Abstract
Colonizing group B Streptococcus (GBS) capsular polysaccharide (CPS) type IV isolates were recovered from vaginal and rectal samples obtained from 97 (8.4%) nonpregnant women of 1,160 women enrolled in a U.S. multicenter GBS vaccine study from 2004 to 2008. Since this rate was much higher than the rate of prevalence of 0.4 to 0.6% that we found in previous studies, the isolates were analyzed by using surface protein profile identification, pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST) to characterize them and identify trends in DNA clonality and divergence. Of the 101 type IV isolates studied, 53 expressed α and group B protective surface (BPS) proteins, 27 expressed BPS only, 20 expressed α only, and 1 had no detectable surface proteins. The isolates spanned three PFGE macrorestriction profile groups, groups 37, 38, and 39, of which group 37 was predominant. The isolates in group 37 expressed the α and BPS proteins, while those in groups 38 and 39 expressed the α protein only, with two exceptions. MLST studies of selective isolates from the four protein profile groups showed that isolates expressing α,BPS or BPS only were of a new sequence type, sequence type 452, while those expressing α only or no proteins were mainly of a new sequence type, sequence type 459. Overall, our study revealed a limited diversity in surface proteins, MLST types, and DNA macrorestriction profiles for type IV GBS. There appeared to be an association between the MLST types and protein expression profiles. The increased prevalence of type IV GBS colonization suggested the possibility that this serotype may emerge as a GBS pathogen.
Group B Streptococcus (GBS) (Streptococcus agalactiae) is a leading cause of neonatal infection in the United States, with maternal vaginal or rectal colonization often resulting in the transmission of GBS to the infant during the perinatal period (8, 23). GBS isolates are classified according to nine capsular polysaccharide (CPS) types: types Ia, Ib, and II to VIII and the recently proposed type IX (9, 15, 21, 23, 46, 52). Isolates that do not express any of the known CPS types are designated nontypeable (NT) (2, 6, 21, 40). In addition to CPS, GBS may express one or more surface-localized proteins, including the α and β components of the c protein (24); the alpha-like R proteins, specifically R1, R4(Rib), and R1,R4 (also known as Alp3) (14, 17, 19, 30, 40); and the group B protective surface (BPS) protein (12). Certain protein profiles are associated with each capsular polysaccharide CPS type (2), for example, the c(α only) protein with types Ia and II, c(α + β) with type Ib, and R4(Rib) with type III (2, 14). BPS, expressed by fewer than 3% of colonizing isolates, can be found alone or with another protein in type Ia, II, and V isolates (12, 14).
In the United States, the predominant serotypes over the past 2 decades, constituting 70 to 75% of all GBS isolates, have been type Ia, type III, and the more recently emerged type V (14, 15, 20, 52). The remaining isolates consisted primarily of types Ib and II, with types IV, VI, VII, and VIII making up a small fraction of the isolates. We found type IV to represent between 0.4 and 0.6% of colonizing GBS isolates (14, 15), but only rare type IV isolates were found in invasive GBS disease during that same time period (14, 43, 52).
In contrast to the previously low percentage of type IV isolates reported for the United States, recent studies in the United Arab Emirates, Turkey, and Zimbabwe showed large proportions of type IV isolates among their GBS isolates. In the United Arab Emirates, type IV was the predominant serotype among colonized pregnant women, representing 26.3% of the GBS isolates (1). In eastern Turkey, it was the second most common serotype, at 8.3%, among colonizing isolates (10), and in Zimbabwe, it was the fourth most common serotype, comprising 5.1% of GBS isolates from colonized pregnant women and 4.0% of all GBS isolates from various sites, including blood and cerebrospinal fluid (CSF), from hospitalized patients (36).
Immunization studies of humans (3, 28) and protection studies with mice (37) have shown the potential of vaccines against the common GBS serotypes to prevent invasive neonatal GBS disease through the vaccination of pregnant women (3, 28). The GBS strains described here are from a phase II randomized, double-blinded clinical trial of a GBS serotype III-tetanus toxoid (CPS III-TT) vaccine to prevent the vaginal acquisition of GBS type III in nonpregnant women in three areas of the United States: Pittsburgh (PA), Georgia, and Texas (S. Hillier, unpublished data). Because we found type IV isolates for almost 10% of these patients, we examined the type IV isolates for surface proteins and clonality.
Pulsed-field gel electrophoresis (PFGE) was used in this analysis because it is a widely used method that can further characterize GBS isolates within particular CPS type and/or protein profile groups (2, 4, 6, 48). Multilocus sequence typing (MLST) was performed in order to assess the general relatedness of strains within and across laboratories (25, 50). Together, the discriminatory power of PFGE and the objectivity of MLST gave insight into the GBS type IV population genetic structure and the identification of emerging clones (2, 5, 13, 18, 19).
MATERIALS AND METHODS
Bacterial isolates.
GBS isolates from vaginal and rectal cultures of 1,160 nonpregnant women in the vaccine trial were received at the University of Minnesota GBS Molecular Reference Laboratory from January 2004 to May 2008. Of the 1,160 patients, 797 visited the clinics for a screening visit only. The remaining 363 patients made between 1 and 11 visits over a period of 18 months. Vaginal and rectal samples were taken and cultured. GBS isolates were isolated, extracted, and serotyped for each visit, with three GBS colonies picked per positive culture.
Storage and growth.
GBS isolates were subcultured onto 5% sheep blood agar plates (BAPs) and grown at 37°C overnight in Todd-Hewitt broth (BD, Sparks, MD) for serotyping or for 48 h on BAPs for PFGE analysis (5). The isolates were stored at −30°C in Todd-Hewitt broth with 2% defibrinated sheep blood (14, 17, 40).
Serotyping and detection of surface proteins.
The capsular polysaccharide type and protein profile were determined for each isolate by using HCl extraction and Ouchterlony double immunodiffusion with rabbit antisera monospecific for CPS type or surface protein as we have previously described (14, 17, 24, 40). Proteins under study in our laboratory included c(α), c(β), R1, R4(Rib), R1,R4 (known as Alp3), and BPS (47). HCl extracts were concentrated 3-fold and retested when a reaction was weak or inconclusive. Prototype strains for the surface proteins were the ones used in our previous studies; strain 3139 (S. Bergner-Rabinowitz, Israel), which did not express any of these proteins, was the CPS type IV prototype (14, 17, 24).
DNA preparation.
At least one isolate per patient colonized with GBS type IV was studied by PFGE. DNA samples were prepared by using a rapid PFGE procedure described previously by our laboratory (5) and digested with the low-frequency restriction enzyme SmaI. The DNA fragments were resolved by using PFGE. Included in each gel was a lambda molecular size standard and isolate 89-022 (Ib/α+β), the internal standard described previously by our laboratory (2, 5, 6).
DNA macrorestriction analysis.
After staining with ethidium bromide, gels were photographed under UV light with a Kodak DC290 digital camera. Kodak 1D digital image analysis software was used to determine the molecular sizes of the bands. The band patterns were then compared to numbered prototypes by using our previously described method (2, 6), adapted from the one described previously by Tenover et al. (48). An isolate with a band pattern identical to that of a prototype was given the same number designation, and those with one to two, three, four, and five band differences were considered subtypes and were given, in addition, the designations a, b, c, and d, respectively. A band difference was counted as the presence or absence of a band (6). If an isolate displayed six or more band differences, it was considered unrelated to the prototype (2, 6). The first isolate with a new PFGE profile was designated the prototype to which all subsequent isolates were compared.
MLST.
Representative isolates studied by MLST were selected from the major surface protein profiles and PFGE macrorestriction profile groups found among these isolates. Isolates were chosen within each group to represent a wide time period. Extraction of DNA and partial sequencing of seven housekeeping genes were carried out as previously described (25). Alleles were given number classifications by comparing them to existing sequences in the online MLST database (http://pubmlst.org/sagalactiae/). The determination of sequence types (STs) based on the unique set of alleles for each isolate and examination of relationships between STs were also accomplished by using the database.
RESULTS
Of the 1,160 patients in the multicenter vaccine study, 97 (8.4%) were colonized at least once with a type IV isolate. For 93 of the 97 patients, only one isolate per patient was included in this analysis because the protein profiles and PFGE profiles were the same for each culture site and visit. The remaining four patients were counted twice, one time for each unique protein profile or PFGE profile identified among their type IV isolates. Of the 101 type IV isolates included in the study, 63 were from vaginal cultures and 38 were from rectal cultures; 50 were from patients with screening visits only and 51 were from patients who had 1 to 10 GBS-positive visits. For these 51 patients, the average number of culture-positive visits was 5, and 17 of the 51 patients (33%) had seven or more culture-positive visits. The majority of the patients were from Pittsburgh (n = 78; 80.4%), 16 (16.5%) were from Georgia, and 3 (3.1%) were from Texas. These numbers reflected the distribution of patients with all GBS isolates in the vaccine study: 857 (73.9%) from Pittsburgh, 243 (20.9%) from Georgia, and 60 (5.2%) from Texas (data not shown).
When digested by SmaI, the type IV isolates in this study gave clear, consistent band patterns, each comprised of 7 to 10 bands. All type IV isolates differed by at least six bands compared to our existing PFGE prototypes 1 to 36, so a new PFGE profile group, group 37, was established, followed by profile groups 38 and 39. PFGE profile group 37 was the predominant profile among type IV isolates in each geographical region (range of 62.5% to 84.0%), with Pittsburgh representing the region with the highest frequency. PFGE group 39 was the next predominant profile (range of 11.1% to 37.5%), with clinic sites in Georgia having the highest frequency (data not shown).
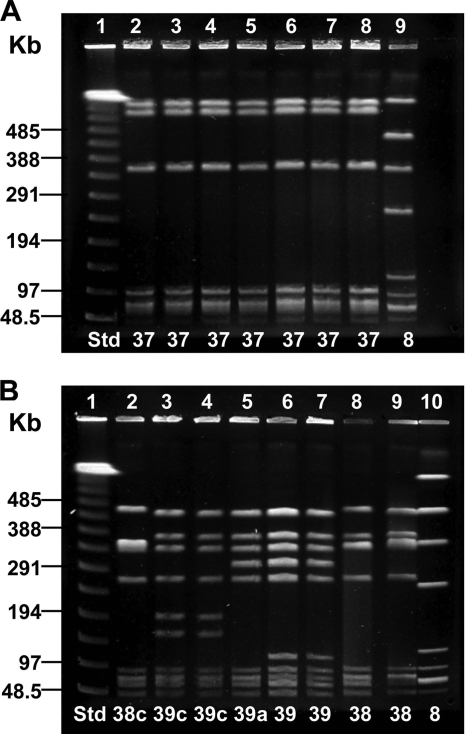
The examples of PFGE profiles from type IV isolates in Fig. 1 illustrated the wide diversity of profiles seen among IV/c(α only) isolates in comparison to profiles of IV/α,BPS or IV/BPS isolates. In Fig. 1A, all of the IV/α,BPS and IV/BPS isolates exhibiting PFGE profile 37 were from different patients. Of the IV/α,BPS and IV/BPS isolates, 89% displayed a PFGE profile identical to the one shown here. In Fig. 1B, the IV/c(α only) isolates, from seven different patients, displayed five unique band patterns. A variety of band patterns within PFGE profile groups 38 and 39 was typical of the IV/c(α only) isolates studied. Studied by MLST, the isolates with BPS only or α,BPS in lanes 2, 4, 6, and 7 of Fig. 1A were of ST 452, while isolates expressing α only in lanes 3 and 6 to 9 of Fig. 1B were of ST 459.
FIG. 1.
SmaI macrorestriction analysis by PFGE of 15 CPS type IV isolates with α-only, BPS, and α,BPS proteins. Lane designations are along the top of the gel, and PFGE profiles are along the bottom. “Std” refers to the lambda molecular size standard. (A) Seven type IV isolates with BPS and α,BPS protein profiles. Lane 1, lambda standard; lanes 2 to 5, IV/BPS; lanes 6 to 8, IV/α,BPS; lane 9, Ib/c(α+β) (internal standard). Lane 6 is the type 37 prototype. Isolates in lanes 2, 4, 6, and 7 were ST 452. (B) Eight CPS type IV isolates with the α protein. Lane 1, lambda standard; lanes 2 to 9, type IV/c(α only); lane 10, type Ib/c(α+β) (internal standard). Lanes 6 and 7 are duplicates of the type 39 prototype; lane 8 is the type 38 prototype. Isolates in lanes 3 and 6 to 9 were of ST 459.
The distribution of the 101 type IV isolates in this study by protein profile, PFGE profile, and sequence type is presented in Table 1. Four different surface protein profiles were observed. Of all isolates, 53 (52.5%) expressed both the α and BPS proteins. The second most common surface protein profile was BPS only (27 isolates; 26.7%), followed by α only (20 isolates; 19.8%). No proteins were detected for one isolate (1.0%). The high prevalence of the BPS protein in the type IV isolates is worth noting, since this protein was less common among isolates of other serotypes (data not shown).
TABLE 1.
Protein profile, PFGE profile, and sequence type distribution of 101 CPS type IV isolates from 97 patients in a 2004-2008 GBS vaccine study in the United States
| Protein profile | PFGE profile group | No. of isolates | No. of isolates analyzed by MLST | Sequence type (no. of isolates)a |
|---|---|---|---|---|
| α,BPS | 37 | 45 | 4 | 452 (4) |
| 37a | 6 | 2 | 452 (2) | |
| 37b | 2 | 1 | 452 (1) | |
| BPS only | 37 | 26 | 2 | 452 (2) |
| 37a | 1 | 0 | ||
| α only | 37 | 1 | 1 | 452 (1) |
| 38 | 3 | 2 | 459 (2) | |
| 38c | 1 | 0 | ||
| 39 | 5 | 2 | 459 (2) | |
| 39a | 4 | 0 | ||
| 39b | 1 | 0 | ||
| 39c | 4 | 1 | 459 (1) | |
| 39d | 1 | 0 | ||
| None | 39 | 1 | 1 | 459 (1) |
The numbers in parentheses represent the numbers of isolates with the specified sequence type.
As shown in Table 1, these isolates were distributed into two sequence types and three PFGE groups. PFGE profile group 37, the most common macrorestriction group, contained 81 (80.2%) of the 101 isolates; 4 isolates (4.0%) were in group 38, and 16 isolates (15.8%) were in group 39. The 16 isolates studied by MLST were found to be either ST 452 or 459 and were cleanly divided into the two groups according to their PFGE profile groups, with all 10 isolates in group 37 corresponding to ST 452 and all six isolates in groups 38 and 39 exhibiting ST 459. Neither ST had been described before this study. ST 452 was found to have the allelic profile 5-25-4-3-2-3-3, a single-locus variant (SLV) of STs 24 and 468, the latter being a new ST from an invasive type IV isolate from Minnesota (P. Ferrieri and R. Creti, unpublished data). Sequence type 459 was found to have the profile 1-1-3-1-41-12-2, an SLV of STs 136 and 196. Allele 41, found at the sdhA locus of ST 459, was newly described here (data not shown).
An association was observed between the BPS protein, ST 452, and PFGE profile group 37 (Table 1). All 53 isolates with α,BPS proteins and all 27 isolates with the BPS protein only had DNA patterns identical or highly related to the PFGE prototype 37 profile, and nine isolates with these protein profiles were of ST 452. Similarly, 16 of the 21 isolates with the α protein only or no detectable surface proteins were classified as being of profile 39 or its subgroups, and four were classified as being of profile 38 or its subgroups; five of these α-only isolates were of ST 459.
Seventy-two of the 81 isolates (88.9%) within PFGE profile group 37 were identical to the PFGE profile 37 prototype, while only 9 (11.1%) belonged to either subgroup 37a or 37b. Isolates in PFGE profile group 39 showed more diversity than those in group 37 since only 6 of the 16 isolates (37.5%) were identical to the group 39 prototype strain, while the remaining 10 isolates (62.5%) were distributed throughout subgroups 39a to 39d. PFGE group 38 was too small for meaningful comparisons.
DISCUSSION
It was of particular interest to study the vaginal/rectal type IV isolates from this population because the 8.4% rate of isolation among this group of patients is more than a 10-fold increase compared to rates found in our previous studies of colonizing GBS isolates from mother-infant pairs and nonpregnant women in various regions of the United States from 1993 to 2002 (14, 15, 23). Based on this evolution of a relatively high prevalence of type IV colonization, it is possible that type IV may emerge as an important pathogen, much like type V in the 1990s (11, 20, 52). Further evidence of this possibility is the increase that we have found in the prevalence of type IV in invasive GBS disease in Minnesota, from 0.7% in 1993 to 1999 to 1.8% in 2000 to 2008 (16). Although this is a small increase, it is significant since in the last 8 years, type IV has been found mostly in infants with early-onset or late-onset disease compared to previous years, where type IV invasive disease was seen primarily in adults (16).
The results from the present study of type IV isolates by DNA macrorestriction and MLST suggested that, in general, there was an association between the sequence type, CPS/protein profile, and PFGE patterns. This is supported by findings that except for one isolate, all ST 459 isolates expressed the α protein only and were in PFGE profile groups 38 and 39, and, with one exception, ST 452 isolates expressed the α,BPS or BPS protein and were in PFGE profile group 37.
The protein profile appeared to be associated with PFGE profile diversity, evidenced by the wider variety of PFGE profiles for IV/α-only isolates than for those with α,BPS or BPS. However, this diversity of PFGE profiles among isolates with the α protein only may be attributed to additional factors, such as the length of time since the original clone appeared, genetic evolution of the individual serotypes (4, 11, 13), or lateral gene transfer (49).
Prior to this study, type IV isolates had been examined by PFGE only on a very small scale. Two Italian studies analyzed type IV isolates using PFGE, one finding three distinct PFGE groups among six isolates (42) and the other finding three groups among three isolates (19). A study in France examined two type IV isolates and found them to be in the same PFGE group (41). In Norway, six distinct band patterns were found among six invasive type IV isolates (44), but a grouping method was not applied. Due to technical limitations of comparing these previously reported PFGE profiles, we could not ascertain any resemblance to our DNA profiles.
The new sequence types 452 and 459, exhibited by the type IV isolates in this study, have allelic similarities to STs in clonal complexes (CCs) 23 and 1, respectively. CC 23 has typically included, among others, STs 23 and 24, which exhibit 4 and 6 allelic similarities to ST 452, respectively (19, 34). CC 1 has included STs 1, 2, and 196, which have 4, 5, and 6 similarities to ST 459, respectively (7, 19, 29). CC 1 and CC 23 have been found to be among the major clonal complexes associated with human GBS invasive disease and colonization (26, 32).
Several sequence types of CPS type IV isolates have been reported in the literature from 2005 to 2009, the most common being sequence type 196 (19, 22, 29, 32, 35, 51). Among the type IV invasive isolates from Minnesota collected between 1996 and 2007 were STs 196 and 291 as well as the new sequence types 452 and 459 described here plus a new sequence type, ST 468 (P. Ferrieri and R. Creti, unpublished data). In contrast to STs of more recently isolated type IV isolates, type IV reference strain 3139 from the 1970s was designated ST 2 (Ferrieri and Creti, unpublished).
In comparing the type IV isolates from our current study to those examined by other studies in our laboratory from 1995 to 2000, we found that they differed in surface protein expression and PFGE profiles. Prior to this study, our type IV isolates expressed the R1,R4 (Alp3) protein and, like the much older prototype strain 3139 (IV/none), had PFGE profiles resembling profile group 4 (our unpublished data), usually seen with type V isolates (2). Compared to PFGE profile 4, the new PFGE profiles 37, 38, and 39 differed by more than six bands, as measured by the size of the DNA fragments.
The type IV isolates in this study were discovered to belong to three PFGE groups with three different protein profiles very soon after the first appearance of CPS type IV in three geographically distant regions in the United States. This finding is in contrast to our previous observations with type V, PFGE profile group 4, isolates, whose DNA macrorestriction patterns very gradually shifted, over a period of several years, from being identical to the O profile described previously by others (11) to evolving into subgroups of profile 4 (2). Despite differences, the type IV isolates presented here were highly clonal, evidenced by the finding that all 80 type IV isolates with α,BPS or BPS belonged to PFGE group 37, associated with ST 452. Although 20 of the 21 isolates with α-only or no proteins showed greater diversity, they spanned only two closely related PFGE groups (groups 38 and 39), associated with ST 459.
PFGE and MLST, although sometimes viewed as interchangeable, were each shown to have unique benefits. PFGE appeared to have greater discriminatory power than MLST, and it could be a valuable molecular epidemiological tool in the investigation of clusters of type IV disease or colonization. Using MLST, it was found that the type IV isolates had two unique sequence types, while PFGE demonstrated that many slight DNA profile variations existed within each sequence type. However, MLST was a more objective method that allowed us to compare our results with those of other laboratories and to see allelic relationships among sequence types.
GBS conjugate vaccines that are being developed to prevent invasive disease may protect only against serotypes Ia, Ib, II, III, and V. Although a pentavalent vaccine that includes the more common serotypes is expected to provide protection for a high percentage of GBS cases (3, 20, 38), the global presence of type IV and the possibility of shifts in serotype predominance due to serotype replacement, capsular switch, or changes in bacterial virulence factors (1, 9, 31, 40, 49) necessitate ongoing surveillance of types IV and VI to VIII and the newly proposed type IX (46). Because of the emergence of invasive disease due to type IV in Minnesota, Boston, and other regions of the United States (16, 27, 39, 45), monitoring of type IV colonization and disease in the population is warranted. If the prevalence of type IV in invasive disease significantly increases, this polysaccharide and/or associated GBS virulence proteins should be included in vaccine design (33, 49).
Acknowledgments
This work was supported by contract N01-AI-25495 from the National Institutes of Health, Bethesda, MD.
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Amin, A., Y. M. Abdulrazzaq, and S. Uduman. 2002. Group B streptococcal serotype distribution of isolates from colonized pregnant women at the time of delivery in United Arab Emirates. J. Infect. 45:42-46. [DOI] [PubMed] [Google Scholar]
- 2.Amundson, N. R., A. E. Flores, S. L. Hillier, C. J. Baker, and P. Ferrieri. 2005. DNA macrorestriction analysis of nontypeable group B streptococcal isolates: clonal evolution of nontypeable and type V isolates. J. Clin. Microbiol. 43:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, C. J., and M. S. Edwards. 2003. Group B streptococcal conjugate vaccines. Arch. Dis. Child. 88:375-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, J. A., and P. Ferrieri. 2001. Rapid pulsed-field gel electrophoresis method for group B streptococcus isolates. J. Clin. Microbiol. 39:3006-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, J. A., A. E. Flores, C. J. Baker, S. L. Hillier, and P. Ferrieri. 2002. Improved methods for typing nontypeable isolates of group B streptococci. Int. J. Med. Microbiol. 292:37-42. [DOI] [PubMed] [Google Scholar]
- 7.Bohnsack, J. F., A. Whiting, M. Gottschalk, D. M. Dunn, R. Weiss, P. H. Azimi, J. B. Philips III, L. E. Weisman, G. G. Rhoads, and F.-Y. C. Lin. 2008. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. academic centers from 1995 to 1999. J. Clin. Microbiol. 46:1285-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2007. Perinatal group B streptococcal disease after universal screening recommendations—United States, 2003-2005. MMWR Morb. Mortal. Wkly. Rep. 56:701-705. [PubMed] [Google Scholar]
- 9.Cieslewicz, M. J., D. Chaffin, G. Glusman, D. Kasper, A. Madan, S. Rodrigues, J. Fahey, M. R. Wessels, and C. E. Rubens. 2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 73:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekin, I. H., and K. Gurturk. 2006. Characterization of bovine and human group B streptococci isolated in Turkey. J. Med. Microbiol. 55:517-521. [DOI] [PubMed] [Google Scholar]
- 11.Elliott, J. A., K. D. Farmer, and R. R. Facklam. 1998. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J. Clin. Microbiol. 36:2115-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdogan, S., P. K. Fagan, S. R. Talay, M. Rohde, P. Ferrieri, A. E. Flores, C. A. Guzman, M. J. Walker, and G. S. Chhatwal. 2002. Molecular analysis of group B protective surface protein, a new cell surface protective antigen of group B streptococci. Infect. Immun. 70:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasola, E., C. Livdahl, and P. Ferrieri. 1993. Molecular analysis of multiple isolates of the major serotypes of group B streptococci. J. Clin. Microbiol. 31:2616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrieri, P., C. J. Baker, S. L. Hillier, and A. E. Flores. 2004. Diversity of surface protein expression in group B streptococcal colonizing & invasive isolates. Indian J. Med. Res. 119:191-196. [PubMed] [Google Scholar]
- 15.Ferrieri, P., S. L. Hillier, M. A. Krohn, D. Moore, L. C. Paoletti, and A. E. Flores. 2004. Characterization of vaginal & rectal colonization with multiple serotypes of group B streptococci using multiple colony picks. Indian J. Med. Res. 119:208-212. [PubMed] [Google Scholar]
- 16.Ferrieri, P., R. Lynfield, and A. E. Flores. 2008. Invasive disease due to group B Streptococcus serotype IV, abstr. P185, p. 284. Abstr. XVII Lancefield Int. Symp. Streptococci Streptococcal Dis., Porto Heli, Greece.
- 17.Flores, A. E., and P. Ferrieri. 1996. Molecular diversity among the trypsin resistant surface proteins of group B streptococci. Zentralbl. Bakteriol. 285:44-51. [DOI] [PubMed] [Google Scholar]
- 18.Foxman, B. 2007. Contributions of molecular epidemiology to the understanding of infectious disease transmission, pathogenesis, and evolution. Ann. Epidemiol. 17:148-156. [DOI] [PubMed] [Google Scholar]
- 19.Gherardi, G., M. Imperi, L. Baldassarri, M. Pataracchia, G. Alfarone, S. Recchia, G. Orefici, G. Dicuonzo, and R. Creti. 2007. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy. J. Clin. Microbiol. 45:2909-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, A. Schuchat, and the Maryland Emerging Infections Program. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 21.Henrichsen, J., P. Ferrieri, J. Jelinkova, W. Kohler, and W. R. Maxted. 1984. Nomenclature of antigens of group B streptococci. Int. J. Syst. Bacteriol. 34:500. [Google Scholar]
- 22.Hery-Arnaud, G., G. Bruant, P. Lanotte, S. Brun, B. Picard, A. Rosenau, N. van der Mee-Marquet, P. Rainard, R. Quentin, and L. Mereghetti. 2007. Mobile genetic elements provide evidence for a bovine origin of clonal complex 17 of Streptococcus agalactiae. Appl. Environ. Microbiol. 73:4668-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hickman, M. E., M. A. Rench, P. Ferrieri, and C. J. Baker. 1999. Changing epidemiology of group B streptococcal colonization. Pediatrics 104:203-209. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, D. R., and P. Ferrieri. 1984. Group B streptococcal Ibc protein antigen: distribution of two determinants in wild-type strains of common serotypes. J. Clin. Microbiol. 19:506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M.-S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. M. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, N., K. A. Oliver, J. Barry, R. M. Harding, N. Bisharat, B. G. Spratt, T. Peto, and D. W. Crook. 2006. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B Streptococcus is independent of capsular serotype. Clin. Infect. Dis. 42:915-924. [DOI] [PubMed] [Google Scholar]
- 27.Jordan, H. T., M. M. Farley, A. Craig, J. Mohle-Boetani, L. H. Harrison, S. Petit, R. Lynfield, A. Thomas, S. Zansky, K. Gershman, B. A. Albanese, W. Schaffner, and S. J. Schrag. 2008. Revisiting the need for vaccine prevention of late-onset neonatal group B streptococcal disease: a multistate, population-based analysis. Pediatr. Infect. Dis. J. 27:1057-1064. [DOI] [PubMed] [Google Scholar]
- 28.Kasper, D. L., L. C. Paoletti, M. R. Wessels, H.-K. Guttormsen, V. J. Carey, H. J. Jennings, and C. J. Baker. 1996. Immune response to type III group B streptococcal polysaccharide-tetanus toxoid conjugate vaccine. J. Clin. Invest. 98:2308-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lartigue, M.-F., G. Hery-Arnaud, E. Haguenoer, A.-S. Domelier, P.-O. Schmit, N. van der Mee-Marquet, P. Lanotte, L. Mereghetti, M. Kostrzewa, and R. Quentin. 2009. Identification of Streptococcus agalactiae isolates from various phylogenetic lineages by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 47:2284-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindahl, G., M. Stålhammar-Carlemalm, and T. Areschoug. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipsitch, M. 1999. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg. Infect. Dis. 5:336-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luan, S.-L., M. Granlund, M. Sellin, T. Lagergård, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. N. Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B Streptococcus vaccine by multiple genome screen. Science 309:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning, S. D., A. C. Springman, E. Lehotzky, M. A. Lewis, T. S. Whittam, and H. D. Davies. 2009. Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J. Clin. Microbiol. 47:1143-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martins, E. R., M. A. Pessanha, M. Ramirez, J. Melo-Cristino, and the Portuguese Group for the Study of Streptococcal Infections. 2007. Analysis of group B streptococcal isolates from infants and pregnant women in Portugal revealing two lineages with enhanced invasiveness. J. Clin. Microbiol. 45:3224-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moyo, S. R., J. A. Maeland, and K. Bergh. 2002. Typing of human isolates of Streptococcus agalactiae (group B streptococcus, GBS) strains from Zimbabwe. J. Med. Microbiol. 51:595-600. [DOI] [PubMed] [Google Scholar]
- 37.Paoletti, L. C., J. Pinel, A. K. Rodewald, and D. L. Kasper. 1997. Therapeutic potential of human antisera to group B streptococcal glycoconjugate vaccines in neonatal mice. J. Infect. Dis. 175:1237-1239. [DOI] [PubMed] [Google Scholar]
- 38.Paoletti, L. C., M. A. Rench, D. L. Kasper, D. Molrine, D. Ambrosino, and C. J. Baker. 2001. Effects of alum adjuvant or a booster dose on immunogenicity during clinical trials of group B streptococcal type III conjugate vaccines. Infect. Immun. 69:6696-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puopolo, K. M., and L. C. Madoff. 2007. Type IV neonatal early-onset group B streptococcal disease in a United States hospital. J. Clin. Microbiol. 45:1360-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramaswamy, S. V., P. Ferrieri, A. E. Flores, and L. C. Paoletti. 2006. Molecular characterization of nontypeable group B streptococcus. J. Clin. Microbiol. 44:2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rolland, K., C. Marois, V. Siquier, B. Cattier, and R. Quentin. 1999. Genetic features of Streptococcus agalactiae strains causing severe neonatal infections, as revealed by pulsed-field gel electrophoresis and hylB gene analysis. J. Clin. Microbiol. 37:1892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savoia, D., C. Gottimer, C. Crocilla, and M. Zucca. 2008. Streptococcus agalactiae in pregnant women: phenotypic and genotypic characters. J. Infect. 56:120-125. [DOI] [PubMed] [Google Scholar]
- 43.Shet, A., and P. Ferrieri. 2004. Neonatal and maternal group B streptococcal infections: a comprehensive review. Indian J. Med. Res. 120:141-150. [PubMed] [Google Scholar]
- 44.Skjaervold, N. K., K. Bergh, and L. Bevanger. 2004. Distribution of PFGE types of invasive Norwegian group B streptococci in relation to serotypes. Indian J. Med. Res. 119:201-204. [PubMed] [Google Scholar]
- 45.Skoff, T. H., M. M. Farley, S. Petit, A. S. Craig, W. Schaffner, K. Gershman, L. H. Harrison, R. Lynfield, J. Mohle-Boetani, S. Zansky, B. A. Albanese, K. Stefonek, E. R. Zell, D. Jackson, T. Thompson, and S. J. Schrag. 2009. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990-2007. Clin. Infect. Dis. 49:85-92. [DOI] [PubMed] [Google Scholar]
- 46.Slotved, H.-C., F. Kong, L. Lambertsen, S. Sauer, and G. L. Gilbert. 2007. Serotype IX, a proposed new Streptococcus agalactiae serotype. J. Clin. Microbiol. 45:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, B. L., A. Flores, J. Dechaine, J. Krepela, A. Bergdall, and P. Ferrieri. 2004. Gene encoding the group B streptococcal protein R4, its presence in clinical reference laboratory isolates & R4 protein pepsin sensitivity. Indian J. Med. Res. 119:213-220. [PubMed] [Google Scholar]
- 48.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. DeBoy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. B. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urwin, R., and M. C. J. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 51.van der Mee-Marquet, N., L. Fourny, L. Arnault, A.-S. Domelier, M. Salloum, M.-F. Lartigue, and R. Quentin. 2008. Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J. Clin. Microbiol. 46:2906-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaleznik, D. F., M. A. Rench, S. Hillier, M. A. Krohn, R. Platt, M.-L. T. Lee, A. E. Flores, P. Ferrieri, and C. J. Baker. 2000. Invasive disease due to group B streptococcus in pregnant women and neonates from diverse population groups. Clin. Infect. Dis. 30:276-281. [DOI] [PubMed] [Google Scholar]