V | I | I | ||||||||||||||||||||||
| Sw/Korea/SCJ09/09c | N | S | A | T | V | I | D | N | |||||||||||||||||||||||
| Sw/Korea/SCJ10/09c | N | S | A | T | V | I | D | N | |||||||||||||||||||||||
| Sw/Korea/SCJ24/09d | S | M | A | T | V | V | T | N | I | D | |||||||||||||||||||||
| Sw/Korea/SCJ33/10d | S | M | A | T | V | V | T | N | I | D | |||||||||||||||||||||
The sequence of the A/California/04/2009 (CA/04/09) virus was used as the consensus for comparison while two reported swine pandemic isolates from Canada (Sw/AL/OTH33-8/09) and Singapore (Sw/SP/M168/09) were also included. Amino acid 1 is the first amino acid residue after the signal peptide. CA, California; AL, Alberta; Sp, Singapore.
Group I swine isolates.
Group II swine isolates.
Group III swine isolates.
Few synonymous and nonsynonymous mutations could also be observed in three segments (PB2, PA, and NP) of the swine isolates which have not been previously implicated with replication, pathogenicity, or virulence. However, all defined molecular markers in viral proteins, such as PB2 (11, 13, 22, 25, 37), PB1 (46), M2 (15), and NS1 (34), have been relatively conserved similar to their pandemic (H1N1) influenza virus (CA/04/09) ancestor (Table 1).
Simian immunodeficiency virus antibody prevalence from field samples.
To investigate the prevalence of swine infection with the pandemic (H1N1) 2009 influenza viruses and provide an approximate time frame as to when the transmission began, pig sera collected since April 2009 from swine-producing provinces in Korea were tested by HI assays using the Sw/Korea/SCJ01/09 virus. For cross-reference and to ensure specificity of serum reactivity, a natural Korean swine H1N1 virus (Sw/Korea/CAN01/04) was also used in the serological testing. It is of note that a national vaccination program was conducted in October 2005 (29); thus, the reference swine strain would help interpret false-positive results. Of the 1,621 pig sera surveyed, 60 (3.7%) had HI antibody specific for the novel pandemic virus, while 49 (3.0%) sera were positive for the Korean swine H1N1 virus (Table 3). Dual-positive samples (i.e., samples positive for both viruses) were also noted in 219 (13.58%) swine sera. Serologic surveillance indicated that initial introduction of the pandemic (H1N1) 2009 virus into pigs occurred possibly between the months of June and July, which drastically increased by early fall (August to September). On the other hand, seropositivity against Sw/Korea/CAN01/04 relatively decreased from April to December (Table 3). When the dual-positive sera from August to December (72 swine sera) were reprocessed and further segregated by HI titers, more seropositive samples (>40 HI units) were obtained against Sw/Korea/SCJ01/09 compared to the seroreactivity against Sw/Korea/CAN01/04 HI titers (≤40 HI units) (Table 4). This result suggests that the higher proportion of the reactive sera were probably due to exposure with pandemic (H1N1) 2009 influenza viruses.
TABLE 3.
Serologic reactivity to swine H1N1 viruses of pig sera collected from different swine farms in Chungbuk Province, South Koreaa
| Mo rangeb | No. of sera tested | No. of sera (%) positive for H1N1 | No. of sera (%) positive for: |
||
|---|---|---|---|---|---|
| Sw/Korea/CAN01/04 | Sw/Korea/SCJ01/09 | Dual infection | |||
| Apr-May | 555 | 157 (28.3) | 34 (6.1) | — | 123 (22.2) |
| June-July | 217 | 33 (15.2) | 7 (3.2) | 2 (1) | 24 (11) |
| Aug-Sept | 369 | 47 (12.7) | — | 25 (6.8) | 22 (5.9) |
| Oct-Dec | 480 | 91 (19) | 8 (1.7) | 33 (6.9) | 50 (10.4) |
| Total | 1,621 | 328 (20.2) | 49 (3) | 60 (3.7) | 219 (13.5) |
Hemagglutination inhibition (HI) assays were performed as previously described (21). Pig sera were tested for the presence of antibody against a 2004 Korean H1N1 swine isolate (29) or a swine pandemic (H1N1) 2009 influenza virus isolate (Sw/Korea/SCJ01/09) using 0.5% turkey RBC. Limit of detection for the HI assays was set to <20 HI units. —, no cross-reactivity.
Apr, April; Aug, August; Sept, September; Oct, October; Dec, December.
TABLE 4.
HI titers of pandemic H1N1 2009 influenza virus-positive and Sw/Kor/CAN01/04 virus-positive sera collected from August to Decembera
| Range of HI titers of positive sera | No. of sera positive for: |
|
|---|---|---|
| Sw/Korea/CAN01/04 | Sw/Korea/SCJ01/09 | |
| 20-40 | 27 | 14 |
| 80-160 | 26 | 20 |
| 320-640 | 17 | 28 |
| 1,280-2,560 | 2 | 10 |
| Total | 72 | 72 |
Limit of detection for the HI assays was set to <20 HI units, and HI titers are expressed as the reciprocal of the highest dilution of serum that inhibits 8 HA units of virus (e.g., as 80 vs 1:80).
To confirm the specificity of the HI results, virus neutralization tests were conducted. Results showed that neutralizing antibodies against the pandemic (H1N1) 2009 virus correlated with the titers obtained from the HI assays, which reached up to 1,280 (Table 4). In contrast, lower neutralizing antibody titers (at least a 2- to 4-fold difference) were observed in the swine sera when the Korean Sw/Korea/CAN01/04 H1N1 virus was used (data not shown).
Animal studies.
Molecular analysis suggests that the swine pandemic (H1N1) 2009 influenza isolates are undergoing the process of genetic evolution, albeit slowly. To determine whether the noted molecular changes have an impact on the overall phenotype of the virus, groups of SPF ferrets were i.n. instillated with ∼105.0 TCID50 of Korea/CJ01/09, Sw/Korea/SCJ01/09, or Sw/Korea/CAN01/04 virus.
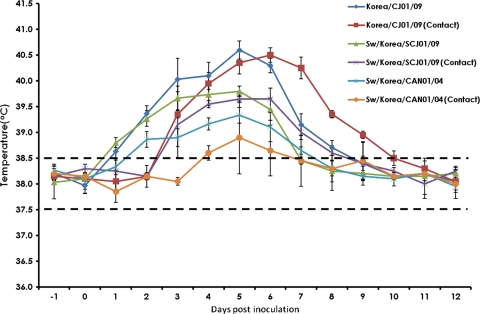
Infection of ferrets with Korea/CJ01/09 and Sw/Korea/SCJ01/09 viruses generally induced more severe flu-like symptoms (e.g., sneezing, nasal discharge, and immobility) than Sw/Korea/CAN01/04-infected ferrets throughout the course of experiment (data not shown). Constantly more elevated body temperatures were also recorded in ferrets infected with the pandemic viruses (Fig. 2). Shedding of all test viruses through the nasal route started at 1 dpi, peaked at 2 dpi, and persisted up to 6 dpi (Table 5). In all the days sampled, the human and swine pandemic (H1N1) 2009 influenza isolates had comparable replication kinetics that were consistently higher than nasal wash titers obtained from Sw/Korea/CAN01/04-inoculated ferrets (6.5 and 6.63 versus 5.75 log10 EID50/ml, respectively, during peak titers; P < 0.05). However, none of the inoculated viruses could be detected in rectal swabs (data not shown). One of the naïve “contact” ferrets (those that have been exposed to experimentally inoculated ferrets) of Sw/Korea/SCJ01/09 was already positive for virus transmission as early as 1 day postcontact (dpc), while the other contact host was able to acquire the virus at 2 dpc. Both the contact ferrets of Korea/CJ01/09 were also positive for detection at 2 dpc. In contrast, the Sw/Korea/CAN01/04 virus was transiently detected in one of the contact ferrets at 2 dpc, lasting only up to 3 days. All experimentally inoculated and positive contact ferrets were seroconverted (data not shown).
FIG. 2.
Monitoring of body temperatures in infected and contact ferrets. Mean body temperatures of ferrets experimentally infected with Korea/CJ01/09, Sw/Korea/SCJ01/09, or the recent Korean swine H1N1 (Sw/Korea/CAN01/04) isolate, including naïve contact animals, were monitored daily starting 1 day before inoculation and continuing up to 12 days postinfection. Ranges of normal body temperatures are indicated as broken lines. Standard error bars are shown.
TABLE 5.
Viral titers from nasal excretions of tested H1N1 viruses in infected and contact ferretsa
| Day | Viral titersb (SD) for: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Korea/CJ01/09 |
Sw/Korea/SCJ01/09 |
Sw/Korea/CAN01/04 |
||||||
| Inoculated | Contact | Inoculated | Contact 1 | Contact 2 | Inoculated | Contact 1 | Contact 2 | |
| 0 | — | — | — | — | — | — | — | — |
| 1 | 5.38 (0.5) | — | 5.25 (0.3) | — | — | 4.50 (0.4) | — | — |
| 2 | 6.50 (0.4)c | — | 6.63 (0.3)c | 2.25 | — | 5.38 (0.3)c | — | — |
| 3 | 4.75 (0.3) | 2.25 (0.4) | 5.00 (0.4) | 4.25 | 1.5 | 5.13 (0.5) | 2.25 | — |
| 4 | 3.50 (0.5) | 4.25 (0.4) | 3.67 (0.3) | 3.25 | 3.75 | 3.50 (0.0) | 1.75 | — |
| 5 | 2.17 (0.3) | 3.50 (0.5) | 2.33 (0.3) | 2.75 | 3.25 | 2.17 (0.3) | 1.25 | — |
| 6 | 1.50 (0.7) | 2.25 (0.4) | 1.75 (0.4) | 1.5 | 2.75 | 1.25 (0.4) | — | — |
| 7 | — | 1.25 (0.4) | — | — | 1.25 | — | — | — |
| 8 | — | — | — | — | — | — | — | — |
| 9 | — | — | — | — | — | — | — | — |
Groups of ferrets were inoculated i.n. with 105 TCID50 of respective viruses in 1 ml of sterile PBS under isoflurane anesthesia. Animals were infected in an enhanced biosafety level 3 (ABSL-3+) containment facility. Virus titrations were done in 11-day-old embryonated chicken eggs calculated by the method of Reed and Muench (32) and expressed as log10 EID50/ml. Dashes indicate no detectable titers.
Mean viral titers from nasal washes.
P < 0.05; compared peak virus titers of ferrets infected with the pandemic viruses (Korea/CJ01/09 and Sw/Korea/SCJ01/09) relative to Sw/Korea/CAN01/04-infected ferrets.
All inoculated viruses could be recovered only in respiratory tissues tested (data not shown). Consistent with nasal wash viral titers, lower tissue titers could also be detected in Sw/Korea/CAN01/04-infected hosts compared to those inoculated with the Korea/CJ01/09 or Sw/Korea/SCJ01/09 isolate. However, Sw/Korea/CAN01/04 could not yield tissue viral titers beyond the limit of detection in tonsils and lungs at 5 dpi. Experimental inoculation of respective viruses in mice recapitulated results similar to those observed in ferrets. Infection with the pandemic (H1N1) isolates (Korea/CJ01/09 and Sw/Korea/SCJ01/09) indicated relatively higher lung viral titers than the Sw/Korea/CAN01/04 virus in mice (data not shown).
DISCUSSION
April 2009 marked the emergence and global spread of a novel influenza A (H1N1) virus in humans, causing the first human influenza pandemic of the 21st century (6, 7). Detailed genomic sequence analysis of the pandemic (H1N1) 2009 influenza virus reveals that it contains a unique reassortment of genes from viruses of North American and Eurasian pigs, establishing their swine origins (12, 39). Since the emergence of the novel pandemic virus, notifications from different countries on natural infections in swine herds have been increasing considerably, including sporadic transmissions in breeding turkeys and in a few domesticated pet animals (2, 28). We describe here the first detection and isolation of pandemic (H1N1) 2009-like influenza viruses from swine lung tissue samples randomly collected from different swine farms in South Korea.
A pig farm in Alberta, Canada, was the first confirmed case of animal infection with the pandemic virus in May 2009 (16), and experts predicted that such infection of animal farms would be more likely as the current pandemic progresses globally. It is particularly difficult to estimate when the pandemic (H1N1) 2009 virus was first introduced into Korean swine herds. However, our serological surveillance provided evidence that sporadic human-to-swine transmission may have started between June and July, which drastically increased during the following months. Although dual-positive specimens were also noted in some of the pig sera tested (at 20 to 40 HI titers), most evidently in April and May (Table 2), this cross-reactivity might be induced by vaccination and/or previous exposure to Sw/Korea/CAN01/04-like or other H1-like viruses in some pigs. We had previously shown that double-vaccinated hosts (mice, ferrets, and miniature pigs) could induce only ≤40 serum antibody titers against the pandemic (H1N1) 2009 influenza virus (30).
The latest update in South Korea has already tallied about 132 cumulative numbers of human fatal cases (8). To evaluate the genetic origins of swine pandemic (H1N1) 2009 influenza viruses in this report, we included in our analysis those human pandemic isolates from patients with respiratory diseases coming from Chungbuk National University Hospital, located in the same province where the swine isolates were collected. All of the collected swine isolates phylogenetically fall under the pandemic (H1N1) 2009 influenza virus lineage with no apparent evidence of reassortment. Although there were no significant differences, phylogenies of the surface genes form distinct nodes, suggesting at least three independent introductions of pandemic viruses into pigs: one group of isolates (group I) was more closely related to a human pandemic virus from Singapore (Singapore/GP2711/09), whereas later groups of isolates (group II and group III) share the same root with local Korean human pandemic viruses (Fig. 1). Though limited, evidence has suggested that most human-to-animal transmissions occurred following direct contact (28, 45). Thus, we could also predict that the swine infections in this report may also be epidemiologically linked to human infections, although the precise source or origin of infection is relatively unknown.
None of the pandemic (H1N1) 2009 viruses isolated from the Canadian swine herd were identical (41), even though they presumably came from a single point of infection (16). Weingartl et al. noted high proportions of nonsynonymous-to-synonymous nucleotide substitutions for both the H1 and N1 genes (41). In contrast, our swine isolates appear to be relatively genetically stable, with viruses within each group having high homology. Of the three groups of swine isolates, group I has the most consensus mutations, most notably around a position (Tyr91) which is a component of the receptor binding site (RBS) of the H1 protein, the major surface antigen. It is noteworthy that 94Glu lies within the phylogenetically important region D (PIR D) (amino acids 94 to 97 in H1 numbering) of the H1 protein (Table 2) (9). PIR D corresponds to the trimer interface on H3 hemagglutinin which is involved in the pH-induced conformational change of the molecule (42). Most neutralizing antibodies against influenza HA recognize epitopes in the hypervariable regions that surround the RBS and interfere with binding to host cells (10). Thus, modifications on these sites might provide escape from immune recognition, which is critical if the virus is to successfully infect and be established in new hosts. Though it is tempting to speculate that the observed genetic modifications were due to passage through pigs, we could not rule out the possibility that these arose during the passage through humans or cumulatively through both.
Experimental infection of representative human (Korea/CJ01/09) and swine (Sw/Korea/SCJ01/09) pandemic isolates in ferrets still epitomizes disease severity and efficient transmission of pandemic (H1N1) 2009 influenza viruses in this host (26, 30), although Sw/Korea/SCJ01/09 appears to be more readily transmitted to naïve contact ferrets. Both Korea/CJ01/09 and Sw/Korea/SCJ01/09 induced higher viral titers (through the nasal route and respiratory tissues) and were more transmissible than the Sw/Korea/CAN01/04 swine virus in ferrets (Table 5). These results might imply that the pandemic viruses isolated from swine herds still have the potential to return to circulation in humans. To date, there has been no evidence yet that pigs play a significant role in the epidemiology and spread of the pandemic (H1N1) 2009 influenza virus in humans. Unfortunately, challenge studies were not carried out in pigs due to time constraints and the unavailability of an expansive biocontainment facility. Thus, accompanied by a lack of pertinent clinical data available associated with the swine isolates, we could not predict how these viruses behave in swine. Whether the viruses were newly transmitted to pigs or have been serially infecting pigs is also a matter that is difficult to presume. We could only infer that following transmission, subsequent infections may have been subclinical since there have been no reports of outbreaks in swine farms prior to this report. Thus, further studies shall shed more light on these concerns.
What is certain in this report is that we provide evidence demonstrating widespread human-to-swine transmission of the pandemic (H1N1) 2009 influenza virus in Korea. Feared for their potential as genetic mixers for human and animal influenza viruses (4, 17, 23, 33), pigs could provide an alternative opportunity to generate more virulent variants or novel viruses. It is therefore imperative that human-to-animal or possibly animal-to-human transmissions of the pandemic (H1N1) 2009 influenza virus, especially events involving pig herds, be closely monitored and minimized.
Acknowledgments
This research work was supported in part by a Top Brand Project grant from Korea Research Council of Fundamental Science and Technology, Korea Research Institute of Bioscience and Biotechnology (KRIBB) Initiative Program (NTM1300811), by grant 2009-0076343 from the National Research Foundation of Korea, and by grant Z-AD14-2009-13 from the National Veterinary Research and Quarantine Service (NVRQS), South Korea.
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221-241. [DOI] [PubMed] [Google Scholar]
- 2.American Veterinary Medical Association. 2009. Public health. 2009 H1N1 flu virus. American Veterinary Medical Association, Schaumburg, IL. http://www.avma.org/public_health/influenza/new_virus/default.asp.
- 3.Brookes, S. M., R. M. Irvine, A. Nunez, D. Clifford, S. Essen, I. H. Brown, K. Van Reeth, G. Kuntz-Simon, W. Loeffen, E. Foni, L. Larsen, M. Matrosovich, M. Bublot, J. Maldonado, M. Beer, and G. Cattoli. 2009. Influenza A (H1N1) infection in pigs. Vet. Rec. 164:760-761. [DOI] [PubMed] [Google Scholar]
- 4.Castrucci, M. R., I. Donatelli, L. Sidoli, G. Barigazzi, Y. Kawaoka, and R. G. Webster. 1993. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology 193:503-506. [DOI] [PubMed] [Google Scholar]
- 5.Chang, H. K., J. H. Park, M. S. Song, T. K. Oh, S. Y. Kim, C. J. Kim, H. Kim, M. H. Sung, H. S. Han, Y. S. Hahn, and Y. K. Choi. 2008. Development of multiplex RT-PCR assays for rapid detection and subtyping of influenza type A viruses from clinical specimens. J. Microbiol. Biotechnol. 18:1164-1169. [PubMed] [Google Scholar]
- 6.Cohen, J., and M. Enserink. 2009. Swine flu. After delays, WHO agrees: the 2009 pandemic has begun. Science 324:1496-1497. [DOI] [PubMed] [Google Scholar]
- 7.Dawood, F. S., S. Jain, L. Finelli, M. W. Shaw, S. Lindstrom, R. J. Garten, L. V. Gubareva, X. Xu, C. B. Bridges, and T. M. Uyeki. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605-2615. [DOI] [PubMed] [Google Scholar]
- 8.European Centre for Disease Prevention and Control. 2009. ECDC daily update. Pandemic (H1N1) 2009. European Centre for Disease Prevention and Control, Stockholm, Sweden. http://ecdc.europa.eu/en/healthtopics/Documents/091222_Influenza_AH1N1_Situation_Report_0900hrs.pdf. Accessed 31 December 2009.
- 9.Fanning, T. G., and J. K. Taubenberger. 1999. Phylogenetically important regions of the influenza A H1 hemagglutinin protein. Virus Res. 65:33-42. [DOI] [PubMed] [Google Scholar]
- 10.Fleury, D., B. Barrere, T. Bizebard, R. S. Daniels, J. J. Skehel, and M. Knossow. 1999. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat. Struct. Biol. 6:530-534. [DOI] [PubMed] [Google Scholar]
- 11.Gabriel, G., B. Dauber, T. Wolff, O. Planz, H. D. Klenk, and J. Stech. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. U. S. A. 102:18590-18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 2009. Antigenic and genetic characteristics of swine-origin 2009 A (H1N1) influenza viruses circulating in humans. Science 325:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 15.Holsinger, L. J., D. Nichani, L. H. Pinto, and R. A. Lamb. 1994. Influenza A virus M2 ion channel protein: a structure-function analysis. J. Virol. 68:1551-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howden, K. J., E. J. Brockhoff, F. D. Caya, L. J. McLeod, M. Lavoie, J. D. Ing, J. M. Bystrom, S. Alexandersen, J. M. Pasick, Y. Berhane, M. E. Morrison, J. M. Keeniside, S. Laurendeau, and E. B. Rohonczy. 2009. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can. Vet. J. 50:1153-1161. [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, T., J. N. Couceiro, S. Kelm, L. G. Baum, S. Krauss, M. R. Castrucci, I. Donatelli, H. Kida, J. C. Paulson, R. G. Webster, and Y. Kawaoka. 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 72:7367-7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh, Y., K. Shinya, M. Kiso, T. Watanabe, Y. Sakoda, M. Hatta, Y. Muramoto, D. Tamura, Y. Sakai-Tagawa, T. Noda, S. Sakabe, M. Imai, Y. Hatta, S. Watanabe, C. Li, S. Yamada, K. Fujii, S. Murakami, H. Imai, S. Kakugawa, M. Ito, R. Takano, K. Iwatsuki-Horimoto, M. Shimojima, T. Horimoto, H. Goto, K. Takahashi, A. Makino, H. Ishigaki, M. Nakayama, M. Okamatsu, K. Takahashi, D. Warshauer, P. A. Shult, R. Saito, H. Suzuki, Y. Furuta, M. Yamashita, K. Mitamura, K. Nakano, M. Nakamura, R. Brockman-Schneider, H. Mitamura, M. Yamazaki, N. Sugaya, M. Suresh, M. Ozawa, G. Neumann, J. Gern, H. Kida, K. Ogasawara, and Y. Kawaoka. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J. K., P. Seiler, H. L. Forrest, A. M. Khalenkov, J. Franks, M. Kumar, W. B. Karesh, M. Gilbert, R. Sodnomdarjaa, B. Douangngeun, E. A. Govorkova, and R. G. Webster. 2008. Pathogenicity and vaccine efficacy of different clades of Asian H5N1 avian influenza A viruses in domestic ducks. J. Virol. 82:11374-11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange, E., D. Kalthoff, U. Blohm, J. P. Teifke, A. Breithaupt, C. Maresch, E. Starick, S. Fereidouni, B. Hoffmann, T. C. Mettenleiter, M. Beer, and T. W. Vahlenkamp. 2009. Pathogenesis and transmission of the novel swine-origin influenza virus A/H1N1 after experimental infection of pigs. J. Gen. Virol. 90:2119-2123. [DOI] [PubMed] [Google Scholar]
- 21.Lee, J. H., P. N. Pascua, M. S. Song, Y. H. Baek, C. J. Kim, H. W. Choi, M. H. Sung, R. J. Webby, R. G. Webster, H. Poo, and Y. K. Choi. 2009. Isolation and genetic characterization of H5N2 influenza viruses from pigs in Korea. J. Virol. 83:4205-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Z., H. Chen, P. Jiao, G. Deng, G. Tian, Y. Li, E. Hoffmann, R. G. Webster, Y. Matsuoka, and K. Yu. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 79:12058-12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, W., R. E. Kahn, and J. A. Richt. 2008. The pig as a mixing vessel for influenza viruses: human and veterinary implications. J. Mol. Genet. Med. 3:158-166. [PMC free article] [PubMed] [Google Scholar]
- 24.Maines, T. R., A. Jayaraman, J. A. Belser, D. A. Wadford, C. Pappas, H. Zeng, K. M. Gustin, M. B. Pearce, K. Viswanathan, Z. H. Shriver, R. Raman, N. J. Cox, R. Sasisekharan, J. M. Katz, and T. M. Tumpey. 2009. Transmission and pathogenesis of swine-origin 2009 A (H1N1) influenza viruses in ferrets and mice. Science 325:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehle, A., and J. A. Doudna. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U. S. A. 106:21312-21316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munster, V. J., E. de Wit, J. M. van den Brand, S. Herfst, E. J. Schrauwen, T. M. Bestebroer, D. van de Vijver, C. A. Boucher, M. Koopmans, G. F. Rimmelzwaan, T. Kuiken, A. D. Osterhaus, and R. A. Fouchier. 2009. Pathogenesis and transmission of swine-origin 2009 A (H1N1) influenza virus in ferrets. Science 325:481-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann, G., T. Noda, and Y. Kawaoka. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.OIE. 2009. World Animal Health Information Database, WAHID Weekly Disease Information: vol. 22, no. 19 (2 May 2009), no. 32 (31 July 2009), no. 35 (27 August 2009), and no. 39 (24 September 2009), no. 40 (29 September 2009), no. 42 (12 October 2009), no. 43 (21 October 2009) no. 44 (23 October 2009), no. 44 (27 October 2009), no. 45 (3 November 2009), no. 49 (30 November 2009), no. 50 (4 December 2009), no. 50 (7 December 2009), no. 50 (10 December 2009), no. 50 (11 December 2009), no. 51 (14 December 2009), and no. 52 (23 December 2009). World Organization for Animal Health (OIE), Paris, France. http://www.oie.int/wahis/public.php?page=weekly_report_index&admin=0. Accessed 31 December 2009.
- 29.Pascua, P. N., M. S. Song, J. H. Lee, H. W. Choi, J. H. Han, J. H. Kim, G. J. Yoo, C. J. Kim, and Y. K. Choi. 2008. Seroprevalence and genetic evolutions of swine influenza viruses under vaccination pressure in Korean swine herds. Virus Res. 138:43-49. [DOI] [PubMed] [Google Scholar]
- 30.Pascua, P. N., M. S. Song, J. H. Lee, K. J. Park, H. I. Kwon, Y. H. Baek, S. P. Hong, J. B. Rho, C. J. Kim, H. Poo, T. S. Ryoo, M. H. Sung, and Y. K. Choi. 2009. Evaluation of the efficacy and cross-protectivity of recent human and swine vaccines against the pandemic (H1N1) 2009 virus infection. PLoS One 4:e8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrière, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 32.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. (London) 27:493-497. [Google Scholar]
- 33.Scholtissek, C. 1995. Molecular evolution of influenza viruses. Virus Genes 11:209-215. [DOI] [PubMed] [Google Scholar]
- 34.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950-954. [DOI] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Stevens, J., A. L. Corper, C. F. Basler, J. K. Taubenberger, P. Palese, and I. A. Wilson. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866-1870. [DOI] [PubMed] [Google Scholar]
- 37.Subbarao, E. K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trifonov, V., H. Khiabanian, B. Greenbaum, and R. Rabadan. 2009. The origin of the recent swine influenza A (H1N1) virus infecting humans. Euro. Surveill. 14(17):pii=19193. [PubMed] [Google Scholar]
- 40.Vaillant, L., R. G. La, A. Tarantola, and P. Barboza. 2009. Epidemiology of fatal cases associated with pandemic H1N1 influenza 2009. Euro. Surveill. 14(33):pii=19309. [DOI] [PubMed] [Google Scholar]
- 41.Weingartl, H. M., Y. Berhane, T. Hisanaga, J. Neufeld, H. Kehler, C. Emburry-Hyatt, K. Hooper-McGreevy, S. Kasloff, B. Dalman, J. Bystrom, S. Alexandersen, Y. Li, and J. Pasick. 2010. Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J. Virol. 84:2245-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White, J. M., and I. A. Wilson. 1987. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J. Cell Biol. 105:2887-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365-394. [DOI] [PubMed] [Google Scholar]
- 44.World Health Organization. 2009. New influenza A (H1N1) virus: global epidemiological situation, June 2009. Wkly. Epidemiol. Rec. 84:249-257. [PubMed] [Google Scholar]
- 45.World Health Organization. 2009. Global alert and response: infection of farmed animals with the pandemic virus, 5 November 2009. Pandemic (H1N1) 2009 briefing note 15. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/swineflu/notes/briefing_20091105/en/index.html.
- 46.Zamarin, D., M. B. Ortigoza, and P. Palese. 2006. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J. Virol. 80:7976-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]