Abstract
Corynebacterium falsenii was described in 1998 as a new Corynebacterium species. We give the first detailed description of a clinically significant Corynebacterium falsenii bacteremia occurring in an infant while on vancomycin therapy.
CASE REPORT
A 13-month-old male was referred to our emergency department with a 2-day history of expanding erythema from a pimple over the left upper medial thigh, associated with fevers and inconsolability. Due to clinical concern for Fournier's gangrene, he was taken urgently to the operating room (OR), where extensive necrotic debris was removed, and further debrided during repeated OR visits. The wound culture grew methicillin-resistant Staphylococcus aureus, Streptococcus agalactiae (beta-hemolytic group B streptococcus), and Bacteroides tectus. Various combinations of vancomycin, clindamycin, gentamicin, and metronidazole were administered, with a good clinical response. A vancomycin goal trough (10 to 15 μg/ml) was achieved. The infant was completing treatment with intravenous vancomycin monotherapy via a central line when a drop in the hematocrit value was noted, requiring a transfusion. A return visit to the OR revealed a hematoma within the wound, leading to an evacuation with washout and wound vacuum placement. Postoperatively, a vancomycin trough was suboptimal (7 μg/ml), and the vancomycin dose was increased by 10%.
The following day, the infant was noted to have temperatures to 101.3 °F, with tachycardia at 132 beats/min and mild tachypnea at 36 breaths/min. Subsequent fevers up to 104.1 °F were recorded, associated with intermittent diaphoresis and tachycardia. A physical exam was notable for a soft systolic murmur unchanged from baseline, a clean wound site, and the absence of a rash. Laboratory investigation revealed a normal white blood cell count (9.1 × 109/liter), hemoglobin level (12.4 g/dl), platelet count (348 × 109/L), and erythrocyte sedimentation rate (19 mm/h) but an elevated C-reactive protein level of 8.6 mg/dl (normal, <0.1 mg/dl). Prior to empirical administration of piperacillin-tazobactam, blood cultures were obtained peripherally and from a central line. All indwelling lines (the central line and a Foley catheter) were removed; the central line catheter tip was submitted for culture. Subsequent to these measures, the infant defervesced.
Gram-positive bacilli resembling diphtheroids were recovered from both the peripheral and central line cultures after 1 day of incubation on an automated blood culture instrument. Piperacillin-tazobactam therapy was discontinued, but vancomycin administration was maintained. The following day, Gram-positive diphtheroids (<15 CFU) were also recovered from the central catheter tip culture. A peripheral blood culture obtained 2 days later was negative for growth. A repeat vancomycin trough level was 15.8 μg/ml; subsequent levels ranged from 8.7 to 10.4 μg/ml. The infant completed a further 2 weeks of vancomycin therapy with no recurrence of fevers or symptoms.
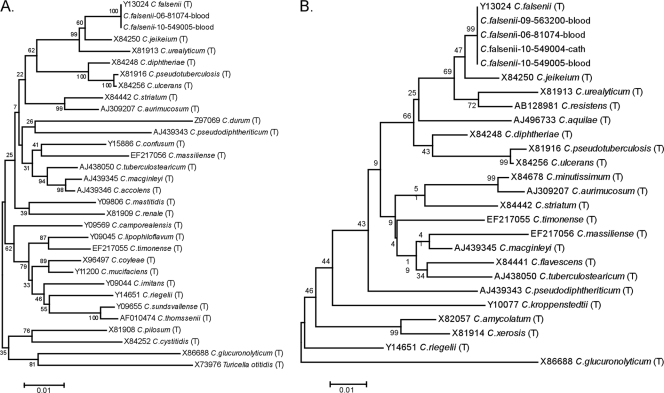
Blood and catheter tip cultures were plated on routine sheep blood agar and incubated at 37°C in an atmosphere of 5% CO2. After 48 to 72 h of incubation, bacterial colonies evolved from pinpoint growth to raised, slightly yellow, smooth, nonhemolytic colonies; the microscopic morphology was consistent with that of Gram-positive diphtheroids. The organism was catalase positive with a slow but positive urease reaction. A zone of inhibition was noted when the organism was tested by disk diffusion against a 10-μg penicillin disk. Phenotypic identification using the API Coryne kit, v.3.0, using the currently available apiweb software and database (bioMerieux, Inc., Durham, North Carolina) yielded different results for representative isolates from the blood and catheter tip cultures. The API numerical profile (and percent identification) favored Corynebacterium propinquum (89.7%) for the blood culture isolate but was consistent with C. urealyticum (92.6%) for the catheter tip isolate. The isolates were subsequently identified as Corynebacterium falsenii, with 100% sequence identity to the C. falsenii type strain, at a reference laboratory (ARUP Laboratories, Salt Lake City, UT), using small subunit rRNA gene sequencing (MicroSEQ 500 16S rRNA genes bacterial identification kit; Applied Biosystems by Life Technologies, Carlsbad, CA) and SmartGene database analysis (SmartGene, Inc., Raleigh, NC). Further analyses with GenBank (BLAST) (1) and RDP (Seqmatch) (5) confirmed the SmartGene results showing C. falsenii as the closest match (100%), with Corynebacterium jeikeium as the second-best match (98.3 to 98.8%). Unrooted neighbor-joining trees (Fig. 1) were generated and analyzed for stability with the bootstrap method (1,000 replicates) using the MEGA4 software program (12). Susceptibility testing by the broth microdilution method read at 24 h (and again at 48 h for β-lactams) (3) indicated the organism was susceptible to several antibiotic agents, including vancomycin (Table 1).
FIG. 1.
Neighbor-joining trees of 16S rRNA gene sequences showing the phylogenetic relationships among our clinical isolates and type strains (T) of other Corynebacterium spp. Sequences are indicated with GenBank accession numbers and were trimmed to the length of the shortest sequence. (A) Nearly full length sequences (1,404 nt) showing relationships among a broad distribution of corynebacteria and related species. (B) 5′ 16S sequences (435 nt) showing the ability of partial sequences to differentiate C. falsenii from other clinically important and recently described Corynebacterium species isolates.
TABLE 1.
Antimicrobial susceptibility results for C. falsenii isolate
| Drug | MIC (μg/ml) | Interpretation | CLSI 2006 M-45A MIC breakpoint interpretations (μg/ml)a |
||
|---|---|---|---|---|---|
| S | I | R | |||
| Ceftriaxone | 8 | Resistant | ≤1 | 2 | ≥4 |
| Clindamycin | 0.5 | Susceptible | ≤0.5 | 1-2 | ≥4 |
| Doxycycline | 0.25 | Susceptible | ≤4 | 8 | ≥16 |
| Erythromycin | 2 | Resistant | ≤0.5 | 1 | ≥2 |
| Gentamicin | ≤0.12 | Susceptible | ≤4 | 8 | ≥16 |
| Linezolid | 0.5 | Susceptible | ≤2 | ||
| Meropenem | 0.5 | Susceptible | ≤4 | 8 | ≥16 |
| Penicillin | 1 | Susceptible | ≤1 | 2 | ≥4 |
| Rifampin | ≤0.5 | Susceptible | ≤1 | 2 | ≥4 |
| Trimethoprim- sulfamethoxazole | ≥4/76 | Resistant | ≤2/38 | ≥4/76 | |
| Vancomycin | 1 | Susceptible | ≤4 | ||
MIC breakpoints as per CLSI document M-45A, 2006 (3): S, susceptible; I, intermediate; R, resistant.
Corynebacterium species are part of the normal human flora and are isolated from the skin, mucous membranes, and gastrointestinal tract. Since 1995, at least 20 species (or taxa consistent with the genus), recovered from human clinical specimens, have been described for Corynebacterium (2). Corynebacterium falsenii sp. nov. was first reported and characterized as such in 1998 (11). Based on 16S rRNA gene sequencing data, the closest phylogenetic relative was Corynebacterium jeikeium, with a genealogical distinctness of approximately 2% sequence divergence. Of note here, the 16S rRNA gene sequence similarities between C. falsenii sp. nov. and C. urealyticum and C. propinquum have been reported to be 97% and 93.5%, respectively (11). The isolates described here (10-549004 and 10-549005) showed only ∼98% identity to the type strain of C. jeikeium, which is generally considered to be below the threshold for species-level identification of corynebacteria (99%) (4). A GenBank comparison of nearly full length 16S rRNA gene sequence from isolate 10-549005 confirmed the identification as C. falsenii (99.9% identical to the type strain over 1,444 nucleotides [nt]) (Fig. 1A) and illustrates the utility of sequencing only the 5′ 500 bp for identification of this species (Fig. 1B).
Previously found among other animals, including eagles (6) and storks (7), C. falsenii is recognized as a potential human pathogen given its isolation from sterile sites, including blood (2, 11). However, to our knowledge, clinical and microbiological data confirming the ability of this species to cause invasive disease have not been published. We report the first case of Corynebacterium falsenii bacteremia in an infant, with isolation from two sets of blood cultures, including isolation from a central line and from the central line catheter tip. There are several observations of interest, microbiological and clinical, discussed herein.
Phenotypic species-level identification of corynebacteria, including C. falsenii, can be problematic due to overlapping, weak, and/or delayed biochemical reactions between various species (2, 11). Importantly, of all the corynebacteria associated with human specimens, C. falsenii and C. xerosis are the only nonlipophilic species to have yellow pigmentation. The two species can be distinguished phenotypically by the dry colonial morphology observed for the latter and genotypically by 16S rRNA gene sequencing (Fig. 1). In our case, the organism had several features consistent with reported expectations for C. falsenii, including yellow-pigmented smooth colonies and delayed reactions for urease and glucose fermentation. But identification of C. falsenii was confounded by misidentification by the API Coryne kit; this species was not in the available API Coryne database. At least two API numeric profiles have been observed for isolates confirmed to be C. falsenii (2, 9), although presently the database cannot warn users of this possibility. Interestingly, one of our numeric profiles differed by one number from the API numeric profile reported in a Canadian study (2), and this correlated with delayed glucose fermentation, described for C. falsenii. The absence of C. falsenii from the API Coryne kit database could contribute to the underrecognition and underreporting of clinically relevant isolates of this species.
Also of interest is that our patient developed bacteremia while on vancomycin therapy. Similar to C. jeikeium, C. falsenii is expected to be susceptible to vancomycin, although, unlike its close relative, C. falsenii has not been noted to have the inherent broad antimicrobial resistance seen in C. jeikeium (2, 13, 14). Our isolates tested susceptible to vancomycin; hence, it remains a concern how the bacteremia developed while the patient was ostensibly on appropriate therapy. At times the vancomycin trough level was noted to be suboptimal; still, this should have been adequate since it was well above the assayed MIC. More likely, the apparent breakthrough bacteremia may be due to the fact that corynebacterium species can form biofilms that are refractory to the activity of vancomycin (10). The relatively low colony count from the catheter tip culture might be explained by specimen processing factors and/or partial effects of the patient's antibiotic regimen. It is noted that while some vancomycin-resistant corynebacterium species have been reported, vancomycin remains the most active agent against corynebacterium-like organisms (14). Standardized guidelines for susceptibility testing of Corynebacterium species became available in 2006 (3). Where comparison to agents tested in earlier work (albeit those based on guidelines for staphylococci [11]) was possible, our C. falsenii isolate was similarly susceptible to penicillin and gentamicin but differed in its resistance to erythromycin.
Finally, while corynebacterial bloodstream infections are associated primarily with central venous catheters (8, 10), it has been suggested that central venous catheter removal may not be necessary to eradicate infection (10). In our case, the patient was febrile, diaphoretic, and tachycardic, and his symptoms resolved once the central line was removed.
Interestingly, a database search at ARUP Laboratories revealed at least 2 other independent isolates of C. falsenii from blood cultures submitted from California (female, 73 years, 06-81074; Fig. 1A and B) and Nebraska (female, 65 years, 09-563200; Fig. 1B). The latter organism displayed an antibiotic susceptibility profile nearly identical to that seen here (Table 1) except that it was susceptible to both erythromycin and trimethoprim-sulfamethoxazole (data not shown). In summary, the data presented here on bacteremia caused by C. falsenii support the postulation that this organism is a cause of clinically significant disease.
Acknowledgments
We thank Keith Simmon and Sue Slechta, ARUP Institute for Clinical Pathology, for assistance with full 16S rRNA gene sequencing.
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard, K. A., C. Munro, D. Wiebe, and E. Ongsansoy. 2002. Characteristics of rare or recently described Corynebacterium species recovered from human clinical material in Canada. J. Clin. Microbiol. 40:4375-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline. CLSI document M45-A. CLSI, Wayne, PA.
- 4.Clinical and Laboratory Standards Institute. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. CLSI document MM18-A. CLSI, Wayne, PA.
- 5.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity, and J. M. Tiedje. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández-Garayzábal, J. F. R. Egido, A. I. Vela, V. Briones, M. D. Collins, A. Mateos, R. A. Hutson, L. Dominguez, and J. Goyache. 2003. Isolation of Corynebacterium falsenii and description of Corynebacterium aquilae sp. nov., from eagles. Int. J. Syst. Evol. Microbiol. 53:1135-1138. [DOI] [PubMed] [Google Scholar]
- 7.Fernández-Garayzábal, J. F., A. I. Vela, R. Egido, R. A. Hutson, M. P. Lanzarot, M. Fernández-Garcia, and M. D. Collins. 2004. Corynebacterium ciconiae sp. nov., isolated from the trachea of black storks (Ciconia nigra). Int. J. Syst. Evol. Microbiol. 54:2191-2195. [DOI] [PubMed] [Google Scholar]
- 8.Funke, G., A. von Gravenitz, J. E. Clarridge III, and K. A. Bernard. 1997. Clinical microbiology of Coryneform bacteria. Clin. Microbiol. Rev. 10:125-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funke, G., and K. A. Bernard. 2007. Coryneform gram-positive rods, p. 485-514. In E. J. Baron, J. H. Jorgensen, M. L. Landry, M. A. Pfaller, and P. R. Murray (ed.), Manual of clinical microbiology, 9th ed., vol. 1. ASM Press, Washington, DC.
- 10.Ghide, S., Y. Jiang, R. Hachem, A. M. Chaftari, and I. Raad. 2010. Catheter-related Corynebacterium bacteremia: should the catheter be removed and vancomycin administered? Eur. J. Clin. Microbiol. Infect. Dis. 29:153-156. [DOI] [PubMed] [Google Scholar]
- 11.Sjödén, B., G. Funke, A. Izquierdo, E. Akervall, and M. D. Collins. 1998. Description of some coryneform bacteria isolated from human clinical specimens as Corynebacterium falsenii sp. nov. Int. J. Syst. Bacteriol. 48:69-74. [DOI] [PubMed] [Google Scholar]
- 12.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 13.Traub, W. H., U. Giepel, B. Leonhard, and D. Bauer. 1998. Antibiotic susceptibility testing (agar disk diffusion and agar dilution) of clinical isolates of Corynebacterium jeikeium. Chemotherapy 44:230-237. [DOI] [PubMed] [Google Scholar]
- 14.Williams, D. Y., S. T. Slepak, and V. J. Gill. 1993. Identification of clinical isolates of nondiphtherial Corynebacterium species and their antibiotic susceptibility patterns. Diagn. Microbiol. Infect. Dis. 17:23-28. [DOI] [PubMed] [Google Scholar]