Abstract
Clinical arbovirus screening requires exclusion of a broad range of viruses with as few assays as possible. We present a reverse transcription-PCR (RT-PCR) for the detection of all species of the genus Alphavirus qualified for exclusion screening (limit of detection [LOD], 5 to 100 RNA copies per reaction across all Alphavirus species; detection of viremia down to ca. 10,000 copies per ml).
Alphaviruses (family Togaviridae) cause a range of human and animal diseases. Human infections with Old World alphaviruses, e.g., Sindbis virus (SINV) or Chikungunya virus (CHIKV), are mainly associated with rash and arthritis. New World alphaviruses, e.g., Venezuelan equine encephalitis virus (VEEV), may cause severe encephalitis. The 29 recognized Alphavirus species are restricted geographically and in terms of their vectors (Aedes, Anopheles, Culex, and Mansonia spp.). The example of CHIKV has shown how alphaviruses can rapidly employ new vectors which in turn may occupy new geographic areas, leading to an introduction into novel tropical and temperate regions (1-3, 7). Due to an ever-expanding range of tourism, the probability of encountering alphavirus infections in clinical practice has increased. Due to the high diversity of alphaviruses, generic alphavirus diagnostic tests validated for the diagnostic laboratory are needed. In two previous studies, alphavirus-specific PCR has been developed (6, 8). However, the sensitivity of the assays was tested using either PFU (6, 8) or plasmids containing alphavirus-specific inserts (8). Moreover, the most recent of these assays has been formulated ca. 10 years ago, omitting more recently identified alphavirus strains. In the present study we have developed a reverse transcription-PCR (RT-PCR) assay suitable for a geographically representative panel of pathogenic alphaviruses and evaluated it on original sera of CHIKV patients with known viral loads.
Due to its high conservation, the nsP4 coding region of alphavirus RNA was chosen as the target for genus-specific RT-PCR. Primers used for reverse transcription and amplification were designed using an up-to-date nucleotide sequence database as available at the end of 2009. Based on the available alphavirus sequences, it was necessary to use two antisense primers differing in one nucleotide.
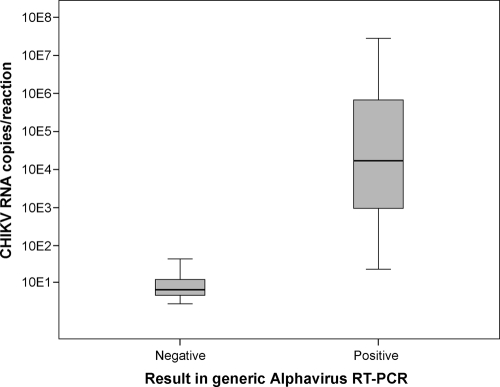
The first amplification reaction mixture (12 μl) contained 1 μl RNA, 0.5 μg bovine serum albumin (BSA), 0.3 μl Superscript III/Platinum Taq enzymes (Invitrogen), 1× buffer, and primers TTTAAGTTTGGTGCGATGATGAAGTC and GCATCTATGATATTGACTTCCATGTT (500 nM each). The second-round mixture (50 μl) contained 1 μl first-round PCR product, 1 μg BSA, 1.6 mM MgCl2, 0.16 mM (each) deoxynucleoside triphosphates (dNTP), and 0.1 μl Platinum Taq (Invitrogen), as well as primers GGTGCGATGATGAAGTCTGGGATGT (200 nM), CTATGATATTGACTTCCATGTTCATCCA (100 nM), and CTATGATATTGACTTCCATGTTCAGCCA (100 nM). First-round RT-PCR involved 15 min at 45°C; 3 min at 95°C; a touchdown cycling element of 10 cycles of 95°C for 20 s, 65°C (−1°C per cycle) for 20 s, and 72°C for 20 s; and a main cycling element of 30 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s. Second-round PCR involved 2 min at 95°C and 40 cycles of 95°C for 20 s, 58°C for 20 s, and 72°C for 20 s. Second-round amplicons (210 bp) were analyzed by electrophoresis in 1.5% agarose gels, stained with ethidium bromide, and subsequently visualized on a UV transilluminator. Plaque titers of cultured alphaviruses were determined in Vero cells, and the alphaviruses were adjusted to 104 PFU per ml and tested. Clear products of approximately 210 bp were visible for all alphaviruses listed in Table 1. In vitro-transcribed RNAs from the same 10 alphaviruses were generated and tested in limiting dilution experiments. The numbers of in vitro transcripts per reaction were 250, 100, 50, 25, 5, and 1, respectively. As shown in Table 1, endpoint sensitivities ranged from 5 to 100 transcripts per reaction. Forty human sera with CHIKV RNA levels previously determined by real-time PCR (4, 5) were extracted with the viral RNA minikit (Qiagen, Hilden, Germany) and tested by the generic alphavirus RT-PCR (Fig. 1). Of 40 samples, 23 were detectable by generic alphavirus RT-PCR (57.5%). The sample with the lowest CHIKV viral load that was still detectable had 1 × 104 CHIKV RNA copies/ml. This corresponded to approximately 24 viral RNA copies per reaction.
TABLE 1.
Virus panel tested with the generic alphavirus PCR
| Virus | Family | Genus | PCR resultb | Limit of detectionc |
|---|---|---|---|---|
| Barmah Forest virus | Togaviridae | Alphavirus | Pos | 50 |
| Chikungunya virus | Togaviridae | Alphavirus | Pos | 25 |
| Mayaro virus | Togaviridae | Alphavirus | Pos | 50 |
| O'nyong-nyong virus | Togaviridae | Alphavirus | Pos | 5 |
| Ross River virus | Togaviridae | Alphavirus | Pos | 100 |
| Semliki Forest virus | Togaviridae | Alphavirus | Pos | 50 |
| Sindbis virus | Togaviridae | Alphavirus | Pos | 50 |
| Eastern equine encephalitis virus | Togaviridae | Alphavirus | Pos | 50 |
| Western equine encephalitis virus | Togaviridae | Alphavirus | Pos | 5 |
| Venezuelan equine encephalitis virus | Togaviridae | Alphavirus | Pos | 100 |
| Rubella virus | Togaviridae | Rubivirus | Neg | NA |
| Other virus speciesa | Neg | NA |
In addition to the Togaviridae, 27 virus species from other virus families were tested using the generic alphavirus PCR. This panel consisted of influenza A and B viruses; human metapneumovirus; human respiratory syncytial virus; human parainfluenza viruses 1 and 2; human astrovirus; Norwalk virus; Sapporo virus; human coronaviruses OC43, NL63, and 229E; human rhinovirus; human enterovirus; human immunodeficiency virus type 1; hepatitis C virus; human parvovirus B19; rotavirus; human adenovirus; hepatitis B virus; cytomegalovirus; herpes simplex virus 1; Epstein-Barr virus; varicella-zoster virus; human herpesvirus 6; human herpesvirus 8; and BK virus.
For the genus Alphavirus, results were determined from virus stock solutions adjusted to 104 PFU/ml; for other viruses, results were determined from PCR-confirmed stored clinical samples. Pos, positive; Neg, negative.
As determined by endpoint dilution of RNA in vitro transcripts covering the full amplicon region exceeding the primer binding sites. NA, not applicable.
FIG. 1.
Box plot of generic alphavirus RT-PCR in 40 sera with known CHIKV RNA viral loads as determined by real-time RT-PCR (4, 5). The box plot was produced using SPSS, version 17. The boxes show the median (bar) and interquartile range (box length). The whiskers represent an extension of the 25th or 75th percentiles by 1.5 times the interquartile range. No outliers were present.
In order to exclude cross-reactivity with other viruses, a set of 29 clinical specimens known to contain related and/or clinically relevant viruses was tested (Table 1). The panel included two samples of rubella virus, representing the most closely related sister genus, Rubivirus, within the family Togaviridae. Alphavirus generic RT-PCR was negative in all tested samples, indicating a high specificity of the assay. It was therefore not considered necessary to increase the detection specificity by using any probe hybridization techniques.
The evident fluctuation of climatic parameters such as the annual minimum temperature in temperate regions and the increase of international trade and travel, as well as changes in vector and virus ecology associated with human intervention, altogether increase the abundance and diversity of arbovirus infections encountered in clinical practice. The diagnosis of alphavirus infections is technically demanding due to the high genetic variability encountered in the genus. The broad range of distinct human-pathogenic viruses precludes the application of single virus species-specific assays. A single broad-range assay for the whole genus Alphavirus can be used in the diagnostic laboratory to exclude alphaviruses in staged diagnostic approaches, provided that the sensitivity of assays is high enough to facilitate exclusion screening with sufficient negative predictive value. The assay presented here provides for the first time a broad-range detection of viruses of the complex genus Alphavirus with evaluated high analytical and clinical sensitivity, compatible with clinical application and exclusion screening.
Acknowledgments
This work was funded by the BMBF (contract no. 01ES0814) and the European Union (contracts 2117575/ArboZooNet, 228292/EVA, and 223498/EMPERIE).
Carmen Poster and Andrea Wuttkopf are acknowledged for technical assistance.
Footnotes
Published ahead of print on 26 May 2010.
REFERENCES
- 1.de Lamballerie, X., E. Leroy, R. N. Charrel, K. Ttsetsarkin, S. Higgs, and E. A. Gould. 2008. Chikungunya virus adapts to tiger mosquito via evolutionary convergence: a sign of things to come? Virology J. 5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gould, E. A., B. Coutard, H. Malet, B. Morin, S. Jamal, S. Weaver, A. Gorbalenya, G. Moureau, C. Baronti, I. Delogu, N. Forrester, M. Khasnatinov, T. Gritsun, X. de Lamballerie, and B. Canard. 2010. Understanding the alphaviruses: recent research on important emerging pathogens and progress towards their control. Antiviral Res. 87:111-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawley, W. A., P. Reiter, R. S. Copeland, C. B. Pumpuni, and G. B. Craig, Jr. 1987. Aedes albopictus in North America: probable introduction in used tires from northern Asia. Science 236:1114-1116. [DOI] [PubMed] [Google Scholar]
- 4.Panning, M., K. Grywna, M. van Esbroeck, P. Emmerich, and C. Drosten. 2008. Chikungunya fever in travellers returning to Europe from the Indian Ocean region, 2006. Emerg. Infect. Dis. 14:416-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panning, M., R. N. Charrel, O. D. Mantke, M. Niedrig, and C. Drosten. 2009. Coordinated implementation of Chikungunya virus reverse transcription-PCR. Emerg. Infect. Dis. 15:469-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeffer, M., B. Proebster, R. M. Kinney, and O. R. Kaaden. 1997. Genus-specific detection of alphaviruses by a semi-nested reverse transcription-polymerase chain reaction. Am. J. Trop. Med. Hyg. 57:709-718. [DOI] [PubMed] [Google Scholar]
- 7.Rezza, G., L. Nicoletti, R. Angelini, R. Romi, A. C. Finarelli, M. Panning, P. Cordioli, C. Fortuna, S. Boros, F. Magurano, G. Silvi, P. Angelini, M. Dottori, M. G. Ciufolini, G. C. Majori, A. Cassone, and the CHIKV Study Group. 2007. Infection with Chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370:1840-1846. [DOI] [PubMed] [Google Scholar]
- 8.Sánchez-Seco, M. P., D. Rosario, E. Quiroz, G. Guzmán, and A. Tenorio. 2001. A generic nested-PCR followed by sequencing for detection and identification of members of the alphavirus genus. J. Virol. Methods 95:153-161. [DOI] [PubMed] [Google Scholar]