Abstract
Clinical isolates that are difficult to identify by conventional means form a valuable source of novel human pathogens. We report on a 5-year study based on systematic 16S rRNA gene sequence analysis. We found 60 previously unknown 16S rRNA sequences corresponding to potentially novel bacterial taxa. For 30 of 60 isolates, clinical relevance was evaluated; 18 of the 30 isolates analyzed were considered to be associated with human disease.
16S rRNA gene sequence analysis is broadly considered the “gold standard” in bacterial identification (6, 29). In daily clinical diagnostics, accurate bacterial identification is essential in judging whether a bacterial isolate is to be considered the causative agent of an infectious disease or merely a colonizer. In our study, we aimed to characterize the bacterial diversity encountered in a diagnostic laboratory by revealing potentially novel, clinically relevant species, according to the current species definition by the Clinical and Laboratory Standards Institute (22).
Routine 16S rRNA gene sequencing is implemented in our laboratory and is a fixed part of our diagnostic algorithms for identification of bacterial isolates (1, 2, 32). We retrospectively reanalyzed 16S rRNA gene sequences collected during 2004 to 2008 to identify potentially novel bacterial taxa of clinical relevance. The Institute of Medical Microbiology (IMM) serves the 850-bed University Hospital of Zurich and surrounding smaller hospitals. Bacterial isolates from blood, cerebrospinal fluid, wounds, joint aspirates, respiratory samples, genitourinary swabs, feces, and urine were recovered by culture on appropriate media according to standard procedures (19). Isolates that could not be identified by phenotypic methods underwent sequencing. 16S rRNA gene analysis was performed as previously described (1). Homology analyses were performed using the SmartGene Integrated Database Network System (IDNS) (24) and NCBI GenBank databases. For the first screening of our large data collection, we selected isolates with sequence homology of <99.0% to members of described taxa, regarding these as potentially novel species; isolates with sequence homology of <95% were regarded as representatives of a novel genus (2). The boundary for novel families was <87.5% homology and, for novel orders, <78.4% 16S rRNA sequence homology (30). After the first screening, we used more stringent cutoff values (<97.5% for species) for taxa with significant interspecies 16S rRNA divergence; i.e., members of the Paenibacillaceae family and the Clostridiales order (6, 25).
During the 5-year study period, 1,663 cultured isolates were subjected to 16S rRNA gene sequence analysis (Table 1). Of those, 60 isolates (0.4‰; see Table S1 in the supplemental material) had a 16S rRNA gene homology of <99% to members of accepted taxa on the date of the first interpretation. A total of 11 of the 60 sequences with a 16S rRNA homology of <99% in the first-time analysis could be allocated to a species established during the study term as a novel species by others: Acinetobacter septicus (16, 20), Brevibacterium ravenspurgense (17), Corynebacterium freiburgense (12), Corynebacterium massiliense (n = 2) (18), C. mastitidis (10, 18), C. pyruviciproducens (26), C. ureicelerivorans (11, 31), Neisseria zoodegmatis (28), Paenibacillus barengoltzii (21), and the reclassified Campylobacter ureolyticus (previously known as Bacteroides ureolyticus) (27).
TABLE 1.
Clinical bacterial isolates with 16S rRNA gene homology < 99% (n = 60)
| Taxonomic group | No. of isolates with indicated 16S rRNA homology |
|
|---|---|---|
| <99% to >95% | <95% | |
| Enteric Gram-negative rods | 0 | 0 |
| Fastidious Gram-negative rods | 1 | 4 |
| Gram-negative cocci | 1 | 0 |
| Gram-negative nonfermenters | 7 | 1 |
| Gram-positive cocci | 12 | 2 |
| Gram-positive rods | 26 | 6 |
| Total | 47 | 13 |
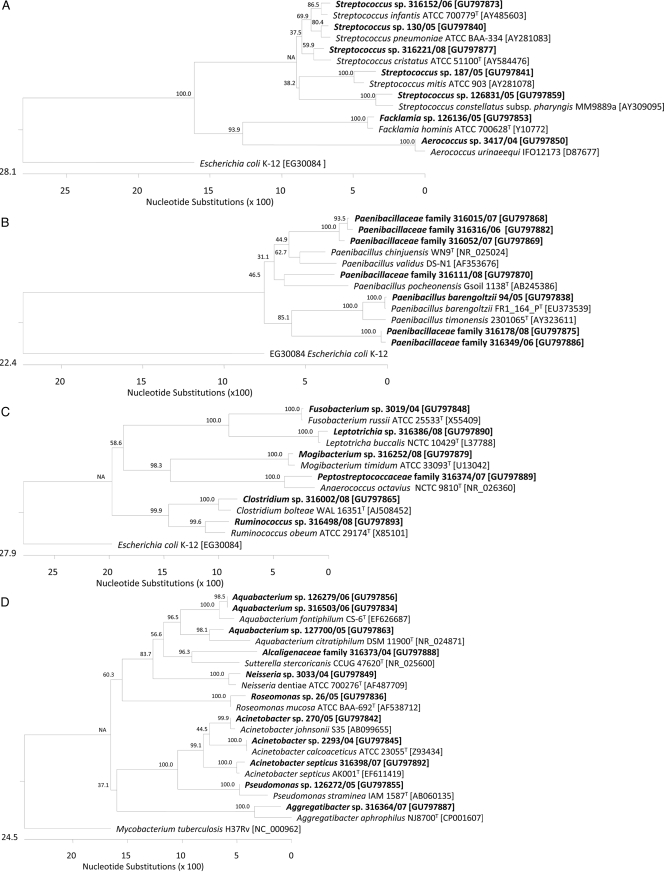
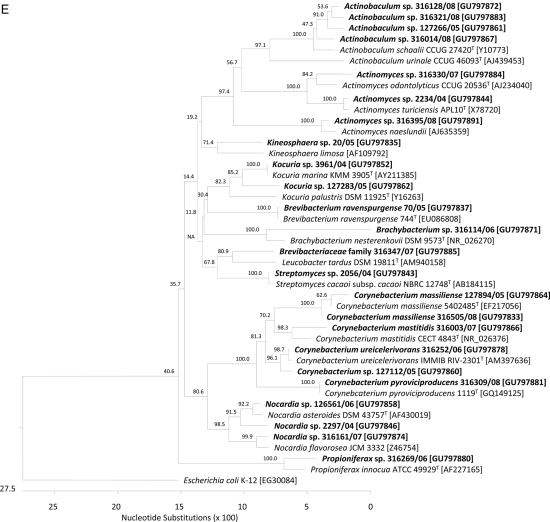
We calculated dendrograms to assess phylogenetic relationships (Fig. 1). Potentially novel streptococcal species clustered with known pathogens. For example, within the streptococci, one isolate (GenBank accession no. GU797873) shared 98.8% sequence homology with S. infantis; another isolate (GU797840) shared 97.8% homology with S. pneumoniae. Within the Bacillales order (Fig. 1B), eight novel Paenibacillus sequence types were recovered: three of them (GU797882, GU797868, and GU797869) were distantly related (<95% sequence homology) to Paenibacillus chinjuensis-P. validus, and two isolates (GU797838 and GU797854) were related to P. timonensis (97.3% and 94.5% sequence homology). Several bacterial families are represented in the Clostridiales order. The novel sequences recovered in the Clostridiales order all belonged to different families (Fig. 1C). We found two representatives of the Fusobacteriales order (Fig. 1C): one isolate (GU797848) was related to Fusobacterium russii (sequence homology 98.8%), and one isolate (GU797890) was most closely related to Leptotrichia buccalis (98.9% sequence homology). Eleven novel sequences belonged to the Pseudomonadales order, and 3 of those (GU797845, GU797842, and GU797892) represented potential novel Acinetobacter spp. (Fig. 1D). The Actinomycetales order (Fig. 1E) comprises 25 potentially novel taxa (data for 24 taxa are given in the figure). Six corynebacterial isolates, i.e., Corynebacterium freiburgense (GU797839), Corynebacterium massiliense (GU797864 and GU797833), C. mastitidis (GU797866), C. ureicelerivorans (GU797878), and C. pyruviciproducens (GU797881), clustered with type strain sequences that were established as novel species during the study period. We recovered three Nocardia spp. (GU797846, GU797858, and GU797874); two of them (GU797846 and GU797858) belonged to the Nocardia asteroides complex and one (GU797874) was related to Nocardia flavorosea. Four potentially novel Actinobaculum spp. (GU797861, GU797867, GU797872, and GU797883/GU797308) were attributed to the same taxonomic clade according to 16S rRNA sequence phylogeny results, with Actinobaculum schaalii as the nearest neighboring species (95.7 to 98.5% sequence homology).
FIG. 1.
Phylogeny of 55 of 60 cultured isolates recovered from clinical specimens with homology of <99% to 16S rRNA gene sequences of members of published taxa. The dendrograms were calculated using CLUSTAL V alignment and a matrix of Jukes-Cantor distances determined by the neighbor-joining method using DNASTAR Lasergene MegAlign 7.0 software. Taxonomic order adherence of 55 taxons identified in this study: (A) Lactobacillales; (B) Bacillales; (C) Fusobacteriales and Clostridiales; (D) Pseudomonadales; (E) Actinomycetales. Study isolates are shown in bold. Species in regular type were selected as (published) type strains of the different taxa. We used Escherichia coli K-12 rrnA [NCBI GenBank accession no. EG30084] and Mycobacterium tuberculosis H37Rv rrs [NC_000962] as outgroups.
To assess the clinical relevance of the microbiological findings, we selected 30 isolates for which sufficient clinical data were available and performed reviews of patient charts (Table 2). We reviewed the patient charts for clinical signs and symptoms of infection, inflammation parameters such as fever, leukocyte count, C-reactive protein (CRP) and procalcitonin levels, radiological and laboratory findings (serology and bacterial culture results), previous infections or bacterial isolates, antibiotic treatment, and clinical diagnosis (see Table S2 in the supplemental material). We established a clinical score incorporating the different parameters mentioned above to determine the likelihood (codified as “yes,” “likely,” “unlikely,” “no”) of an infectious disease in each case and to assess the association of the bacterial isolate found with disease.
TABLE 2.
Subset analysis of microbial and clinical data of 30 patientsa
| Source | 16S rRNA-based identification | NCBI GenBank accession no. | Match with highest % homology | Homology of 1st match |
Polymicrobial etiology | Clinical diagnosis | Clinical relevance | ||
|---|---|---|---|---|---|---|---|---|---|
| No. of mismatches | % | Match length (bp) | |||||||
| Urine | Acinetobacter septicus | GU797892 | Acinetobacter septicus | 0 | 100.0 | 531 | No | Urinary tract infection (urothelial carcinoma) | Yes |
| Peritoneal dialysate | Acinetobacter sp. | GU797845 | Acinetobacter calcoaceticus | 10 | 98.3 | 574 | No | Peritonitis (CAPD) | Yes |
| Urine | Actinobaculum sp. | GU797883, GU797308 | Actinobaculum schaalii | 26 | 96.6 | 768 | No | Ascending urinary tract infection (pigtail catheter) | Yes |
| Blood culture | Actinobaculum sp. | GU797872 | Actinobaculum schaalii | 10 | 98.5 | 641 | No | Urosepsis, urothelial carcinoma | Yes |
| Parotid gland aspirate | Actinomyces sp. | GU797891 | Actinomyces naeslundii | 15 | 97.4 | 573 | Yes | Parotid inflammation | Yes |
| Urine | Gardnerella sp. | GU797857 | Gardnerella vaginalis | 6 | 98.9 | 527 | No | Urinary tract infection | Yes |
| Tissue (hand) | Neisseria zoodegmatis | GU797849 | Neisseria zoodegmatis | 3 | 99.4 | 525 | Yes | Cat bite | Yes |
| Central venous catheter | Paenibacillus barengoltzii | GU797838 | Paenibacillus barengoltzii | 3 | 99.4 | 505 | No | Intravascular catheter-associated infection | Yes |
| Blood culture | Streptococcus sp. | GU797859 | Streptococcus constellatus | 8 | 98.6 | 571 | No | Cholangitis, sepsis | Yes |
| Femur bone | Streptococcus sp. | GU797840 | Streptococcus oralis | 9 | 97.8 | 403 | No | Hip prosthesis infection with soft tissue abscess | Yes |
| Pleural aspirate | Actinomyces sp. | GU797884 | Actinomyces odontolyticus | 17 | 96.6 | 496 | Yes | Anastomosis insufficiency (pneumonectomy) | Likely |
| Blood culture | Burkholderiales order | GU797888 | Sutterella stercoricanis | 54 | 89.8 | 530 | No | Small intestine ischemia | Likely |
| Corneal tissue | Corynebacterium mastitidis | GU797866 | Corynebacterium mastitidis | 0 | 100.0 | 463 | Yes | Chronic blepharitis | Likely |
| Intravenous catheter | Corynebacterium massiliense | GU797833 | Corynebacterium massiliense | 1 | 99.8 | 556 | No | Sepsis (unclear focus of infection) | Likely |
| Spongiosa tissue | Mogibacterium sp. | GU797879 | Mogibacterium timidum | 7 | 98.7 | 538 | Yes | Maxillary bone necrosis | Likely |
| Sputum | Nocardia sp. | GU797874 | Nocardia flavorosea | 8 | 98.4 | 494 | Yes | Upper lobe pneumonia (COPD)b | Likely |
| Deep wound swab | Peptostreptococcaceae family | GU797889 | Anaerococcus octavius | 28 | 94.7 | 530 | Yes | Axillary abscess | Likely |
| Superficial wound | Pseudomonas sp. | GU797855 | Pseudomonas fulva | 10 | 98.1 | 526 | Yes | Ulceration, digit II of right foot (diabetes mellitus) | Likely |
| Contact lens | Kocuria sp. | GU797852 | Kocuria marina | 26 | 96.1 | 668 | Contact lens-associated ceratitis | Unlikely | |
| Hip joint aspirate | Paenibacillaceae family | GU797869 | Paenibacillus chinjuensis | 34 | 92.9 | 480 | Rheumatoid arthritis | Unlikely | |
| Knee joint aspirate | Paenibacillaceae family | GU797870 | Paenibacillus pocheonensis | 35 | 93.8 | 563 | Intravenous drug abuse, hepatitis C | Unlikely | |
| Blood culture | Propioniferax sp. | GU797880 | Propioniferax innocua | 27 | 96.4 | 720 | Aplastic anemia | Unlikely | |
| Blood culture | Aquabacterium sp. | GU797863 | Aquabacterium citratiphilum | 22 | 95.8 | 520 | Fever, AMLc | No | |
| Blood culture | Campylobacter ureolyticus | GU797876 | Campylobacter ureolyticus | 1 | 99.8 | 519 | Fever, neutropenia | No | |
| Bone biopsy | Corynebacterium pyruviciproducens | GU797881 | Corynebacterium pyruviciproducens | 0 | 100.0 | 745 | Open bone fracture | No | |
| Sputum | Kineosphaera sp. | GU797835 | Kineosphaera limosa | 24 | 95.6 | 549 | Chronic bronchitis | No | |
| urine | Brevibacteriaceae family | GU797885 | Leucobacter tardus | 57 | 92.2 | 734 | Urinary tract infection | No | |
| Bursa aspirate | Paenibacillaceae family | GU797882 | Paenibacillus validus | 32 | 93.5 | 493 | Trochanteric bursitis | No | |
| Blood culture | Paenibacillaceae family | GU797875 | Paenibacillus contaminans | 51 | 90.7 | 551 | HIV infection, Pneumocystis jirovecii pneumonia | No | |
| Blood culture | Ruminococcus sp. | GU797893 | Ruminococcus obeum | 22 | 95.9 | 539 | HIV infection | No | |
The likelihood of a relevant infectious disease associated with the microbiological findings was estimated after retrospective patient chart analysis.
COPD, chronic obstructive pulmonary disease.
AML, acute myelogenous leukemia.
Clinical relevance was attributed to 18 isolates. In 10 cases, patient history, laboratory findings, and clinical course following antibiotic therapy guided by the isolate's drug susceptibility testing results were compatible with a pathogenic role for the isolated microorganism. In eight cases, we concluded that the bacterial isolate was likely to have been the cause of an infection. The putative novel disease-associated species mostly belonged to the Actinomycetales order of Gram-positive rods. The recently described Acinetobacter septicus (GU797892) (16), two Actinobaculum spp. (GU797883 and GU797872), and a Gardnerella sp. (GU797857) were each identified as present in samples from patients with urinary tract infections. In one case, we recovered a potentially novel Actinobaculum sequence type (GU797872) from several blood cultures of a patient suffering from urosepsis, underlining the pathogenic potential of species of the Actinobaculum genus in ascending urinary tract infections. A potentially novel Acinetobacter sp. (GU797845), most closely related to Acinetobacter calcoaceticus, was identified in the peritoneal dialysate of a 22-year-old patient with kidney failure. He exhibited infective monobacterial peritonitis as a complication of continuous ambulatory peritoneal dialysis (CAPD). A previously unknown Actinomyces sp. (GU797891) was isolated from a 49-year old female patient suffering from severe, acute, suppurative parotitis. Neisseria zoodegmatis (previously Neisseria CDC EF-4 group [28]) was found in an isolate from a patient with a wound and a history of a cat bite (GU797849). A novel sequence type of Paenibacillus barengoltzii (21) was cultured from a central venous catheter in the jugular vein of a patient with a burn injury (GU797838). A potentially novel Streptococcus sp. (GU797859), belonging to the Streptococcus anginosus group, was found in an isolate from a septic patient suffering from cholangitis. A potentially novel Streptococcus sp. related to Streptococcus oralis (GU797840) was cultured from a hip joint aspirate of a patient with hip prosthesis infection. Bacterial isolates that were found to be irrelevant to a patient's clinical disease entity (12/30) were considered to represent skin flora (e.g., Paenibacillus spp. or Ruminococcus spp.) or to belong to environmental bacteria (e.g., Aquabacterium spp. or Kineosphaera spp.).
Broad-range 16S rRNA gene amplification readily allows the detection of members of as-yet-unknown bacterial taxa (4, 5, 9). Major hypervariable regions are present in the first 500 bp of the roughly 1,600 bp comprising the 16S rRNA gene downstream of the conserved primer target sites (5, 9, 29). Thus, analysis of this part of the gene sequence allows the recognition of potentially novel taxa based on previously established cutoff values of <99% homology for new species and <95% homology for new genera (1-3, 7). While such a general cutoff is appropriate for overall first analysis of large data sets, we note that the boundaries for species definition by 16S rRNA sequence homology may be different for different phyla (13, 25). A less stringent cutoff value (i.e., <99.6% homology) could have been used to delimit different species in bacterial groups such as the Streptococcus mitis group or nonfermenters (13). Conversely, for species belonging to the Paenibacillaceae family and the Clostridiales order, a more stringent cutoff value (i.e., 97.5% homology) is more appropriate (6) and was therefore applied after the first selection performed with the 99% cutoff value.
In 2008, the fraction of bacterial isolates submitted for molecular identification was 0.8%. Previous investigations reported rates of 0.5% to 1% for a similar study setup (7) and a rate of 14% for isolates restricted to aerobic Gram-positive rods (2). Gram-positive rods and Gram-negative cocci are overrepresented in the group of sequenced isolates in our comparative analysis of phenotypic and 16S rRNA-based identification methods. Of the 1,663 (3.7%) sequences determined during the study period, 60 were judged to be representatives of potentially novel species or novel genera. A recent review (29), summarizing 16S rRNA gene-based studies published from 2001 to 2007, calculated that 215 unique sequences recovered during this period from human specimens represented potentially novel species. Of the 215, 29 belonged to novel genera. During our study, the number of 16S rRNA sequences deposited in the NCBI nucleotide database increased by a factor of 15. The SmartGene IDNS 16S rRNA database, which is a curated database derived from the NCBI repository, increased in size by a factor of 4. Despite this increase in the number of sequences deposited, the recovery of sequences with <99% homology to members of established taxa in our data set during 2004 to 2008 remained relatively constant at between 2.4% and 5.1%. This finding may reflect the fact that many of the sequences deposited in public databases were the outcome of large-scale ecological or environmental (metagenomic) sequencing projects and did not include sequences of clinical laboratory isolates.
Bacterial taxonomic classification has advanced differently in various taxonomic groups (25): Phenotypic methods readily allow species determination below the resolution of 16S rRNA-based sequence analysis in studies of enteric Gram-negative bacteria (14, 15). In contrast, the Actinomycetales order is a rich but poorly investigated group (2). For example, within the Corynebacterium genus, 18 novel species were validly described from 2004 to 2009. A total of 5 of these, namely, Corynebacterium freiburgense (12), Corynebacterium pyruviciproducens (26), C. mastitidis (10, 18), C. massiliense (18), and C. ureicelerivorans (11, 31), were also identified in our study.
When we calculated phylogenetic trees based on partial 16S rRNA sequences (Fig. 1), we found that differentiation was numerically strong (as measured by nucleotide substitutions of base pairs) in the Actinomycetales, Clostridiales, Fusobacteriales, and Pseudomonales orders whereas it was less profound in the Lactobacillales order and, more specifically, in the Streptococcaceae family. Regarding potentially novel Streptococcus spp., further molecular analysis of additional loci (by, e.g., sodA, rpoB, and recA sequence homology) would be required to determine exact phylogenetic relationships (8, 23).
In summary, out of 1,663 bacterial isolates subjected to 16S rRNA sequencing during a 5-year period, we recovered 60 clinical bacterial isolates that were indicative of the presence of putative novel bacterial species. Of these 60 isolates, 9 were established as novel pathogens in the literature during the period of the study. A total of 18 (60%) isolates showed clinical relevance in a subset analysis of 30 of the 60 isolates. Isolates with clinical implications are mostly representatives of genera that comprise known pathogens (i.e., Streptococcus spp., Actinobaculum spp., Actinomyces spp., and Neisseria spp.). Our findings underline the importance of 16S rRNA gene sequencing in routine identification algorithms designed to recognize novel pathogens in the diagnostic laboratory.
Supplementary Material
Acknowledgments
We thank the laboratory technicians for their dedicated help.
The study was supported by the University of Zurich.
Footnotes
Published ahead of print on 14 July 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bosshard, P. P., S. Abels, M. Altwegg, E. C. Böttger, and R. Zbinden. 2004. Comparison of conventional and molecular methods for identification of aerobic catalase-negative gram-positive cocci in the clinical laboratory. J. Clin. Microbiol. 42:2065-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosshard, P. P., S. Abels, R. Zbinden, E. C. Böttger, and M. Altwegg. 2003. Ribosomal DNA sequencing for identification of aerobic gram-positive rods in the clinical laboratory (an 18-month evaluation). J. Clin. Microbiol. 41:4134-4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosshard, P. P., R. Zbinden, S. Abels, B. Böddinghaus, M. Altwegg, and E. C. Böttger. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 44:1359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böttger, E. C. 1996. Approaches for identification of microorganisms. ASM News 62:247-250. [Google Scholar]
- 5.Böttger, E. C. 1989. Rapid determination of bacterial ribosomal RNA sequences by direct sequencing of enzymatically amplified DNA. FEMS Microbiol. Lett. 53:171-176. [DOI] [PubMed] [Google Scholar]
- 6.Clarridge, J. E., III. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drancourt, M., P. Berger, and D. Raoult. 2004. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J. Clin. Microbiol. 42:2197-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drancourt, M., V. Roux, P. E. Fournier, and D. Raoult. 2004. rpoB gene sequence-based identification of aerobic Gram-positive cocci of the genera Streptococcus, Enterococcus, Gemella, Abiotrophia, and Granulicatella. J. Clin. Microbiol. 42:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Garayzabal, J. F., M. D. Collins, R. A. Hutson, E. Fernandez, R. Monasterio, J. Marco, and L. Dominguez. 1997. Corynebacterium mastitidis sp. nov., isolated from milk of sheep with subclinical mastitis. Int. J. Syst. Bacteriol. 47:1082-1085. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Natal, M. I., J. A. Saez-Nieto, S. Valdezate, R. H. Rodriguez-Pollan, S. Lapena, F. Cachon, and F. Soriano. 2009. Isolation of Corynebacterium ureicelerivorans from normally sterile sites in humans. Eur. J. Clin. Microbiol. Infect. Dis. 28:677-681. [DOI] [PubMed] [Google Scholar]
- 12.Funke, G., R. Frodl, K. A. Bernard, and R. Englert. 2009. Corynebacterium freiburgense sp. nov., isolated from a wound obtained from a dog bite. Int. J. Syst. Evol. Microbiol. 59:2054-2057. [DOI] [PubMed] [Google Scholar]
- 13.Janda, J. M., and S. L. Abbott. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janda, J. M., and S. L. Abbott. 2006. The family Enterobacteriaceae: taxonomic considerations, p. 7-14. In J. M. Janda (ed.), The enterobacteriaceae, 2nd ed. ASM Press, Washington, DC.
- 15.Johnson, J. R. 2000. Shigella and Escherichia coli at the crossroads: Machiavellian masqueraders or taxonomic treachery? J. Med. Microbiol. 49:583-585. [DOI] [PubMed] [Google Scholar]
- 16.Kilic, A., H. Li, A. Mellmann, A. C. Basustaoglu, M. Kul, Z. Senses, H. Aydogan, C. W. Stratton, D. Harmsen, and Y. W. Tang. 2008. Acinetobacter septicus sp. nov. association with a nosocomial outbreak of bacteremia in a neonatal intensive care unit. J. Clin. Microbiol. 46:902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mages, I. S., R. Frodl, K. A. Bernard, and G. Funke. 2008. Identities of Arthrobacter spp. and Arthrobacter-like bacteria encountered in human clinical specimens. J. Clin. Microbiol. 46:2980-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merhej, V., E. Falsen, D. Raoult, and V. Roux. 2009. Corynebacterium timonense sp. nov. and Corynebacterium massiliense sp. nov., isolated from human blood and human articular hip fluid. Int. J. Syst. Evol. Microbiol. 59:1953-1959. [DOI] [PubMed] [Google Scholar]
- 19.Murray, P. R., and E. J. Baron. 2007. Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 20.Nemec, A., M. Musilek, M. Vaneechoute, E. Falsen, and L. Dijkshoorn. 2008. Lack of evidence for “Acinetobacter septicus” as a species different from Acinetobacter ursingii? J. Clin. Microbiol. 46:2826-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osman, S., M. Satomi, and K. Venkateswaran. 2006. Paenibacillus pasadenensis sp. nov. and Paenibacillus barengoltzii sp. nov., isolated from a spacecraft assembly facility. Int. J. Syst. Evol. Microbiol. 56:1509-1514. [DOI] [PubMed] [Google Scholar]
- 22.Petti, C. A., P. P. Bosshard, M. E. Brandt, J. E. Clarridge III, T. V. Feldblyum, P. Foxall, M. R. Furtado, N. Pace, and G. W. Procop. 2006. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing: approved guideline, vol. MM18-A. Clinical and Laboratory Standards Institute (CLSI), Wayne, PA.
- 23.Poyart, C., G. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simmon, K. E., A. C. Croft, and C. A. Petti. 2006. Application of SmartGene IDNS software to partial 16S rRNA gene sequences for a diverse group of bacteria in a clinical laboratory. J. Clin. Microbiol. 44:4400-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stackebrandt, E. 2006. Defining taxonomic ranks, p. 29-57. In M. Dworkin and S. Falkow (ed.), The prokaryotes: a handbook on the biology of bacteria, 3rd ed. Springer, New York, NY.
- 26.Tong, J., C. Liu, P. Summanen, H. Xu, and S. M. Finegold. 2010. Corynebacterium pyruviciproducens sp. nov., a pyruvic acid producer. Int. J. Syst. Evol. Microbiol. 60:1135-1140. [DOI] [PubMed] [Google Scholar]
- 27.Vandamme, P., L. Debruyne, E. De Brandt, and E. Falsen. 2 October 2009, posting date. Reclassification of Bacteroides ureolyticus as Campylobacter ureolyticus comb. nov. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] [DOI] [PubMed]
- 28.Vandamme, P., B. Holmes, H. Bercovier, and T. Coenye. 2006. Classification of Centers for Disease Control Group Eugonic Fermenter (EF)-4a and EF-4b as Neisseria animaloris sp. nov. and Neisseria zoodegmatis sp. nov., respectively. Int. J. Syst. Evol. Microbiol. 56:1801-1805. [DOI] [PubMed] [Google Scholar]
- 29.Woo, P. C., S. K. Lau, J. L. Teng, H. Tse, and K. Y. Yuen. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908-934. [DOI] [PubMed] [Google Scholar]
- 30.Yarza, P., M. Richter, J. Peplies, J. Euzeby, R. Amann, K. H. Schleifer, W. Ludwig, F. O. Glockner, and R. Rossello-Mora. 2008. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31:241-250. [DOI] [PubMed] [Google Scholar]
- 31.Yassin, A. F. 2007. Corynebacterium ureicelerivorans sp. nov., a lipophilic bacterium isolated from blood culture. Int. J. Syst. Evol. Microbiol. 57:1200-1203. [DOI] [PubMed] [Google Scholar]
- 32.Zbinden, A., E. C. Bottger, P. P. Bosshard, and R. Zbinden. 2007. Evaluation of the colorimetric VITEK 2 card for identification of gram-negative nonfermentative rods: comparison to 16S rRNA gene sequencing. J. Clin. Microbiol. 45:2270-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.