Abstract
Parachlamydia acanthamoebae is an obligately intracellular bacterium that infects free-living amoebae and is a potential human pathogen in hospital-acquired pneumonia. We examined whether the presence of P. acanthamoebae is related to the presence of Acanthamoeba in an actual hospital environment and assessed the in vitro survival of P. acanthamoebae. Ninety smear samples were collected between November 2007 and March 2008 (trial 1, n = 52) and between October 2008 and February 2009 (trial 2, n = 38) from the floor (dry conditions, n = 56) and sink outlets (moist conditions, n = 34) of a hospital. The prevalences of P. acanthamoebae DNA in the first and second trials were 64.3% and 76%, respectively. The prevalences of Acanthamoeba DNA in the first and second trials were 48% and 63.1%, respectively. A statistical correlation between the prevalence of P. acanthamoebae and that of Acanthamoeba was found (trial 1, P = 0.011; trial 2, P = 0.022), and that correlation increased when samples from just the dry area (floor smear samples, P = 0.002) were analyzed but decreased when samples from a moist area were analyzed (P = 0.273). The in vitro experiment showed that, without Acanthamoeba, P. acanthamoebae could not survive in dry conditions for 3 days at 30°C or 15 days at 15°C. Thus, both organisms were coincidentally found in an actual hospital environment, with the presence of Acanthamoeba having a significant effect on the long-term survival of P. acanthamoebae, suggesting that this potential human pathogen could spread through a hospital environment via Acanthamoeba.
Chlamydiae, which are obligate intracellular bacterial pathogens, have been reclassified as the order Chlamydiales, which includes four families: Chlamydiaceae, Parachlamydiaceae, Waddliaceae, and Simkaniaceae (5). The family Chlamydiaceae is well known to have a broad range of distribution in animals and humans and to be the causative agents of human diseases (4, 20, 24, 31, 36). This family includes two major human pathogens: Chlamydophila pneumoniae, a causal agent of common respiratory infection and also suspected of being involved in some chronic diseases, such as asthma and atherosclerosis (6), and Chlamydia trachomatis, responsible for sexually transmitted disease and preventable blindness (33).
Parachlamydiaceae, Waddliaceae, and Simkaniaceae have recently been recognized as chlamydiae that exhibit a wide range of distribution in the natural environment, such as in rivers and in soil (9). All these species can grow and survive dependently within the free-living amoeba Acanthamoeba, the most abundant genus of amoeba (1, 9, 11, 12). Parachlamydia acanthamoebae and Simkania negevensis have been associated with lower respiratory tract infections (3, 27), and Waddlia chondrophila, which was originally isolated from an aborted bovine fetus, is considered a potential abortogenic agent (9). There is accumulating evidence supporting the pathogenic role of P. acanthamoebae in humans (3, 9, 13). Several studies have reported that parachlamydial DNA was detected by PCR in mononuclear cells of sputa and in bronchoalveolar lavage samples from a patient with bronchitis (10). Other studies have suggested that P. acanthamoebae may cause inhalation pneumonia and be responsible for hospital-acquired pneumonia in HIV-infected patients and organ transplant recipients receiving immunosuppressive therapy (7, 8, 14). Thus, P. acanthamoebae, presumably spreading through amoebae, is emerging as a potential etiological agent of hospital-acquired pneumonia. However, the overlap between the distribution of P. acanthamoebae and that of Acanthamoeba in hospitals and the survival of the bacteria in harsh conditions without Acanthamoeba remain unknown. We examined whether the prevalence of P. acanthamoebae correlates with that of Acanthamoeba in a hospital in Sapporo, Japan. We also examined the in vitro survival of P. acanthamoebae and its requirement for Acanthamoeba.
P. acanthamoebae Bn9 (ATCC VR-1476) was purchased from the American Type Culture Collection. The bacteria were propagated in an amoeba cell culture system according to methods described previously (17). The numbers of infective progeny were determined according to the procedure described below. Free-living amoebae, Acanthamoeba castellanii C3 (ATCC 50739), were purchased from the American Type Culture Collection. Amoebae were maintained in PYG broth (0.75% [wt/vol] peptone, 0.75% [wt/vol] yeast extract, and 1.5% [wt/vol] glucose) at 30°C (35). The numbers of infective progeny of P. acanthamoebae were determined by a procedure known as the amoeba infectious unit (AIU) assay using coculture with amoebae as described previously (28).
Ninety smear samples were obtained from the floor (n = 56) and sink outlets (n = 34) of a hospital (Hokkaido University Hospital) containing approximately 900 beds (number of outpatients, approximately 3,000 per day; number of hospital patients, approximately 1,000 per day) in a 12-story building located in the central area of Sapporo, Japan, from November 2007 to March 2008 (trial 1, n = 52) and from October 2008 to February 2009 (trial 2, n = 38). The samples were collected by wiping an approximately 1-m2 area on the floor or sink outlet with sterilized gauze (approximately 25 cm2) moistened with Page's amoeba saline (PAS) (30). Each piece of gauze was then vortexed for 60 s in 20 ml sterilized PAS containing 0.05% (vol/vol) Tween 80, and the suspension was then centrifuged at 2,100 × g for 20 min. Pellets were resuspended in 200 μl PAS and then used for P. acanthamoebae culture and DNA extraction.
One-half of the resuspended smear pellet (100 μl) was used for P. acanthamoebae culture. Culture detection was performed by a method based on amoeba lysis described previously (18). In brief, serially diluted sample solution was added to 100 μl of amoeba suspension containing 1 × 105 A. castellanii C3 cells in 1 well of a 96-well microplate and incubated for up to 10 days at 30°C in a normal atmosphere. The microplate was read daily to determine the highest dilution of bacteria that led to amoeba lysis.
The DNA extraction was performed using the UltraClean soil DNA extraction kit (MBL, Carlsbad, CA) according to the manufacturer's instructions. The primers used in PCR amplification were as follows: PacI (5′-GAG GTG AAG CAA ATC CCA AA-3′) and Pac2 (5′-CTC CTT GCG GTT AAG TCA GC-3′) for amplification of P. acanthamoebae 16S rRNA (191 bp), JDP1 (5′-GGC CCA GAT CGT TTA CCG TGA A-3′) and JDP2 (5′-TCT CAC AAG CTG CTA GGG AGT CA-3′) for amplification of 18S rRNA from Acanthamoeba species (423 to 551 bp) (34), and Bac11 (5′-GAG GAA GGT GGG GAT GAC GT-3′) and Bac12 (5′-AGG CCC GGG AAC GTA TTC AC-3′) for amplification of bacterial 16S rRNA excluding the order Chlamydiales (216 bp) (38). The primers for amplification of P. acanthamoebae 16S rRNA were designed based on GenBank cDNA sequences (accession number NR026357) using the program Primer 3 (http://frodo.wi.mit.edu/primer3/input.htm). To overcome inhibition of PCR amplification by humic acid, bovine serum albumin (BSA) was added to each reaction mixture according to methods described previously (23, 39). The quality of extracted DNA was confirmed by PCR amplification using universal primers that target bacterial 16S rRNA, which is conserved across a broad spectrum of bacteria. All smear samples (n = 90) yielded PCR amplicons of the expected size and were then used in specific P. acanthamoebae and Acanthamoeba PCRs. Search results returned by the BLAST program showed that the primers used for each PCR were specific for P. acanthamoebae and Acanthamoeba detection. The amount of template DNA used in each PCR was 2 μl. The total reaction volume was 25 μl and consisted of 200 μM each deoxynucleoside triphosphate (dNTP), 10 μM BSA, 1× commercial reaction buffer, and 0.625 U Taq DNA polymerase (New England Biolabs, Herts, United Kingdom). The thermal cycling profile involved an initial denaturation step at 94°C for 10 min; 35 cycles, each consisting of 30 s of denaturation at 94°C and 30 s of annealing at 60°C for P. acanthamoebae 16S rRNA, 60°C for Acanthamoeba species 18S rRNA, and 52°C for bacterial 16S rRNA; and a 45-s extension at 72°C. The amplified products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. The presence of amplified target genes in randomly selected positive specimens was confirmed by direct oligonucleotide sequencing of the PCR products (Macrogen, Seoul, South Korea). As a quality control for each PCR, diluted DNA extracted from the P. acanthamoebae Bn9 or A. castellanii C3 strain was used in each amplification. As a negative control, DNA-free water (Sigma) was also used in amplification. To prevent contamination, the preparation of the PCR mixture was performed in a separate room. Alignment analysis and the construction of a phylogenetic tree for P. acanthamoebae amplicons (n = 4) with previously reported Parachlamydiaceae sequences were performed using Genetyx-Mac software (version 10.1) and the neighbor-joining method in MEGA software (version 4). The detection limits of the PCR for P. acanthamoebae 16S rRNA and Acanthamoeba 18S rRNA were examined by using DNA extracted from the sterilized gauze moistened with PAS that had been spiked with defined numbers of P. acanthamoebae AIU and Acanthamoeba cells, respectively. The detection limits of the PCR targeted to P. acanthamoebae and Acanthamoeba were 102 AIU and 10 cells, respectively.
The procedure for monitoring bacterial viability was as follows. A bacterial solution of 100 μl containing approximately 107 to 108 AIU prepared with PAS was placed into wells of a 24-well plate with (moist conditions) or without (dry conditions) 900 μl PAS, and the plates incubated for up to 28 days at 15°C or 30°C in a normal atmosphere. At 0, 3, 7, 15, and 28 days after inoculation, the supernatant in each well was collected and centrifuged at 3,500 × g for 30 min. In the case of the samples for moist conditions, supernatants were directly transferred to a centrifuge tube. For the dry condition samples, the wells were washed with 900 μl PAS and this was then transferred to a centrifuge tube. The resulting bacterial pellet was resuspended in 100 μl PAS. The numbers of infective progeny as a marker of bacterial viability in the solution were determined by the AIU assay as described above. The bacterial membrane integrity as a possible indicator for bacterial viability was also confirmed with fluorescence microscopy by using a Live/Dead reduced biohazard viability/cytotoxicity kit (Molecular Probes, Eugene, OR), according to the manufacturer's instructions.
The correlation between the frequency of P. acanthamoebae and that of Acanthamoeba spp. was analyzed by Fisher's exact test. The influence of floor level on the prevalence of both organisms was also analyzed by a two-way analysis of variance (ANOVA) test. Comparison of bacterial numbers in the in vitro experiment was assessed by an unpaired t test. A P value of less than 0.05 was considered significant.
Prevalences of P. acanthamoebae and Acanthamoeba in a hospital environment.
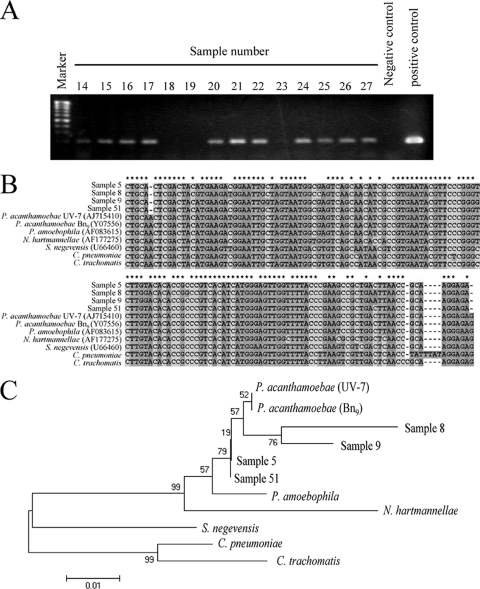
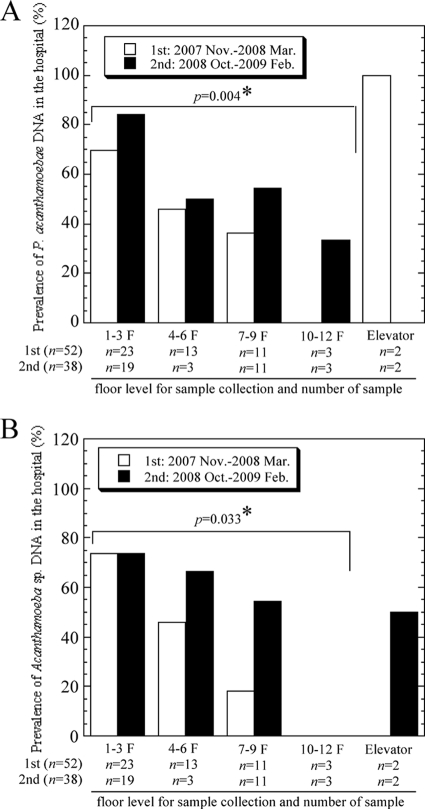
In total, 90 smear samples were collected from a hospital environment and assessed. The smear samples were randomly obtained from the floor and sink outlets. Figure 1 A shows a representative image of PCR amplification for P. acanthamoebae. The presence of amplified target genes was confirmed by direct sequencing with homology analysis. The DNA sequences obtained from P. acanthamoebae amplification products were mostly identical to previously reported P. acanthamoebae sequences (concordance rate, 96.3%) (Fig. 1B), and the sequences of amplicons formed a cluster on the phylogenetic tree under the family Parachlamydiaceae (Fig. 1C). As shown in Table 1, the prevalences of P. acanthamoebae DNA in the first and second trials were 64.3% and 76%, respectively. The prevalences of Acanthamoeba DNA in the first and second trials were 48% and 63.1%, respectively. A statistical correlation between the prevalence of P. acanthamoebae and that of Acanthamoeba was found (trial 1, P = 0.011; trial 2, P = 0.022). No difference in prevalence between the sampling periods was observed, and the correlation became much higher when floor smear samples were separately analyzed (P = 0.002). Interestingly, the prevalence of both organisms in the hospital statistically significantly decreased toward the upper floors (Fig. 2). Successful culture of P. acanthamoebae was not achieved.
FIG. 1.
Detection and analysis of P. acanthamoebae DNA in smear samples obtained from a hospital environment. (A) Representative results of agarose gel electrophoresis of P. acanthamoebae PCR products amplified from smear samples. (B) Alignment analysis of four P. acanthamoebae amplicons (samples 5, 8, 9, and 51) with previously reported Parachlamydiaceae sequences was performed using Genetyx-Mac software (version 10.1). Asterisks represent conserved sequences in all P. acanthamoebae PCR amplicons. Dots and unmarked positions represent base mismatches and additional bases in the alignment, respectively. P. amoebophila, Protochlamydia amoebophila; N. hartmannellae, Neochlamydia hartmannellae. (C) Construction of a phylogenetic tree for P. acanthamoebae amplicons (samples 5, 8, 9, and 51) with previously reported Parachlamydiaceae sequences was performed using the neighbor-joining method in MEGA software (version 4). Strain names are in parentheses. The accession numbers assigned by DDBJ were as follows: sample 5, uncultured Parachlamydia sp. HUHP-5, AB565471; sample 8, HUHP-8, AB565472; sample 9, HUHP-9, AB565473; sample 51, HUHP-51, AB565474.
TABLE 1.
Correlation between prevalence of P. acanthamoebae and that of Acanthamoeba spp. in the hospital environment
| Sampling period or site | Result for Acanthamoeba sp. DNA (n) | No. (%) of samples for P. acanthamoebae DNA testing: |
Pa | |
|---|---|---|---|---|
| Positive | Negative | |||
| November 2007-March 2008 (1st trial; n = 52) | Positive (25) | 18 (64.3) | 7 (29.2) | 0.011* |
| Negative (27) | 10 (35.7) | 17 (70.8) | ||
| October 2008-February 2009 (2nd trial; n = 38) | Positive (24) | 19 (76) | 5 (38.4) | 0.022* |
| Negative (14) | 6 (24) | 8 (61.6) | ||
| Sink outlet (moist condition; n = 34)b | Positive (11) | 6 (42.9) | 5 (25.0) | 0.273 |
| Negative (23) | 8 (57.1) | 15 (75.0) | ||
| Floor (dry condition; n = 56)b | Positive (39) | 32 (82.1) | 7 (41.2) | 0.002* |
| Negative (17) | 7 (17.9) | 10 (58.8) | ||
Correlation between the frequency of P. acanthamoebae and that of Acanthamoebae was analyzed by Fisher's exact test. Asterisks indicate statistically significant results.
Mixture of 1st and 2nd trials.
FIG. 2.
Influence of floor level on the prevalences of P. acanthamoebae (A) and Acanthamoeba (B). Asterisks indicate statistical significance.
In vitro survival of P. acanthamoebae without Acanthamoeba.
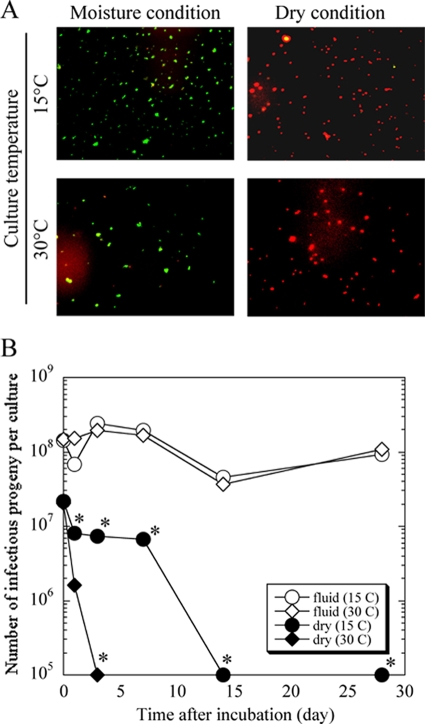
To evaluate the in vitro survival of P. acanthamoebae under conditions lacking amoebae, P. acanthamoebae was incubated without amoebae in moist and dry conditions. As shown in Fig. 3 B, the numbers of P. acanthamoebae progeny in moist conditions, regardless of culture temperature, were stably maintained for up to 28 days. The numbers of P. acanthamoebae progeny grown in dry conditions rapidly decreased, with the bacterial numbers declining more rapidly at 30°C than at 15°C. We also confirmed by Live/Dead staining analysis that P. acanthamoebae membrane integrity as a possible indicator for viability in moist conditions was maintained during this culture period but not in dry conditions (Fig. 3A).
FIG. 3.
Representative images of Live/Dead staining in P. acanthamoebae (A) and numbers of P. acanthamoebae infective progeny in cultures without amoebae (B). (A) The bacteria at 28 days after incubation were stained with a Live/Dead staining kit. Green, membrane integrity stable (possibly viable bacteria); red, dead bacteria. Magnification, ×400. (B) A bacterial solution of 100 μl containing approximately 107 to 108 AIU prepared with PAS was inoculated into the wells of a 24-well plate with (moist conditions) or without (dry conditions) 900 μl of PAS and incubated for up to 28 days at 15°C or 30°C in a normal atmosphere. The numbers of infective progeny as a marker of bacterial viability in the solution were determined using the AIU assay. The data represent the average AIU counts ± standard deviations. *, P < 0.05.
The emergence of P. acanthamoebae as a novel pathogenic agent of respiratory tract infections in hospital environments has important implications for preventing and controlling hospital-acquired diseases (3, 8, 9, 10, 14). Several in vitro studies have shown that P. acanthamoebae grew and multiplied in free-living Acanthamoeba amoebae, which are distributed in a broad range of natural environments but also in the water systems of hospitals (1, 12-15). Whether P. acanthamoebae could survive in an actual environment within Acanthamoeba spp., which are considered a protective reservoir for bacteria from multiple environmental stresses, remains to be seen. We hence attempted to determine the effects on P. acanthamoebae prevalence and survival due to the interaction of P. acanthamoebae with Acanthamoeba in the hospital environment.
The use of standard culture techniques to detect microorganisms in complex biological samples obtained from hospital surfaces, such as the floor and sink outlets, is limited when the target population is in a minority because most microorganisms in the environment shift to a nonculturable stage. Thus, the molecular technique of PCR, which can be applied to analyze the entire microorganism community, was used for this study. Since it is known that humic acid, which is ubiquitous in natural environments, inhibits PCR amplification (23, 29), BSA was added to each reaction mixture according to methods described previously (23, 29). No amplification of bacterial 16S rRNA or amoeba 18S rRNA was observed from our smear samples without the addition of BSA (data not shown).
Although the prevalences of P. acanthamoebae and Acanthamoeba in the hospital environment were very high and there was a significant correlation between the prevalence of P. acanthamoebae and that of Acanthamoeba, how these organisms came to be inside a hospital remains unknown. Interestingly, the prevalences of both organisms statistically changed depending on floor level, with the prevalences on lower floor levels much higher than those on upper levels. It was also confirmed that the prevalences of both organisms in a total of 75 soil samples collected from public parks (75 places) located in an approximately 10-km2 area around the hospital, which is located in the central area of Sapporo, were 72% (54/75) and 92% (69/75), respectively, and that a significant correlation between the prevalence of P. acanthamoebae and that of Acanthamoeba species was found, as in the hospital experiments (data not shown). Taken together, these results imply that P. acanthamoebae, which is a potential human pathogen, can enter from outside a hospital and spread throughout the hospital courtesy of an Acanthamoeba host. Meanwhile, we could not isolate P. acanthamoebae using an amoeba coculture system, but Thomas et al. have reported that Chlamydia-related organisms in 200 samples obtained from a water system could be detected using an amoeba coculture method with amoeba strain A. castellanii ATCC 30010C (37). Although the exact reason for the contradiction between our results and their findings remains unknown, it is possible that usage of different amoeba strains is associated with these results. In addition, as mentioned above, the preparation of the PCR mixture was performed in a separate room to prevent DNA contamination, so the possibility of a false positive is negligible.
Nevertheless, it is not clear how the bacteria are able to survive the oligotrophic conditions of a hospital environment. The finding that a correlation between the prevalence of P. acanthamoebae and that of Acanthamoeba was much stronger in dry conditions provided a hint, suggesting a difference in this bacterium's stability depending on habitat. As expected, the in vitro experiment indicated that the stability of the bacteria under artificial dry conditions in the absence of amoebae was not preserved compared to that in moist conditions, suggesting that the protection and survival of P. acanthamoebae through Acanthamoeba in the hospital environment would be limited, particularly in dry conditions. In contrast, P. acanthamoebae grown under moist conditions persistently maintained viability for the culture period, suggesting that P. acanthamoebae would be relatively stable in moist environments without the requirement for amoebae for an extended period of time.
The mechanism by which P. acanthamoebae can survive in moist environments without the requirement for amoebae remains unknown. As stated above, the order Chlamydiales includes the pathogenic chlamydiae, such as Chlamydia pneumoniae and Chlamydia trachomatis, and the environmental chlamydia P. acanthamoebae. These similarly exhibit reduced central metabolic and biosynthetic pathways and are auxotrophic for most amino acids and nucleotides (18). A notable exception is the tricarboxylic acid (TCA) cycle, which is incomplete in the pathogenic chlamydiae (18). Admittedly, the detection of P. acanthamoebae by PCR is not definitive proof of the actual presence of viable P. acanthamoebae organisms. Culturing is the gold standard for detection of P. acanthamoebae from samples; therefore, this was also performed using a method based on amoeba lysis as described previously (15). However, there were no positive cultures from the smear samples collected from the locations determined as PCR positive because of contamination with fungi and other bacteria. Further efforts are needed to demonstrate and separate viable organisms in an actual hospital environment.
We demonstrated that there is an overlap between the presence of P. acanthamoebae and Acanthamoeba in the hospital environment. Acanthamoeba spp. are distributed in a broad range of natural environments (2, 21, 22, 27, 29, 32) and have been classified into 15 different genotypes (T1 to T15) (13, 17, 19). It has been reported that the T4 genotype is frequently present in the natural environment and that 90% of isolates belong to the T4 genotype, which is the genotype most likely to exhibit strong virulence against humans (22, 26). Whether there is a possible variation that supports growth of P. acanthamoebae among Acanthamoeba genotypes remains to be seen. The elimination of Acanthamoeba may be important for preventing the emergence of P. acanthamoebae infections in hospitals.
In conclusion, this study demonstrated that the presence of Acanthamoeba had a significant effect on the presence of P. acanthamoebae in the hospital environment, suggesting that P. acanthamoebae could stably survive and spread in hospitals via free-living Acanthamoeba amoebae.
Acknowledgments
This study was supported in part by grants-in-aid for scientific research from KAKENHI, the Suhara Memorial Foundation, and the Akiyama Foundation and a research grant from the Institute for Fermentation, Osaka, Japan.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Amann, R., N. Springer, W. Schönhuber, W. Ludwig, E. N. Schmid, K. Müller, and R. Michel. 1997. Obligate intracellular bacterial parasites of Acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Birtles, R. J., T. J. Rowbotham, C. Storey, T. J. Marrie, and D. Raoult. 1997. Chlamydia-like obligate parasite of free-living amoebae. Lancet 349:925-926. [DOI] [PubMed] [Google Scholar]
- 4.Bodetti, T. J., and P. Timms. 2000. Detection of Chlamydia pneumoniae DNA and antigen in the circulating mononuclear cell fraction of humans and koalas. Infect. Immun. 68:2744-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush, R. M., and K. D. E. Everett. 2001. Molecular evolution of the Chlamydiaceae. Int. J. Syst. Evol. Microbiol. 51:203-220. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, L. A., and C. C. Kuo. 2003. Chlamydia pneumoniae and atherosclerosis. Semin. Respir. Infect. 18:48-54. [DOI] [PubMed] [Google Scholar]
- 7.Casson, N., J. M. Entenza, N. Borel, A. Pospischil, and G. Greud. 2008. Murine model of pneumonia caused by Parachlamydia acanthamoebae. Microb. Pathog. 45:92-97. [DOI] [PubMed] [Google Scholar]
- 8.Casson, N., and G. Greub. 2006. Resistance of different Chlamydia-like organisms to quinolones and mutations in the quinoline resistance-determining region of the DNA gyrase A- and topoisomerase-encoding gene. Int. J. Antimicrob. Agents 27:541-544. [DOI] [PubMed] [Google Scholar]
- 9.Corsaro, D., and G. Greub. 2006. Pathogenic Potential of novel Chlamydiae and diagnostic approached to infections due to these obligate intracellular bacteria. Clin. Microbiol. Rev. 19:283-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corsaro, D., D. Venditti, and M. Valassina. 2002. New parachlamydial 16S rDNA phylotypes detected in human clinical samples. Res. Microbiol. 153:563-567. [DOI] [PubMed] [Google Scholar]
- 11.Fritsche, T. R., M. Horn, M. Wagner, R. P. Herwig, K. Schleifer, and R. K. Gauton. 2000. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 66:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gast, R. J. 2001. Development of an Acanthamoeba specific reverse dot-blot and the discovery of a new ribotype. J. Eukaryot. Microbiol. 48:609-615. [DOI] [PubMed] [Google Scholar]
- 13.Greub, G. 2009. Parachlamydia acanthamoebae, an emerging agent of pneumonia. Clin. Microbiol. Infect. 15:18-28. [DOI] [PubMed] [Google Scholar]
- 14.Greub, G., I. Boyadjiev, B. L. Scola, O. Raoult, and C. Martin. 2003. Serological hint suggesting that Parachlamydiaceae are agents of pneumonia in polytraumatized intensive care patients. Ann. N. Y. Acad. Sci. 990:311-319. [DOI] [PubMed] [Google Scholar]
- 15.Greub, G., and D. Raoult. 2002. Crescent bodies of Parachlamydia acanthamoeba and its life cycle within Acanthamoeba polyphaga: an electron micrograph study. Appl. Environ. Microbiol. 68:3076-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greub, G., B. L. Scola, and D. Raoult. 2003. Parachlamydia acanthamoeba is endosymbiotic or lytic for Acanthamoeba polyphage depending on the incubation temperature. Ann. N. Y. Acad. Sci. 990:628-634. [DOI] [PubMed] [Google Scholar]
- 17.Hewett, M. K., B. S. Robinson, P. T. Monis, and C. P. Saint. 2003. Identification of a new Acanthamoeba 18S rRNA gene sequence type corresponding to the species Acanthamoeba jacobsi Sawyer, Nerad and Visvesvara, 1992 (Lobosea: Acanthamoebidae). Acta Protozool. 42:325-329. [Google Scholar]
- 18.Horn, M., A. Collingro, S. Schmitz-Esser, C. L. Beier, U. Purkhold, B. Fartmann, P. Brandt, G. J. Nyakatura, M. Droege, D. Frishman, T. Rattei, H. W. Mewes, and M. Wagner. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728-730. [DOI] [PubMed] [Google Scholar]
- 19.Horn, M., T. R. Fritsche, R. K. Gauton, K. H. Schleifer, and M. Wagner. 1999. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryohilus. Environ. Microbiol. 1:357-367. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, M., N. White, P. Giffard, and P. Timms. 1999. Epizootiology of Chlamydia infections in two free-range koala populations. Vet. Microbiol. 65:255-264. [DOI] [PubMed] [Google Scholar]
- 21.Khan, N. A. 2003. Pathogenesis of Acanthamoeba infections. Microb. Pathog. 34:277-285. [DOI] [PubMed] [Google Scholar]
- 22.Khan, N. A. 2006. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol. Rev. 30:564-595. [DOI] [PubMed] [Google Scholar]
- 23.Kreader, C. A. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutlin, A., P. M. Roblin, S. Kumar, S. Kohlhoff, T. Bodetti, P. Timms, and M. R. Hammerschlag. 2007. Molecular characterization of Chlamydophila pneumoniae isolates from Western barred bandicoots. J. Med. Microbiol. 56:407-417. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman, D., S. Kahane, and M. G. Friedman. 1997. Pneumonia with serological evidence of acute infection with the Chlamydia-like microorganism “Z”. Am. J. Respir. Crit. Care Med. 156:578-582. [DOI] [PubMed] [Google Scholar]
- 26.Maghsood, A. H., J. Sissons, M. Rezaian, D. Nolder, D. Warhurst, and N. A. Khan. 2005. Acanthamoeba genotype T4 from the UK and Iran and isolation of the T2 genotype from clinical isolates. J. Med. Microbiol. 54:755-759. [DOI] [PubMed] [Google Scholar]
- 27.Marciano-Cabral, F., R. Puffenbauer, and G. A. Cabral. 2000. The increasing importance of Acanthamoeba infections. J. Eukaryot. Microbiol. 47:29-36. [DOI] [PubMed] [Google Scholar]
- 28.Matsuo, J., Y. Hayashi, S. Nakamura, M. Sato, Y. Mizutani, M. Asaka, and H. Yamaguchi. 2008. Novel Parachlamydia acanthamoebae quantification method based on coculture with amoebae. Appl. Environ. Microbiol. 74:6397-6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mergeryan, H. 1991. The prevalence of Acanthamoeba in the human environment. Rev. Infect. Dis. 13(Suppl. 5):S390-S391. [DOI] [PubMed] [Google Scholar]
- 30.Page, F. C. 1988. A new key to freshwater and soil gymnamoebae. Freshwater Biological Association, Ambleside, United Kingdom.
- 31.Peeling, R. W., and R. C. Brunham. 1996. Chlamydiae as pathogens: new species and new issues. Emerg. Infect. Dis. 2:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Zaragoza, S. 1994. Ecology of free-living amebae. Crit. Rev. Microbiol. 20:225-241. [DOI] [PubMed] [Google Scholar]
- 33.Schachter, J. 1988. The intracellular life of Chlamydia. Curr. Top. Microbiol. Immunol. 138:109-139. [PubMed] [Google Scholar]
- 34.Schroeder, J. M., G. C. Booton, J. Hay, I. A. Niszl, D. V. Seal, M. B. Markus, P. A. Fuerst, and T. J. Byers. 2001. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J. Clin. Microbiol. 39:1903-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster, F. L. 2002. Cultivation of pathogenic and opportunistic free-living amebas. Clin. Microbiol. Rev. 15:342-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storey, C., M. Lusher, P. Yates, and S. Richmond. 1993. Evidence for Chlamydia pneumoniae of non-human origin. J. Gen. Microbiol. 139:2621-2626. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, V., K. Herrera-Rimann, D. S. Blanc, and G. Greub. 2006. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 72:2428-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Widjojoatmodjo, M. N., A. C. Fluit, and J. Verhoef. 1994. Rapid identification of bacteria by PCR-single-strand conformation polymorphism. J. Clin. Microbiol. 32:3002-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]