Abstract
The objective of this study was to determine the reliability of the real-time PCR assay for determining the group B Streptococcus (GBS) status of women in labor. In this prospective study we compared the results of culture and PCR testing of vaginal and rectal samples collected by nursing staff when women were in labor. Patients' charts were also reviewed to obtain relevant information about pregnancy risk factors. Our results demonstrated a sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of 90.5%, 96.1%, 86.4%, and 97.4%, respectively, for rapid PCR. Of the 196 women evaluated, 29 (14.8%) presented with unknown GBS status, 11 (37.9%) of whom received unnecessary intrapartum antibiotics. The rapid real-time PCR test was robust and was able to reliably detect the presence of GBS in women in labor within 1 h of specimen submission to the laboratory. We recommend that the rapid PCR test be targeted to women who present in labor with unknown GBS status. In cases where the laboratory does not offer 24-h availability of testing, sample collection followed by PCR testing the next morning is still valuable and provides reliable results 24 to 48 h faster than culture and will aid appropriate decision-making regarding continuing or stopping antibiotics for neonates of women with unknown GBS status.
Streptococcus agalactiae, the group B Streptococcus (GBS) species, colonizes the genitourinary and/or gastrointestinal tract of 10 to 40% of pregnant women (2, 8, 11, 14, 16, 18, 20, 22, 24). GBS causes early-onset sepsis (EOS) in newborns and can be responsible for severe morbidity and mortality in infected neonates (2, 8, 11, 14, 24). Early-onset sepsis in neonates due to GBS has been shown to be preventable if intrapartum antimicrobial prophylaxis is given to mothers who carry GBS (5, 9, 19). Providing that there has been at least 2 h of exposure to the prophylactic antibiotics during labor, the risk of GBS colonization and EOS in neonates born to mothers who carry GBS is significantly reduced (9). Current American and Canadian guidelines recommend that routine rectal/vaginal screening be performed on pregnant women at 35 to 37 weeks of gestation to determine their GBS status (20, 22). If the screening or urine cultures during pregnancy indicate that GBS is present, or if the women have previously given birth to an infant with invasive GBS disease, intrapartum prophylaxis should be given to the mother (20, 22). If the GBS status is unknown and the woman is in labor, risk factors are used to determine whether intrapartum antibiotics for GBS should be administered or not (20).
Although the rate of GBS neonatal EOS decreased remarkably after the introduction of intrapartum prophylaxis recommendations, a sizable number of newborns still develop GBS disease, particularly preterm infants (15). This is significant since women who deliver preterm usually do not get the chance to be screened for GBS at 35 to 37 weeks of gestation, and therefore, their GBS status is mostly unknown at labor. Also of concern are reports indicating that as many as two-thirds of infants who developed GBS EOS were born to mothers who were negative for GBS upon prenatal screening (19). On the other hand, evidence shows that the increasing use of intrapartum prophylactic antibiotics has shifted the profile of causative agents of EOS toward Gram-negative organisms (15). Moreover, the administration of maternal intrapartum antibiotics has been linked to the emergence of resistant strains of bacteria in recent years (3, 23).
Such data reveal the inadequate use of prophylactic antibiotics despite screening at 35 to 37 weeks of gestation. By following current GBS recommendations, a considerable portion of pregnant women are still treated inappropriately, either as a result of the failure to identify a subset of pregnant women who are colonized with GBS at delivery or by providing unnecessary prophylactic antibiotics to a portion of GBS-negative women. One study showed that the risk-based approach exposes 65 to 85% of women with risk factors who are GBS negative to antibiotics inappropriately (14).
While culture has always been the standard method for GBS detection, this method also carries some limitations. Using culture as a diagnostic tool, it usually takes 24 to 72 h for results to become available. Furthermore, even when culture is performed as a screening tool during weeks 35 to 37 of pregnancy, difficulties such as the availability of results at the time of labor or change in the status of GBS colonization can be encountered. Also, women with no prenatal care and ones who deliver preterm do not have access to prenatal GBS screening (21).
There is a need for a rapid test that would facilitate in-labor testing for GBS so that women who present with unknown GBS status can be given intrapartum antibiotics only if they carry GBS. A rapid PCR test has been approved by both the FDA and Health Canada for in-labor testing for GBS, but its position in everyday GBS practice remained unclear (i.e., to replace current culture screening or to provide the test for all or a subset of women in labor).
The objectives of this study were to determine whether the real-time PCR assay was robust enough when samples were collected by the routine nursing staff (and not study personnel) and whether the turnaround time when offered to women in labor was fast enough to still provide value in diagnostic laboratories that were not functional 24 h a day.
MATERIALS AND METHODS
Study design.
This study was reviewed and approved by the ethics and research boards at the University of Manitoba and St. Boniface General Hospital, and all relevant clinical study standard operation procedures were followed. The St. Boniface General Hospital Women and Child program handles about 5,000 births per year. Two hundred five consecutive women who provided informed consent were enrolled between July 2004 and July 2005. Data from nine patients with incomplete charts/missing information were excluded from the study.
Specimen processing and culturing.
A combined rectal/vaginal specimen was collected into the Copan Venturi Transystem and sent immediately to the laboratory through the pneumatic tube system. If the sample was received afterhours, when the technologist was not available, the rectal/vaginal swab was stored at 4°C until the following day. In the laboratory the specimen was eluted from the swab, and 50 μl was removed for PCR testing. The swab and 500 μl of the remaining sample were set up for culture by adding 5 ml of selective Todd-Hewitt broth containing 15 μg/ml nalidixic acid and 10 μg/ml colistin to the sample tube. The broth enrichment culture was incubated overnight at 35°C and subcultured onto tryptic soy agar medium containing 5% sheep blood (BA) followed by overnight incubation at 35°C. Any suspected GBS colonies were further evaluated by using group B latex agglutination to confirm the identity of GBS. All confirmed GBS isolates were stocked and frozen at −70°C.
Rapid PCR testing.
The real-time PCR IDI-StrepB assay (Somagen, Edmonton, AB, Canada) was used to detect GBS in combined rectal/vaginal specimens taken from women in labor. The genetic target of this assay is the cfb gene, which encodes a diffusible extracellular protein that is well conserved in all GBS isolates. The kit was used according to the manufacturer's instructions, and PCR was performed directly on the specimens collected (i.e., no culture enrichment prior to PCR). Each sample was processed within 24 h of receipt.
The DNA was extracted from the 50-μl aliquot by physical and chemical lysis according to the manufacturer's instructions. This one-step lysis was performed without the need for additional manipulation/purification of DNA and took less than 15 min. The DNA was stored at −20°C until there were sufficient samples to run a whole kit (usually performed in batches of 50 samples) in a Smart Cycler instrument. One positive-control tube and one negative-control tube were prepared and run for each kit. Amplification and detection were performed as a one-step process and took 20 to 25 min. Overall, the completion of the test from receipt of the sample in the laboratory to the final PCR result took less than 45 min. Where there were discordant PCR and culture results, the broth and plate cultures were reviewed, and the PCR was repeated.
Statistical analysis.
A chi-square test based on the two-tailed Fisher exact method was applied to detect significant differences between variables using GraphPad QuickCalcs online software.
Erythromycin and clindamycin susceptibility of GBS.
To obtain site-specific information, 148 GBS isolates from rectal/vaginal swabs submitted to the St. Boniface General Hospital Microbiology Laboratory between 1 January 2003 and 31 December 2003 (i.e., 1 year prior to the current GBS study) were tested for their susceptibility to erythromycin and clindamycin using the 2003 version of standard Kirby-Bauer (KB) testing and interpretation according to Clinical and Laboratory Standards Institute (CLSI) methods (6).
RESULTS
Of 205 women in labor who gave informed consent and were enrolled consecutively in this study, 196 were included. The average time between admission and delivery was 17 h 20 min (range of 35 min to 5 days 12 h 36 min), with a 95% confidence interval of 14 h 41 min and 19 h 35 min. Average turnaround times from admission to sample acquirement and from sample acquirement to delivery were 3 h 14 min (range of 0 min to 2 days 6 h 45 min) and 13 h 54 min (range of 0 min to 5 days 11 h 46 min), respectively.
The results of GBS culture and PCR were discordant for 10 women at the time of labor. The PCR assay was repeated for four samples that were positive by culture and negative by PCR. One of these four cases became PCR positive. For the three remaining cases, the PCR limit of detection may explain the discordant results. For six samples the intrapartum GBS culture was negative and PCR was positive. Of these six discordant results, one was culture positive after 5 days of incubation, four were from patients who received intrapartum antibiotics (repeat cultures remained negative), and one was nonresolvable.
The results of the GBS real-time PCR and culture at labor are presented in Table 1. In summary, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were 90.5%, 96.1%, 86.4%, and 97.4%, respectively, when real-time PCR was compared to culture in labor (Table 1). Five of the six samples that were GBS culture negative and PCR positive at labor were culture negative at prenatal screening, and likely, these represent true-PCR-positive GBS cases in labor. Taking into account these culture-negative and PCR-positive samples as true GBS-positive samples at labor, the sensitivity, specificity, PPV, and NPV of rapid real-time PCR would have been 91.7%, 99.3%, 97.7%, and 97.3%, respectively.
TABLE 1.
Comparison of the results of real-time PCR and culture for the detection of GBS in women in labor
| Culture result for GBS | No. of women with indicated real-time PCR result for GBSa |
Total no. of women tested | |
|---|---|---|---|
| + | − | ||
| + | 38 | 4 | 42 |
| − | 6 | 148 | 154 |
| Total | 44 | 152 | 196 |
Results of the first PCR (and not the repeat that was done to resolve the discordant data) were used to determine the sensitivity (90.5%), specificity (96.1%), PPV (86.7%), and NPV (97.4%) for in-labor real-time PCR.
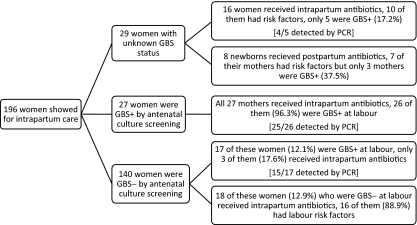
From the total of 196 women included in this study, 167 had known GBS status and 29 had unknown GBS status at the time of delivery. Of the women with known GBS status, 140 presented with GBS-negative and 27 presented with GBS-positive screening results (which is routinely performed at 35 to 37 weeks of gestation) (Fig. 1). In total, 48 (24.5%) women in our study were GBS positive at the time of labor (i.e., culture positive or PCR positive). Evaluating the relationship between having any pregnancy risk factors (previous infant with invasive GBS disease or GBS bacteriuria during the current pregnancy) and/or labor risk factors (temperature > 38°C, rupture of membrane for >18 h, delivery at <37 weeks of gestation) and being a GBS carrier, 28.8% (15/52) of women with risk factors and 22.9% (33/144) of women without any risk factors were GBS positive (P = 0.45). None of the newborns under study developed EOS.
FIG. 1.
Flow diagram providing the group B Streptococcus (GBS) status of women presenting for intrapartum care, their actual GBS status at labor (i.e., culture positive or PCR positive), and the application of prophylactic intrapartum antibiotics as well as the application of postpartum antibiotics.
Overall, 29 (14.8%) women went into labor with unknown GBS status. Of these women, 16 received intrapartum antibiotics based on risk factors and other considerations, but only five were truly GBS positive at labor (i.e., 11 received unnecessary antibiotic prophylaxis for GBS). Also, 8 newborns of these 29 women received postpartum antibiotics, whereas only three of the mothers had GBS colonization (Fig. 1). For all the women admitted for delivery with unknown GBS status, the time from admission to delivery was >2 h 30 min, which indicated sufficient time for the PCR test to be completed.
From 140 women who were GBS negative at screening, 17 (12.1%) were GBS positive at labor, but only three received prophylactic antibiotics (Fig. 1). On the other hand, 18 of these mothers received intrapartum antibiotics based on risk factors, whereas none of them were actually colonized with GBS. Almost all of the women who were GBS positive at screening were also GBS positive at labor (26/27 women).
The majority of women (148/196) enrolled in this study went into labor outside laboratory hours (21:30 to 7:30). This means that at sites such as ours that do not provide 24-h laboratory service, the rapid GBS PCR test would be immediately available only for approximately 25% of women in labor. Twelve of the women who were admitted during laboratory hours had an unknown GBS status. For these women, the rapid PCR test would have been offered, and results would have been ready within 1 h. For the remaining 17 women with unknown GBS status, the results of the rapid PCR test would not have been available immediately.
Testing of 148 rectal/vaginal GBS isolated at this site over the period of a year indicated that for this population, 65.5% of GBS isolates were sensitive and 16.2% were resistant to both erythromycin and clindamycin. Furthermore, 13.5% of isolates were resistant only to erythromycin (and not clindamycin), and 4.7% were resistant only to clindamycin (and not erythromycin). Overall, 20.9% of GBS isolates were clindamycin resistant, and 29.7% were erythromycin resistant.
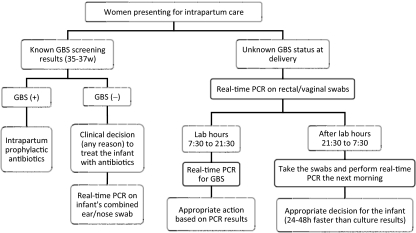
Based on the findings of this study, the algorithm in Fig. 2 was proposed for the use of real-time PCR for GBS detection in pregnant women for laboratories that do not operate 24 h a day.
FIG. 2.
Proposed flowchart for the use of real-time PCR for GBS detection in pregnant women (for laboratories unable to provide 24-h coverage).
DISCUSSION
This study showed that in routine clinical situations, rapid real-time PCR provides robust and accurate results regarding the GBS status of women in labor. Our results demonstrated that for the rapid PCR test, the sensitivity, specificity, PPV, and NPV were 90.5%, 96.1%, 86.4%, and 97.4%, respectively. This rapid test is able to detect nonviable or low-count bacteria (24) and is not affected by the presence of interfering substances such as blood, meconium, or amniotic fluid (8).
Although several studies confirmed the robustness of rapid real-time PCR for GBS detection (1, 8, 10, 12, 24), this method also has some limitations. Real-time PCR requires special laboratory equipment and a designated area as well as technically skilled personnel (10, 24). In addition, specimens are usually batched for processing, which increases the turnaround time of this process (24). A modeling analysis by Daniels et al. revealed that screening at 35 to 37 weeks of gestation using a rapid test is not cost-effective (7). However, two separate studies by Haberland et al. (13) and Goodrich and Miller (12) argued that although more expensive, the increased accuracy and considerably shorter turnaround time of rapid PCR will decrease the mortality and morbidity of GBS EOS and its associated medical costs in the long run.
Another drawback of rapid real-time PCR is its inability to provide antimicrobial susceptibility testing (21). While penicillin is a drug of choice for intrapartum prophylaxis, a portion of the population is allergic to this antibiotic (20). For these women, either erythromycin or clindamycin is recommended (15, 20). However, notable subsets of GBS are resistant to erythromycin, clindamycin, or both. Assessment of GBS isolates submitted to our laboratory over the period of a year indicated that 20.9% of isolates were clindamycin resistant, 29.7% were erythromycin resistant, and 16.2% were resistant to both erythromycin and clindamycin. The susceptibility profile for the GBS strain relevant for each patient would not be available using rapid real-time PCR for GBS screening unless the genes responsible for these resistances are incorporated into the PCR assay.
The fact that GBS colonization of women is transient has been brought up in many studies previously. In a previous study by Boyer et al., 8.5% of women with negative GBS cultures at 26 to 28 weeks of gestation were colonized with GBS at delivery (4). Davies et al. also reported a change in the GBS colonization status from screening to delivery for 9% of women (8). A more recent study by El Helali et al. showed that the GBS status of 11% of women had changed between screening (gestation weeks 35 to 37) and delivery, with 63/818 GBS-negative women becoming positive and 40/115 GBS-positive women becoming negative (10). Our study also revealed a change in the GBS colonization status of 10.8% (18/167) of women: most of them (17/140) were women who acquired GBS colonization after a negative screening (Fig. 1). Such data reveal the need for more considerations regarding the efficacy of GBS screening at 35 to 37 weeks of gestation.
For laboratories that are open 24 h a day, the GBS PCR test would be a valuable test to offer to women who are in labor and whose GBS status is unknown. Such laboratories may even consider using the GBS PCR test for all women in labor with unknown GBS status as well as for those with GBS-negative screening tests in their third trimester who have risk factors. Our data showed that the average time between taking the GBS swabs and delivery is 13 h 54 min. About 155/196 women (79%) delivered later than 4 h after sample acquisition, and numbers were similar for women with an unknown GBS status (23/29; 79.3%). Although 4 h of intrapartum prophylactic antibiotics was considered the ideal duration to give infants the maximum protection against GBS (98.8% protection), de Cueto et al. showed that 2 h of prophylaxis still provides protection to more than 97% of neonates (9). Therefore, 4 h between sample acquirement and delivery will be enough time for the results of rapid PCR to be ready (1 to 2 h) and intrapartum antibiotics to be given for more than 2 h to about 79% of women in delivery.
Natarajan and colleagues took one additional step and used a rapid real-time PCR that they developed for the detection of GBS in infants born to women with unknown GBS status at labor (17). Those authors concluded that rapid PCR performed on swabs from neonates' ear and nose has an excellent NPV (98.9%) and allows earlier discharge from hospital, and no additional tests or antibiotics were needed for half of the neonates born to women with an unknown GBS status (17). This is in agreement with our finding that rapid PCR has an NPV of 98%. Out of 29 women with unknown GBS status at delivery, only 5 were GBS positive. As per 2002 CDC guidelines, the majority of infants born to women with unknown GBS status are monitored in hospitals for 48 h. We suggest that by performing rapid real-time PCR, even in laboratories that do not operate 24 h a day, we would obtain results 24 to 48 h quicker than with standard culture methods, which ultimately results in making an appropriate decision regarding whether to continue or stop the infant's antibiotic therapy. Although our results demonstrate the value of the rapid PCR test for testing rectal/vaginal swabs from women in delivery, its application to ear/nose swabs from neonates also seems promising (17), particularly considering that only about half of neonates born to GBS-positive women are actually GBS carriers (2). In cases where the infant of a mother who was GBS negative by prenatal screening is treated with antibiotics, it may be helpful to do rapid GBS PCR on a combined ear/nose swab, as shown in Fig. 2.
Altogether, our study confirmed that the rapid real-time PCR test is robust and able to reliably detect the presence of GBS in women in labor within 1 h of specimen submission to the laboratory. We recommend that the rapid PCR test be targeted to women who present in labor with an unknown GBS status. In cases where the laboratory does not offer 24 h of testing, rectal/vaginal sample collection followed by PCR testing the next morning is still valuable and provides reliable results 24 to 48 h faster than culture and will help make appropriate decisions regarding continuing or stopping antibiotics for neonates born to women with unknown GBS status.
Acknowledgments
Test kits and partial funding for this study were provided by Somagen.
We acknowledge technologists in the clinical microbiology laboratory at St. Boniface General Hospital for their technical expertise and for all culture testing done for this study. We also acknowledge the support of the St. Boniface Research Centre.
Footnotes
Published ahead of print on 30 June 2010.
REFERENCES
- 1.Atkins, K. L., R. M. Atkinson, A. Shanks, C. A. Parvin, W. M. Dunne, and G. Gross. 2006. Evaluation of polymerase chain reaction for group B Streptococcus detection using an improved culture method. Obstet. Gynecol. 108:488-491. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron, M. G., and D. Ke. 2001. New DNA-based PCR approaches for rapid real-time detection and prevention of group B streptococcal infections in newborns and pregnant women. Expert Rev. Mol. Med. 3:1-14. [DOI] [PubMed] [Google Scholar]
- 3.Bizzarro, M. J., L. M. Dembry, R. S. Baltimore, and P. G. Gallagher. 2008. Changing patterns in neonatal Escherichia coli sepsis and ampicillin resistance in the era of intrapartum antibiotic prophylaxis. Pediatrics 121:689-696. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, K. M., C. A. Gadzala, P. D. Kelly, L. I. Burd, and S. P. Gotoff. 1983. Selective intrapartum chemoprophylaxis of neonatal group B streptococcal early-onset disease. II. Predictive value of prenatal cultures. J. Infect. Dis. 148:802-809. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, K. M., and S. P. Gotoff. 1986. Prevention of early-onset neonatal group B streptococcal disease with selective intrapartum chemoprophylaxis. N. Engl. J. Med. 314:1665-1669. [DOI] [PubMed] [Google Scholar]
- 6.CLSI. 2010. Performance standards for antimicrobial susceptibility testing; twentieth information supplement, CLSI document M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Daniels, J., J. Gray, H. Pattison, T. Roberts, E. Edwards, P. Milner, L. Spicer, E. King, R. K. Hills, R. Gray, L. Buckley, L. Magill, N. Elliman, B. Kaambwa, S. Bryan, R. Howard, P. Thompson, and K. S. Khan. 2009. Rapid testing for group B Streptococcus during labour: a test accuracy study with evaluation of acceptability and cost-effectiveness. Health Technol. Assess. 13:1-154. [DOI] [PubMed] [Google Scholar]
- 8.Davies, H. D., M. A. Miller, S. Faro, D. Gregson, S. C. Kehl, and J. A. Jordan. 2004. Multicenter study of a rapid molecular-based assay for the diagnosis of group B Streptococcus colonization in pregnant women. Clin. Infect. Dis. 39:1129-1135. [DOI] [PubMed] [Google Scholar]
- 9.de Cueto, M., M. J. Sanchez, A. Sampedro, J. A. Miranda, A. J. Herruzo, and M. Rosa-Fraile. 1998. Timing of intrapartum ampicillin and prevention of vertical transmission of group B Streptococcus. Obstet. Gynecol. 91:112-114. [DOI] [PubMed] [Google Scholar]
- 10.El Helali, N., J. C. Nguyen, A. Ly, Y. Giovangrandi, and L. Trinquart. 2009. Diagnostic accuracy of a rapid real-time polymerase chain reaction assay for universal intrapartum group B Streptococcus screening. Clin. Infect. Dis. 49:417-423. [DOI] [PubMed] [Google Scholar]
- 11.Gavino, M., and E. Wang. 2007. A comparison of a new rapid real-time polymerase chain reaction system to traditional culture in determining group B Streptococcus colonization. Am. J. Obstet. Gynecol. 197:388e1-388e4. [DOI] [PubMed] [Google Scholar]
- 12.Goodrich, J. S., and M. B. Miller. 2007. Comparison of culture and 2 real-time polymerase chain reaction assays to detect group B Streptococcus during antepartum screening. Diagn. Microbiol. Infect. Dis. 59:17-22. [DOI] [PubMed] [Google Scholar]
- 13.Haberland, C. A., W. E. Benitz, G. D. Sanders, J. B. Pietzsch, S. Yamada, L. Nguyen, and A. M. Garber. 2002. Perinatal screening for group B streptococci: cost-benefit analysis of rapid polymerase chain reaction. Pediatrics 110:471-480. [DOI] [PubMed] [Google Scholar]
- 14.Ke, D., and M. G. Bergeron. 2001. Molecular methods for rapid detection of group B streptococci. Expert Rev. Mol. Diagn. 1:175-181. [DOI] [PubMed] [Google Scholar]
- 15.Koenig, J. M., and W. J. Keenan. 2009. Group B Streptococcus and early-onset sepsis in the era of maternal prophylaxis. Pediatr. Clin. North Am. 56:689-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohle-Boetani, J. C., A. Schuchat, B. D. Plikaytis, J. D. Smith, and C. V. Broome. 1993. Comparison of prevention strategies for neonatal group B streptococcal infection. A population-based economic analysis. JAMA 270:1442-1448. [PubMed] [Google Scholar]
- 17.Natarajan, G., Y. R. Johnson, F. Zhang, K. M. Chen, and M. J. Worsham. 2006. Real-time polymerase chain reaction for the rapid detection of group B streptococcal colonization in neonates. Pediatrics 118:14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picard, F. J., and M. G. Bergeron. 2004. Laboratory detection of group B Streptococcus for prevention of perinatal disease. Eur. J. Clin. Microbiol. Infect. Dis. 23:665-671. [DOI] [PubMed] [Google Scholar]
- 19.Puopolo, K. M., L. C. Madoff, and E. C. Eichenwald. 2005. Early-onset group B streptococcal disease in the era of maternal screening. Pediatrics 115:1240-1246. [DOI] [PubMed] [Google Scholar]
- 20.Schrag, S., R. Gorwitz, K. Fultz-Butts, and A. Schuchat. 2002. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. MMWR Recomm. Rep. 51:1-22. [PubMed] [Google Scholar]
- 21.Schrag, S. J. 2004. The past and future of perinatal group B streptococcal disease prevention. Clin. Infect. Dis. 39:1136-1138. [DOI] [PubMed] [Google Scholar]
- 22.Shah, V., and A. Ohlsson with the Canadian Task Force on Preventive Health Care. 2001. Prevention of early-onset group B streptococcal (GBS) infection in the newborn: systematic review & recommendations. CTFPHC technical report number 01-6. Canadian Task Force on Preventive Health Care, London, Ontario, Canada.
- 23.Stoll, B. J., N. I. Hansen, R. D. Higgins, A. A. Fanaroff, S. Duara, R. Goldberg, A. Laptook, M. Walsh, W. Oh, and E. Hale. 2005. Very low birth weight preterm infants with early onset neonatal sepsis: the predominance of Gram-negative infections continues in the National Institute of Child Health and Human Development Neonatal Research Network, 2002-2003. Pediatr. Infect. Dis. J. 24:635-639. [DOI] [PubMed] [Google Scholar]
- 24.Wernecke, M., C. Mullen, V. Sharma, J. Morrison, T. Barry, M. Maher, and T. Smith. 2009. Evaluation of a novel real-time PCR test based on the ssrA gene for the identification of group B streptococci in vaginal swabs. BMC Infect. Dis. 9:148. [DOI] [PMC free article] [PubMed] [Google Scholar]