Abstract
The Aptima HIV-1 RNA qualitative assay tested with a WHO-approved HIV type 1 RNA standard in 16- and 32-member pools detected 100% of the pools (1,070 and 2,130 HIV-1 RNA copies/ml/pool, respectively), thus exceeding the FDA-required lower limit of detection. The Aptima test can be used to screen for acute-phase HIV infection.
Since 1999, pooled nucleic acid amplification testing (NAAT) for HIV type 1 (HIV-1) and hepatitis C virus has been used by blood donor screening programs (1, 8, 11). In 2002, the North Carolina Department of Health and Human Services and the University of North Carolina at Chapel Hill evaluated the use of pooled NAAT to screen for acute-phase HIV infection among a routine testing population and concluded that pooling specimens for NAAT is feasible in public health practice (10). The Centers for Disease Control and Prevention (CDC) initiated the Acute HIV-1 Infection (AHI) study in 2006 to investigate the utility of pooled NAAT with third-generation antibody screening to identify acute-phase HIV infection cases in targeted and routine screening programs. The CDC AHI study chose the 16-member pooled HIV-1 NAAT so that test results could be reported within 7 days after specimen collection (9). As part of this study, we evaluated the sensitivity of the Aptima HIV-1 RNA qualitative assay in 16- and 32-member pools.
The Aptima HIV-1 RNA qualitative assay (Gen-Probe Inc., San Diego, CA) was approved by the FDA in October 2006 as an aid in the diagnosis of HIV infection but not for HIV-1 detection in pooled specimens (4). Because no guidelines exist for using a pooled HIV-1 RNA protocol for routine HIV screening, we chose HIV RNA levels for our evaluation that were above and below the FDA blood donor sensitivity requirement of 5,000 copies/ml/specimen in minipools of blood product (5). The Aptima product insert (3) states that when a diluted WHO standard is used, the lowest detection limit is 33 copies/ml/specimen (100% reliability). For our study, the three HIV RNA testing standards were prepared at the CDC laboratory by diluting a WHO standard containing 150,000 HIV-1 RNA copies/ml in HIV-negative plasma to 1,070 (level 1), 2,130 (level 2), and 5,330 (level 3) copies/ml for final concentrations of 67, 133, and 333 copies/ml/16-member pool, respectively (7). These testing standards were requantified in triplicate in three separate runs by using the Roche Cobas Amplicor HIV-1 Monitor test, v1.5 (Roche Molecular Systems, Inc., Pleasanton, CA). Dilution-specific group means and standard deviations were calculated as the mean of means by using least-squares regression and assuming constant variance in all vials (6). Given the imprecision of viral load estimates at low concentrations, we rounded estimates to two significant digits. The HIV-1 RNA levels in the three CDC-prepared standards were 900, 2,500, and 6,100 copies/ml, representing 1:16-diluted concentrations of 56, 156, and 381 copies/ml/16-member pool, respectively (Table 1). Aliquots of each standard were put in coded vials, stored at −70°C, and shipped on dry ice to the New York State Department of Health (NYSDOH) laboratory.
TABLE 1.
Characterization of secondary HIV-1 RNA standards used to construct 16- and 32- member specimen pools
| Testing standard | No. of copies/ml |
|||
|---|---|---|---|---|
| Expected in undiluted standard | Expected in 16-member specimen pool | Measured in undiluted standard | Calculated for 16-member specimen pool | |
| Level 1 | 1,070 | 67 | 900 (200)a | 56 |
| Level 2 | 2,130 | 133 | 2,500 (1,300) | 156 |
| Level 3 | 5,330 | 333 | 6,100 (1,200) | 381 |
Mean and standard deviation calculated by using the mean-of-means model and rounding to 100 copies/ml.
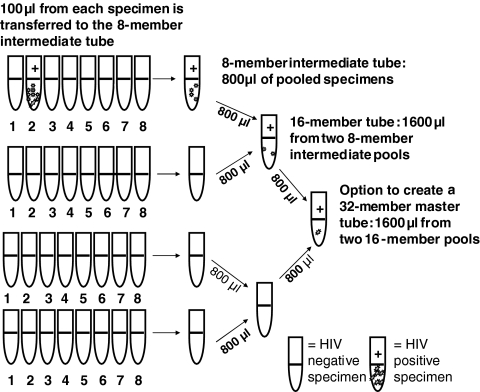
Three technologists performed a two-stage pooling strategy with the Hamilton Microlab AT Plus 2 (Hamilton Company, Reno, NV) where 100 μl is pipetted from eight plasma specimens to create an 800-μl intermediate pool. Two intermediate pools were combined to create a 1.6-ml 16-member pool. Only the level 2 testing standard (2,130 copies/ml) was used to create 32-member pools from two 16-member pools (Fig. 1).
FIG. 1.
Construction of 16- and 32-member plasma pools using an automated two-stage pooling strategy with the Hamilton Microlab AT Plus 2 (Hamilton Company, Reno, NV). One hundred microliters was pipetted from eight plasma specimens to create an 800-μl intermediate pool. Two intermediate pools were combined to create a 1.6-ml 16-member pool. To expand the pool to 32 members, 800 μl was pipetted from two 16-member pools into a single tube for analysis.
According to the common technical specifications of the European directive 98/79/EC for in vitro diagnostics, viral standard dilutions should be tested at least 24 times for a statistically valid 95% detection limit (2). The standards were run in 21 separate runs; the last 3 runs (runs 19 to 21) tested two aliquots of a testing standard, for a total of 24 runs; and retesting was done to verify reactivity (n = 24).
All 16- and 32-member pooling runs were HIV-1 RNA positive, including one invalid run (16-member pool) with a level 2 testing standard that was positive after retesting. The increasing luminometry values (signal-to-cutoff [S/CO] ratios) with increasing RNA input from the three testing standards used in these pools indicated that these low levels of HIV-1 RNA were still within the assay's dynamic range (data not shown). After unblinding of the 16-member pooling results, the level 2 testing standard was chosen for the 32-member pools. Thus, the level 2 testing standard (2,130 copies/ml) in the 32-member pool was the equivalent of the level 1 standard (1,070 copies/ml) in the 16-member pools. Due to cost, the 32-member pools were analyzed in 12 runs and the analyses were not repeated unless the results were reported as nonreactive or invalid (Table 2).
TABLE 2.
Performance of the Aptima HIV-1 RNA qualitative assay with 16- and 32-member specimen pools
| Pool size and testing standard | Mean no. of HIV-1 RNA copies/ml | Mean NYSDOH S/CO ratio (SD) | CV%c |
|---|---|---|---|
| 16 membersa | |||
| Level 1 | 1,070 | 12.4 (5.1) | 40.7 |
| Level 2 | 2,130 | 17.3 (5.4) | 30.9 |
| Level 3 | 5,330 | 22.0 (4.6) | 20.8 |
| 32 members,b level 2 | 2,130 | 13.2 (6.4) | 48.4 |
Each standard was analyzed 24 times, and the analysis was repeated.
The standard was analyzed 12 times, and the analysis was not repeated.
CV%, percent coefficient of variation, analysis of variance.
The 100% rate of HIV-1 RNA detection in the 16-member (1,070 copies/ml of infected donor plasma/pool) and 32-member (2,130 copies/ml of infected donor plasma/pool) pools shows that the pooling assay's sensitivity exceeded that of the FDA-required lower limit of detection (5,000 copies/ml of infected donor plasma/pool) by at least 2-fold. This high detection rate would be predicted in both the 16- and 32-member pooling strategies because of the Aptima assay's level of sensitivity (33 HIV-1 RNA copies/ml of plasma) and the final concentration of HIV-1 RNA used in the 16- and 32-member pools (≥67 HIV-1 RNA copies/ml). Our results demonstrate that using one standard concentration, such as 2,100 copies/ml of infected donor plasma/pool, with 16- or 32-member pools in the Aptima assay should allow most public health laboratories to establish a pooling protocol that will detect HIV infection.
Our evaluation was subject to two limitations: we evaluated only 16- and 32-member pools to allow faster turnaround in reporting test results (9), and we did not conduct an evaluation using multiple reagent lots. Gen-Probe manufactured and released only one lot during our study.
In summary, the NYSDOH laboratory validated the Aptima assay for identifying low levels of HIV-1 RNA in pooled specimens. The level of sensitivity in these pools exceeded the FDA-required lower limit of detection (5,000 copies/ml of infected donor plasma/pool). We previously reported that two laboratories, the NYSDOH laboratory and the Florida Bureau of Laboratories, used the Aptima HIV-1 qualitative assay in the CDC HIV-1 acute-phase infection study for 16-member pools; Aptima had a positive predictive value of 0.9804 (95% confidence interval [CI], 0.9071 to 0.9990) and a specificity of 0.9998 (95% CI, 0.9992 to 1.000) (9). Thus, we encourage public health laboratories to evaluate the use of pooled HIV-1 NAAT to improve the laboratory detection of early and acute-phase HIV-1 infections while reducing the cost of NAAT for the detection of HIV-1 infection.
Acknowledgments
We thank the following for their contributions to this evaluation: Kurt Dunn, Philip Rivenburg, and Thomas Miller at the Wadsworth Center of the NYSDOH; Sally Fordan and David Olanike at the Retrovirology Unit of the Florida Bureau of Laboratories; and the Division of Laboratory Systems, CDC.
The findings and conclusions in this report are ours and do not necessarily represent the official position of the CDC.
The use of trade names in this report is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services or the CDC.
Footnotes
Published ahead of print on 30 June 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Fiebig, E. W., D. J. Wright, B. C. Rawal, P. E. Garrett, R. T. Schumacher, L. Peddada, C. Heldebrant, R. Smith, A. Conrad, S. H. Kleinman, and M. P. Busch. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871-1879. [DOI] [PubMed] [Google Scholar]
- 2.Finney, D. J. 1971. Probit analysis: parallel line analysis, 3rd ed. Cambridge University Press, Cambridge, United Kingdom.
- 3.Gen-Probe Incorporated. 2006. Product insert, Aptima HIV-1 proficiency panel. Gen-Probe Incorporated, San Diego, CA.
- 4.Gen-Probe Incorporated. 2009. Product insert, Aptima HIV-1 RNA qualitative assay. Gen-Probe Incorporated, San Diego, CA.
- 5.Lelie, P. N., A. J. van Drimmelen, H. T. Cuypers, S. J. Best, S. L. Stramer, C. Hyland, J. P. Allain, P. Moncharmont, C. Defer, M. Nübling, A. Glauser, M. S. Cardoso, J. F. Viret, M. H. Lankinen, L. Grillner, W. Urs, J. Coste, V. Schottstedt, B. Masecar, and E. M. Dax. 2002. Sensitivity of HCV RNA and HIV RNA blood screening assays. Transfusion 42:527-536. [DOI] [PubMed] [Google Scholar]
- 6.Neter, J., M. H. Kutner, W. Wasserman, and C. J. Nachtsheim. 1989. Applied linear regression models. Richard D. Irwin, Inc., Homewood, IL.
- 7.NIH AIDS Research and Reference Reagent Program. Reagent: HIV-1 VQA RNA quantification standard. Catalog number 3443, release category D, lot number 200207029. NIH, Bethesda, MD.
- 8.Owen, S. M., C. Yang, T. Spira, C. Y. Ou, C. P. Pau, B. S. Parekh, D. Candal, D. Kuehl, M. S. Kennedy, D. Rudolph, W. Luo, N. Delatoree, S. Masciotra, M. L. Kalish, F. Cowart, T. Barnett, R. Lal, and J. S. McDougal. 2008. Alternative algorithms for human immunodeficiency virus infection diagnosis using tests that are licensed in the United States. J. Clin. Microbiol. 46:1588-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel, P., D. Mackellar, P. Simmons, A. Uniyal, K. Gallagher, B. Bennett, T. Sullivan, A. Kowalski, M. Parker, M. LaLota, P. Kerndt, and P. Sullivan for the CDC AHI Study Group. 2010. Detecting acute human immunodeficiency virus infection using 3 different screening immunoassays and nucleic acid amplification testing for human immunodeficiency virus RNA, 2006-2008. Arch. Intern. Med. 170:66-74. [DOI] [PubMed] [Google Scholar]
- 10.Pilcher, C. D., S. A. Fiscus, T. Q. Nguyen, E. Foust, L. Wolf, D. Williams, R. Ashby, J. O. O'Dowd, J. T. McPherson, B. Stalzer, L. Hightow, W. C. Miller, J. J. Eron, Jr., M. S. Cohen, and P. A. Leone. 2005. Detection of acute infections during HIV testing in North Carolina. N. Engl. J. Med. 352:1873-1883. [DOI] [PubMed] [Google Scholar]
- 11.Stramer, S. L., S. A. Glynn, S. H. Kleinman, D. M. Strong, S. Caglioti, D. J. Wright, R. Y. Dodd, and M. P. Busch for the National Heart, Lung, and Blood Institute Nucleic Acid Test Study Group. 2004. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N. Engl. J. Med. 351:760-768. [DOI] [PubMed] [Google Scholar]