Abstract
BP3385 has been proposed as a diagnostic PCR target for discriminating between Bordetella pertussis and other Bordetella species that also infect humans. Our results demonstrate that this gene is also present in some strains of Bordetella hinzii and Bordetella bronchiseptica.
Bordetella pertussis is an obligate human pathogen and the causative agent of whooping cough. Identification based on traditional culture and biochemical testing is the current diagnostic standard but is hampered by poor sensitivity and a slow turnaround time. Primers targeting a variety of genes have been proposed and evaluated for PCR-based identification strategies (11). These methods provide rapid and highly sensitive detection, but specificity is suboptimal since other species of Bordetella sometimes associated with respiratory disease in humans, including Bordetella bronchiseptica and Bordetella hinzii, may also possess the targeted sequences (4, 7, 9). Based on a comparison of the genomes of 22 isolates of B. bronchiseptica, 1 of human origin, and 10 isolates each of B. pertussis and B. parapertussis, it was suggested that open reading frame BP3385 of B. pertussis may be a highly specific diagnostic target (1). The authors of a subsequent study that included an unspecified number of B. bronchiseptica and B. hinzii isolates similarly concluded that BP3385 is specific for B. pertussis (3).
The goal of the present study was to screen a large and genetically heterogeneous group of Bordetella strains, including many of human origin, to further evaluate the specificity of BP3385 as a diagnostic target for B. pertussis.
A total of 142 Bordetella isolates originating from the United States, Europe, or Australia were evaluated using the previously described BP3385-specific real-time (RT)-PCR assay (3). These included 112 B. bronchiseptica isolates representing 31 unique PvuII ribotypes, 23 isolates of B. hinzii representing 4 PvuII ribotypes, and 7 B. avium isolates representing 6 PvuII ribotypes (5, 6, 8; K. B. Register, unpublished data). Twenty-five B. bronchiseptica isolates and 6 B. hinzii isolates were of human origin. The Tohama strain of B. pertussis, in which BP3385 was identified previously (1), was used as a positive control. The RB50 strain of B. bronchiseptica, which is missing BP3385 (1), was used as a negative control. Bacteria were cultured on Bordet-Gengou agar supplemented with 10% sterile, defibrinated sheep's blood at 37°C for 18 to 36 h. Chromosomal DNA was purified using a commercially available kit (Promega, Madison, WI) and quantified with PicoGreen reagent (Invitrogen, Carlsbad, CA).
Excepting the B. pertussis positive control, only B. bronchiseptica strain PV6 gave unequivocally positive results in the BP3385 RT-PCR assay. Results for an additional 8 isolates of B. bronchiseptica and 18 isolates of B. hinzii were interpreted as inconclusive on the basis of measurable, but elevated, cycle threshold (CT) values (Table 1). All remaining isolates tested gave negative results.
TABLE 1.
Bordetella isolates with positive or inconclusive RT-PCR results for BP3385a
| Species | Isolate or strain | CT | Hybridization to BP3385 probe/approx fragment size (kb) | Amplicon obtained with BP3385-1/BP3385-2 | BP3385 allelic variant |
|---|---|---|---|---|---|
| B. pertussis | Tohama | 26.2 | +/4.7 | + | 1 |
| B. bronchiseptica | RB50 | ND | − | − | |
| B. bronchiseptica | PV6 | 26.1 | +/8.0 | + | 4 |
| B. bronchiseptica | OSU-083 | 42.9 | +/3.8 | + | 5 |
| B. bronchiseptica | OSU-189 | 35.0 | − | ND | |
| B. bronchiseptica | OSU-553 | 34.6 | − | ND | |
| B. bronchiseptica | OSU-585 | 32.9 | − | ND | |
| B. bronchiseptica | ISU-CA90 BB1187 | 37.8 | − | ND | |
| B. bronchiseptica | ISU-CA90 BB135 | 30.1 | − | ND | |
| B. bronchiseptica | ISU-WI91 BB062/T5 | 40.8 | +/3.8 | + | 5 |
| B. bronchiseptica | ISU-WI91 BB064/T3 | 40.9 | +/3.8 | + | 5 |
| B. hinzii | ATCC 51783 | 36.8 | +/7.9 | + | 3 |
| B. hinzii | 4134 | 35.5 | +/7.9 | + | 3 |
| B. hinzii | 4135 | 36.0 | +/11.2 | + | 2 |
| B. hinzii | 4140 | 32.3 | − | ND | |
| B. hinzii | 4159 | 35.5 | +/7.9 | + | 3 |
| B. hinzii | 4161 | 35.3 | +/7.9 | + | 3 |
| B. hinzii | 4449 | 33.3 | − | ND | |
| B. hinzii | 4595 | 35.2 | +/7.9 | + | 3 |
| B. hinzii | OH87 BAL006II | 34.8 | +/11.2 | + | 2 |
| B. hinzii | OH87 BAL007II | 44.2 | − | ND | |
| B. hinzii | OH87 BAL028II | 34.7 | +/7.9 | + | 3 |
| B. hinzii | OH87 BAL030II | 35.2 | +/7.9 | + | 3 |
| B. hinzii | CA90 BAL33 | 35.6 | +/7.9 | + | 3 |
| B. hinzii | CA90 BAL1384 | 38.9 | − | ND | |
| B. hinzii | BAL168II | 35.6 | +/7.9 | + | 3 |
| B. hinzii | F-1 | 33.6 | − | − | |
| B. hinzii | 1277 | 34.4 | − | − | |
| B. hinzii | DBL191 | 34.2 | − | ND |
ND, not determined; +, present; −, absent.
Isolates with either positive or inconclusive RT-PCR results were further evaluated by Southern blotting. Three micrograms of purified DNA from each isolate was digested with EcoRV, for which no recognition sites are present in BP3385, and the resulting fragments were separated on 0.6% agarose gels. Blots were prepared and hybridized under highly stringent conditions, as described previously (5), with a probe encompassing the entire 450-bp BP3385 open reading frame. The probe was amplified from purified B. pertussis Tohama DNA and labeled with digoxigenin by using a commercially available kit (Roche Applied Science, Indianapolis, IN) and PCR primers BP3385-1 (5′-ATGGCACGACCATCCAAATACCA-3′) and BP3385-2 (5′-TTAAGCGTCGGCATCAGGGA-3′). Following brief exposure times (5 to 20 min), films displayed a single fragment hybridizing to the BP3385 probe for B. pertussis, 4 isolates of B. bronchiseptica (including PV6), and 11 isolates of B. hinzii (Table 1). A total of 4 uniquely sized fragments, none of which displayed the same mobility as the B. pertussis BP3385-containing fragment, was detected among all positive isolates. Representative results are included in Fig. 1. When films were overexposed, an additional, faintly detectable fragment of ∼4.2 kb was evident in B. hinzii strains 1277 and F-1.
FIG. 1.
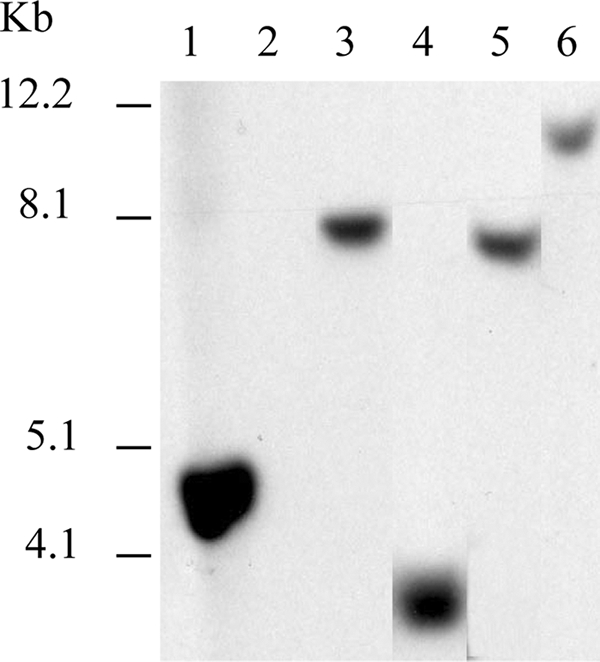
Southern blot with EcoRV-digested Bordetella genomic DNA hybridized to a BP3385 probe. Lanes: 1, B. pertussis Tohama; 2, B. bronchiseptica RB50; 3, B. bronchiseptica PV6; 4, B. bronchiseptica OSU-083; 5, B. hinzii ATCC 51783; and 6, B. hinzii 4135. Relative positions of DNA size markers are indicated to the left.
To further verify the presence of a BP3385 ortholog in B. bronchiseptica and B. hinzii isolates that were positive by Southern blotting, conventional PCR was carried out in a model 9700 thermal cycler (Applied Biosystems, Foster City, CA) using 100-ng samples of purified DNA from the appropriate strains and primers BP3385-1 and BP3385-2. Reaction mixtures included 0.4 μM primers, 1 U of AmpliTaq polymerase (Applied Biosystems, Foster City, CA), 2.5 μl of 10× buffer II (100 mM Tris-HCl, pH 8.3, 500 mM KCl), 2.5 mM MgCl2, 10% dimethyl sulfoxide (DMSO), and 200 μM deoxynucleoside triphosphates (dNTPs) in a final volume of 25 μl. Cycling conditions were 2 min at 95°C, 30 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s, and a final extension step of 72°C for 7 min. Five microliters of each PCR mixture was analyzed by agarose gel electrophoresis in a 3:1 NuSieve gel (Cambrex BioScience, Rockland, ME) containing a 10−4 dilution of GelRed stain (Phenix Research Products, Candler, NC). An amplicon of the size predicted for B. pertussis BP3385 was obtained from each of the 4 isolates of B. bronchiseptica and 11 isolates of B. hinzii previously noted to strongly hybridize with the BP3385 probe, as well as B. pertussis Tohama. No amplicons were obtained when DNA from B. hinzii strain 1277 or F-1 was used, although parallel testing with a 16S rRNA-specific PCR (10) confirmed the integrity of the DNA used as a template. Amplicons were purified using spin columns (Qiagen, Valencia, CA) and sequenced directly (with two reactions from each strand) by fluorescence-based cycle sequencing with AmpliTaq and BigDye Terminator kits on an ABI 377 sequencer at the National Animal Disease Center Genomics Unit. Sequences were analyzed using the Vector NTI suite (Invitrogen).
The sequence we obtained from the BP3385-1/BP3385-2 amplicon from B. pertussis Tohama is identical to the corresponding sequence already reported for both Tohama (GenBank accession no. NC_002929) and B. pertussis isolate ATCC 9797 (3). Four additional allelic variants were identified among the B. hinzii and B. bronchiseptica strains evaluated, with two each found exclusively in one species (Fig. 2). Every variant is associated with an EcoRV fragment of unique size (Table 1), suggesting that strains constituting variant-specific groups may have additional genetic similarities. The predicted amino acid sequences of all variants are distinct from one another. The amino acid sequence predicted for variant 4, found only in B. bronchiseptica strain PV6, most closely approximates the sequence for B. pertussis BP3385 (with 98.0% amino acid identity). The amino acid sequences of the remaining variants are 89.1 to 91.1% identical to the corresponding sequence from B. pertussis.
FIG. 2.
Comparison of B. pertussis, B. bronchiseptica, and B. hinzii BP3385-1/BP3385-2 amplicon sequence variants. The top line shows the sequence obtained from B. pertussis Tohama (BP3385); an identical sequence has also been reported for B. pertussis ATCC 9797 (3). Variants 2 and 3 were found only in B. hinzii, while variants 4 and 5 were found only in B. bronchiseptica. Dashes represent conserved bases; substitutions are indicated by the appropriate letter. Bases in bold indicate nonconservative substitutions. RT-PCR primer sequences are shaded, and the RT-PCR probe sequence is underlined. Nucleotide positions are numbered relative to the BP3385 start codon.
The prevalence of BP3385 reported here for B. hinzii, 47.8%, suggests that the usefulness of this locus as a detection target for B. pertussis is limited. We found its prevalence in B. bronchiseptica to be considerably lower, 3.6%, but additional study may reveal a higher prevalence within specific subpopulations. With the exception of B. bronchiseptica PV6, all B. hinzii and B. bronchiseptica isolates in which we identified a BP3385 homolog are of avian origin. Approximately 70% of the B. hinzii strains we evaluated here were obtained from an avian host, compared to only 15% of the B. bronchiseptica strains tested. It is currently unknown whether B. hinzii isolates arising from particular hosts share unique genetic characteristics. However, sequence data derived from housekeeping genes indicate that most avian B. bronchiseptica strains, including many evaluated here, fall within the phylogenetic lineage from which B. pertussis is proposed to have evolved and which comprises largely human isolates (2; Register, unpublished). Although originally cultured from a pig, B. bronchiseptica PV6 is also in this lineage. A search for BP3385 in additional strains of the lineage may more accurately predict the frequency with which human B. bronchiseptica isolates would be mistakenly identified as B. pertussis when BP3385 is used as a diagnostic target.
Allelic variants 2, 3, and 5 have multiple base mismatches with the BP3385 RT-PCR primer and probe sequences, including the 3′-terminal position of the forward primer (Fig. 2). Although both Southern blotting and sequencing of amplicons obtained by conventional PCR prove the existence of a BP3385 homolog in isolates with these alleles, mispriming at some other locus in their genomes is a more likely explanation for the high CT values obtained with the BP3385 RT-PCR. In contrast, except for a mismatch near the 5′ terminus of the RT-PCR forward primer, the variant 4 sequence corresponding to the RT-PCR probe and primers is identical to that from B. pertussis, thus explaining the lower CT value and positive RT-PCR result obtained with B. bronchiseptica PV6.
Our results demonstrate that BP3385 is not restricted solely to B. pertussis and suggest that diagnostic assays targeting this open reading frame are unlikely to provide a degree of specificity any higher than that achieved with currently used PCR assays. Clinical Bordetella isolates with positive BP3385 RT-PCR results require additional confirmatory testing for definitive identification as B. pertussis. Ongoing Bordetella genome sequencing projects and global genome comparisons will perhaps identify promising alternative targets.
Nucleotide sequence accession numbers.
GenBank accession numbers for sequences obtained in this study are HM189275 to HM189289.
Acknowledgments
We gratefully acknowledge the expert technical assistance of Michael Mullins and Sarah Shore. We are indebted to David Alt and Karen Halloum at the National Animal Disease Center Genomics Unit for DNA sequence data.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 186:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diavatopoulos, D. A., C. A. Cummings, L. M. Schouls, M. M. Brinig, D. A. Relman, and F. R. Mooi. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guthrie, J. L., C. Seah, S. Brown, P. Tang, F. Jamieson, and S. J. Drews. 2008. Use of Bordetella pertussis BP3385 to establish a cutoff value for an IS481-targeted real-time PCR assay. J. Clin. Microbiol. 46:3798-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loeffelholz, M. J., and G. N. Sanden. 2007. Bordetella, p. 803-814. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 5.Register, K. B., A. Boisvert, and M. R. Ackermann. 1997. Use of ribotyping to distinguish Bordetella bronchiseptica isolates. Int. J. Syst. Bacteriol. 47:678-683. [DOI] [PubMed] [Google Scholar]
- 6.Register, K. B., and T. Magyar. 1999. Optimized ribotyping protocol applied to Hungarian Bordetella bronchiseptica isolates: identification of two novel ribotypes. Vet. Microbiol. 69:277-285. [DOI] [PubMed] [Google Scholar]
- 7.Register, K. B., and T. L. Nicholson. 2007. Misidentification of Bordetella bronchiseptica as Bordetella pertussis using a newly described real-time PCR targeting the pertactin gene. J. Med. Microbiol. 56:1608-1610. [DOI] [PubMed] [Google Scholar]
- 8.Register, K. B., R. E. Sacco, and G. E. Nordholm. 2003. Comparison of ribotyping and restriction enzyme analysis for inter- and intraspecies discrimination of Bordetella avium and Bordetella hinzii. J. Clin. Microbiol. 41:1512-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Register, K. B., and G. N. Sanden. 2006. Prevalence and sequence variants of IS481 in Bordetella bronchiseptica: implications for IS481-based detection of Bordetella pertussis. J. Clin. Microbiol. 44:4577-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Register, K. B., and A. G. Yersin. 2005. Analytical verification of a PCR assay for identification of Bordetella avium. J. Clin. Microbiol. 43:5567-5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riffelmann, M., C. H. Wirsing von Konig, V. Caro, and N. Guiso. 2005. Nucleic acid amplification tests for diagnosis of Bordetella infections. J. Clin. Microbiol. 43:4925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]



