Abstract
The interaction of Nipah virus (NiV) nucleocapsid (N) protein with phosphoprotein (P) during nucleocapsid assembly is the essential process in the viral life cycle, since only the encapsidated RNA genome can be used for replication. To identify the region responsible for N-P interaction, we utilized fluorescent protein tags to visualize NiV N and P proteins in live cells and analyzed their cellular localization. N protein fused to monomeric enhanced cyan fluorescence protein (N-ECFP) exhibited a dotted pattern in transfected cells, while P protein fused to monomeric red fluorescent protein (P-mRFP) showed diffuse distribution. When the two proteins were coexpressed, P-mRFP colocalized with N-ECFP dots. N-ECFP mutants with serial amino acid deletions were generated to search for the region(s) responsible for this N-P colocalization. We found that, in addition to the 467- to 496-amino-acid (aa) region reported previously, aa 135 to 146 were responsible for the N-P colocalization. The residues crucial for N-P interaction were further investigated by introducing alanine substitutions into the untagged N protein. Alanine scanning in the region of aa 135 to 146 has revealed that there are distinct regions essential for the interaction of N-P and the function of N. This is the first study to visualize Nipah viral proteins in live cells and to assess the essential domain of N protein for the interaction with P protein.
Nipah virus (NiV) belongs to the genus Henipavirus within the family Paramyxoviridae. It is an emerging zoonotic agent and has been associated with outbreaks of human illness in Malaysia, Singapore, Bangladesh, and India in the 1990s and 2000s (5, 6, 10, 21, 26, 34, 35). NiV has an unusually wide host tropism, with serological evidence of infection in humans, pigs, dogs, cats, horses, and goats (9). Such a broad host range, along with its high mortality rate in humans and lack of antiviral therapy, has led to NiV being classified as a biosafety level 4 agent.
Like the other members of the order Mononegavirales, NiV is an enveloped virus with a negative-sense single-stranded RNA genome. The genome contains six transcriptional units that encode the viral structural proteins nucleocapsid (N), phosphoprotein (P), matrix (M), fusion (F), attachment (G), and RNA-dependent RNA polymerase (L) proteins and nonstructural proteins (V, W and C), in the order 3′-N-P/V/W/C-M-F-G-L-5′. The viral structural proteins all have essential roles throughout the viral life cycle. Of these, the viral core or nucleocapsid that consists of N, P, and L proteins contains all factors necessary for viral transcription and genome replication.
The interaction of N and P proteins is important for the efficient encapsidation of viral RNA in Mononegavirales. In the absence of viral RNA, N binds cellular RNA and no longer is available for involvement in viral replication. To avoid this, P protein binds to N shortly after synthesis and maintains it in a soluble form while enhancing the specificity of N for viral RNA rather than nonspecific cellular RNA (8, 12, 13, 15, 17, 25, 32, 36, 37, 46, 50, 52). Since only the nucleocapsid-coated RNA genome can become the template for viral replication, such interaction is critical for the viral life cycle.
For many paramyxoviruses, domains responsible for N-P interaction have been identified (1, 4, 14, 18, 22, 23, 27-29, 31, 32, 38-42, 44, 45, 48, 52). For NiV, Chan et al. have carried out a protein-blotting protein overlay assay and designated amino acids (aa) 467 to 496 as the core region that binds to the P protein (7). They also have found that NiV N protein can interact with the P protein of Hendra virus, another member of the Henipavirus genus, and vice versa, which indicates the close relationship between the two viruses. Meanwhile, the authors have pointed out that at least one other binding domain should exist in the non-C-terminal region of N protein.
In the present study, we utilized fluorescence-tagged NiV N and P proteins to examine their interaction in live cells. The two proteins merged and exhibited a dotted pattern upon cotransfection. Serial deletion was introduced into the N coding region, and by analyzing their cellular distribution, a region that was indispensable for the dotted distribution was identified. The critical residues for N-P interaction and its function also were investigated.
MATERIALS AND METHODS
Cell culture.
Cos7, Vero, and BHK-T7 cells were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 5% fetal calf serum (FCS), 50 U/ml penicillin, and 50 μg/ml streptomycin (Invitrogen).
Cell lysates.
For immunoprecipitation, transfected and 35S-labeled BHK-T7 cells were washed once with phosphate-buffered saline (PBS), and lysis buffer (5 mM EDTA and 0.5% [wt/vol] Triton X-100 in PBS supplemented with Complete Mini [Roche]) was added. The cells were transferred to 1.5-ml tubes and rotated overnight at 4°C for protein extraction. After centrifugation at 15,000 rpm for 30 min, supernatants were collected and stored at −30°C.
For reporter gene assays, cells were washed once with PBS and passive lysis buffer (Promega) was added. The samples were subjected to one freeze-thaw cycle and incubated at room temperature for 2 h with vigorous shaking.
Plasmid construction.
Monomeric enhanced cyan fluorescence protein (ECFP-A206K; ECFP) and monomeric red fluorescence protein (mRFP) plasmids were kindly provided by Y. Kawaguchi (47) and R. Tsien, respectively.
For the generation of fluorescence-tagged NiV N and P proteins, full open reading frames (ORFs) of N, P, ECFP, and mRFP were amplified by PCR. ECFP and mRFP ORFs were joined in-frame to the C termini of the N and P ORFs, respectively, to generate N-ECFP and mRFP fusion genes and were cloned into a modified pCAGGS vector that contained a unique NotI site at the multiple-cloning site. The resulting pN-ECFP and pP-mRFP plasmid DNAs were purified using the Wizard plasmid purification kit (Promega) and Genopure plasmid midi kit (Roche) according to the manufacturers' instructions. The obtained fusion genes were verified by sequencing using an ABI Prism 3130 genetic analyzer (Applied Biosystems).
Mutagenesis.
To introduce deletion and alanine substitutions, KOD plus (Toyobo) was used with the inverse PCR method according to the manufacturer's instructions. The constructed deletion mutants were NmutA (aa 1 to 109 deleted), NmutB (aa 110 to 146 deleted), NmutC (aa 147 to 253 deleted), NmutD (aa 254 to 360 deleted), NmutE (aa 361 to 466 deleted), NmutF (aa 497 to 532 deleted), and Nmut1 (aa 467 to 496 deleted). NmutB-ECFP was generated by taking advantage of the two in-frame HincII restriction sites; the plasmid was digested with HincII and self ligated. The mutations were verified by sequencing. Alanine substitution mutations were introduced to aa 135 to 146, four residues at a time, with a 2-aa overlap. The sequences of the primers will be provided upon request.
Transfection of plasmid DNA.
Cells were plated onto gelatin-coated 35-mm glass-bottom dishes (Matsunami) or six-well plates (Corning) 1 day prior to transfection. Expression vector pN-ECFP and its mutants (1.25 μg/dish or well) and pP-mRFP (0.8 μg/dish or well) were transfected using Opti-MEM (Invitrogen) and FuGene6 reagent (Roche) or Lipofectamine LTX (Invitrogen) according to the manufacturers' instructions.
Quantification of N-P speckle formation.
To evaluate the dot-like colocalization of the viral proteins, cells with speckles that consisted of N protein fused with ECFP (N-ECFP) and P protein fused with mRFP (P-mRFP) were counted 48 h after transfection by fluorescence microscopy. The percentage of cells with N-ECFP speckles was determined as the number of the cells with visible dots divided by the total number of ECFP- and mRFP-positive cells counted. Fluorescence images were captured with Biozero (Keyence) or a IX70 laser confocal microscope and the Fluoview FV500 system (Olympus).
Antibodies, immunoprecipitation, and immunofluorescence assay (IFA).
For the generation of polyclonal anti-NiV P antibody, aa 249 to 709 of P protein were expressed in Escherichia coli as a glutathione S transferase (GST) fusion protein, and the GST-purified P protein fragment was used as an antigen to immunize the rabbits. The animal experiment was approved by the animal ethics committee in our institute and conducted according to the guidelines for animal experiments of the University of Tokyo.
For immunoprecipitation analysis, six-well cultured BHK-T7 cells were transfected with the plasmids, and 24 h after transfection the cells were labeled with Express 35S-protein labeling mix (Perkin Elmer) for an additional 16 h. The cells were harvested, and the lysates were precleared using protein A-Sepharose at room temperature for 30 min. One microliter of anti-NiV P polyclonal antibody and protein A-Sepharose was added to each precleared sample and incubated at 4°C overnight. The immunoprecipitates absorbed to protein A-Sepharose were washed five times with PBS and suspended in SDS-PAGE sample buffer, boiled for 10 min, and subjected to SDS-PAGE. After electrophoresis, the gel was dried and autoradiographed with an FLA-5100 imaging system (Fujifilm).
IFA was performed as described previously (43). In brief, the transfected cells were fixed with 3% paraformaldehyde (PFA), washed with PBS, and permeabilized with 0.2% Triton X-100-PBS. Anti-NiV N and anti-NiV P antibodies were used as the primary antibodies (1:500 dilution). For secondary antibody, Alexa Fluor 488-conjugated goat anti-rabbit IgG was used (1:500 dilution; Invitrogen). Nuclei were counterstained with Hoechst 33342 (Cambrex). For labeling anti-NiV N and anti-NiV P antibodies, a Zenon rabbit IgG labeling kit (Invitrogen) was used. One μg of anti-NiV N and anti-NiV P antibody was labeled with Zenon Alexa Fluor 488 and Zenon Alexa Flour 555 rabbit IgG labeling reagents, respectively, according to the manufacturer's information.
Minigenome replication assay.
The NiV minigenome encoding luciferase was constructed as described previously (20), with a modification that the firefly luciferase gene was used instead of the chloramphenicol acetyltransferase (CAT) gene. The resulting minigenome fragment was sequenced and cloned into EagI-BsmI sites of pMDB1 (2). One day prior to transfection, BHK-T7 cells were seeded at 1 × 105 cells/well onto 24-well plates (Corning). The cells were transfected with 0.5 μg/well of minigenome plasmid and T7-driven NiV N or its mutants (0.6 μg/well), P (0.4 μg/well), and L (0.2 μg/well) using Lipofectamine LTX reagent (Invitrogen). Twenty- four h posttransfection, the cells were lysed and subjected to a reporter gene assay using Pickagene reagent (Toyo Ink) and a MiniLumat LB9506 luminometer (Berthold) according to the manufacturers' instructions.
RESULTS
Fluorescent protein-tagged N and P proteins colocalize in live cells.
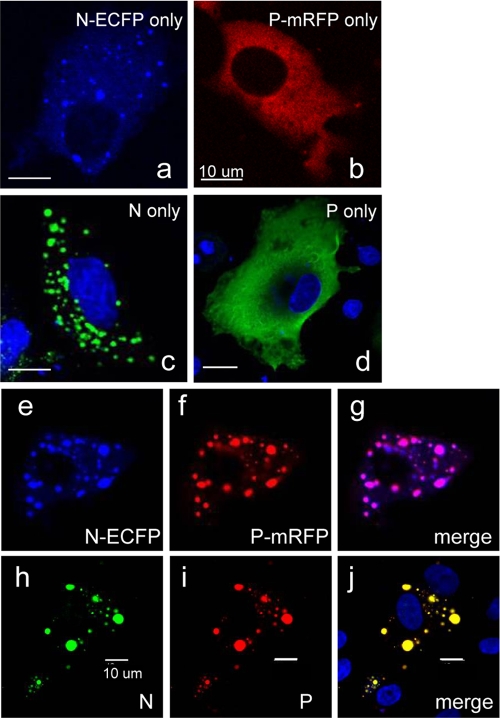
To visualize NiV N and P proteins in live cells, ECFP and mRFP were fused to the C termini of N and P proteins, respectively. ECFP contained an amino acid substitution at position 206 (A206K) that inhibits dimerization (47). When these two proteins were individually expressed under CAG promoter control in BHK cells, they exhibited distinct cellular localization: N-ECFP formed speckles or dot-like localization in the cytoplasm, and P-mRFP showed diffuse distribution throughout the cytoplasm (Fig. 1 a and b). The untagged N and P proteins also showed similar appearances in IFA (Fig. 1c and d), proving that the fluorescence tags did not affect the localization pattern of N and P proteins. The dot-like localization of N protein was also observed in other cell lines, including Cos7, Vero, and 293 cells (data not shown), which is in accordance with previous studies of other members of the Mononegavirales (3, 19). ECFP alone has diffused throughout the cell (data not shown), indicating that N protein itself confers such a distinct localization. When P-mRFP was coexpressed with N-ECFP, it merged completely into N-ECFP speckles (Fig. 1e to g), which indicates that P-mRFP colocalizes with N-ECFP. Again, this colocalization pattern was identical to that of the untagged N and P proteins visualized with IFA (Fig. 1h to j), further confirming that the observed fluorescence pattern reflects the appearance of untagged proteins. No substantial change in the formation or localization of N-ECFP speckles has occurred upon P-mRFP cotransfection.
FIG. 1.
ECFP- and mRFP-tagged NiV proteins exhibit intracellular localization similar to that of untagged proteins. N-ECFP and P-mRFP expression vectors or the untagged N and P expression vectors were transfected to BHK cells. For panels a to d, the cells were transfected with the expression vectors indicated. For panels e to j, the cells were transfected with both N and P expression vectors. Twenty-four h after transfection, the cells were subjected to fluorescence laser-scanning microscopy (a, b, and e to g) or fixed and stained with anti-NiV N (c and h) or anti-NiV P (d and i) antibody. The fixed cells were counterstained with Hoechst 33342 (shown in blue). Panels g and j show the merged images of panels e and f as well as h and i, respectively, indicating that P (-mRFP) completely merges with the dot-like N (-ECFP) fluorescence.
Determination of P-interacting domain in N protein.
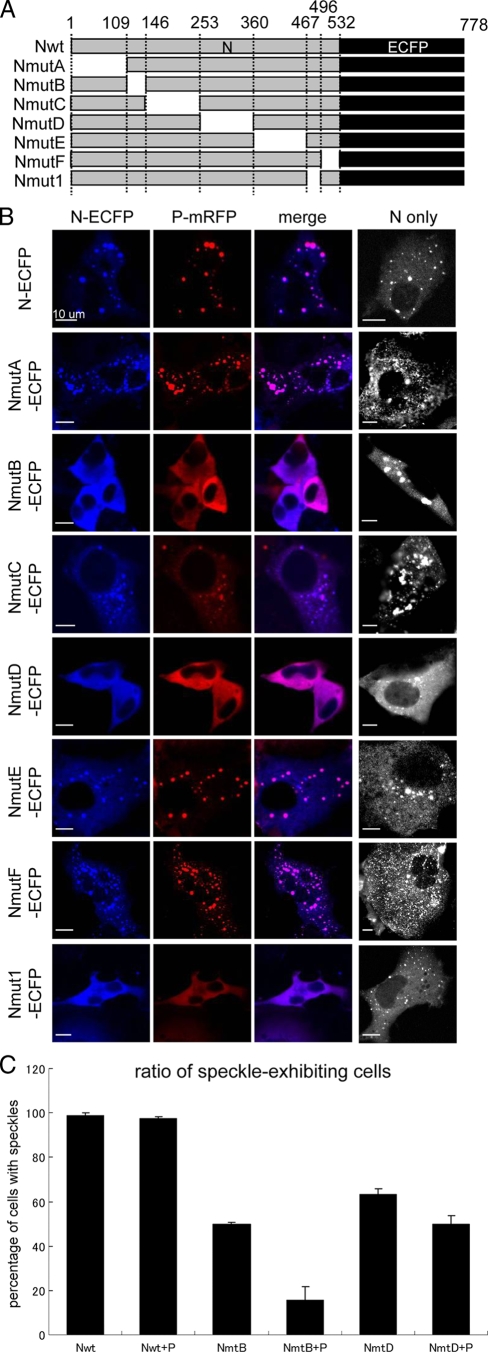
To elucidate the regions in N protein that are required for N-P colocalization in live cells, we constructed serial N-ECFP deletion mutants as shown in Fig. 2 A and coexpressed them with P-mRFP. Nmut1-ECFP, which lacked the region reported by Chan et al. (7), was tested as a control. Cells that expressed Nmut1-ECFP exhibited diffuse distribution with little N-P speckle formation (Fig. 2B). This indicated the loss of N-P interaction and was in accordance with the previous report (7). Similarly, cells that expressed NmutB-ECFP or NmutD-ECFP have dominantly exhibited diffuse distribution patterns (Fig. 2B). Meanwhile, most of the cells that express NmutA-ECFP, NmutC-ECFP, NmutE-ECFP, or NmutF-ECFP, as well as N-ECFP, showed dotted colocalization patterns with P-mRFP (Fig. 2B), suggesting that these regions are dispensable for N-P speckle formation.
FIG. 2.
Effect of deletion mutation of N in N-P colocalization. (A) Schematic representation of the mutation constructs used in this study. The shaded box represents the region expressed in each clone, with the name of the construct shown on its left and the amino acid position indicated at the top. The dark box represents ECFP fused to the C terminus of the N protein. Nwt, full-length N protein. (B) The effect of each deletion mutation on N-P speckle formation. Each mutated N-ECFP was coexpressed with P-mRFP in BHK cells, and the fluorescence was observed at 48 h posttransfection. The rightmost column shows the localization pattern of mutated N-ECFP proteins without the presence of P-mRFP. (C) The ratio of speckled to diffuse distribution patterns in N-P colocalization. BHK cells transfected with the expression vector Nwt, NmutB, or NmutD with or without P-mRFP was counted for the presence of speckle-expressing cells. After 48 h, cells expressing N (and P) were examined for the presence of speckles, and the percentage of cells with N (−P) speckles are shown. Approximately 100 cells were counted at a time; the counting was performed three times on different fields, and the averages are shown. The total number of cells counted was set to 100%.
The loss of the N-P speckles in NmutB-ECFP and NmutD-ECFP may have resulted from the inhibition of N-P colocalization or the disruption of N-N interaction, since N-ECFP itself formed speckles when expressed alone (Fig. 1). Thus, the number of cells that exhibited dotted and diffuse N localization patterns was counted. As shown in Fig. 2C, the deletion of regions B and D similarly caused a decrease in the number of speckle-expressing cells when expressed alone, but the deletion of region B significantly reduced the number of cells exhibiting N-P speckles when P-mRFP was coexpressed (Fig. 2C).
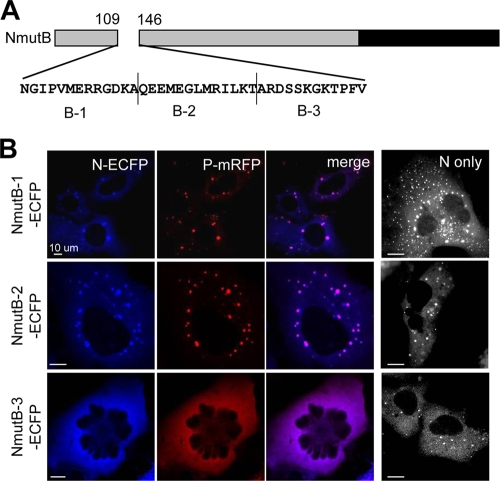
Mutant B contained a deletion that spanned 36 residues, thus we divided region B further into three regions (designated B-1, B-2, and B-3) (Fig. 3A), and the cellular distribution of each deletion mutant was analyzed. As shown in Fig. 3B, mutant B-3 showed a substantial loss of N-P dotted colocalization. This indicated that the 12 aa in region B-3 were important for the dotted N-P colocalization.
FIG. 3.
Region responsible for N-P speckle formation. (A) Schematic representation and amino acid sequence of region B. The region was divided into three 12-aa domains named B-1, B-2, and B-3. (B) Subcellular distribution of NmutB-1, NmutB-2, and NmutB-3. Each mutant was coexpressed with P-mRFP, and their subcellular localization was observed. The rightmost column shows the cells expressing the N mutants only.
Alanine scanning.
To map more precisely the residues responsible for N-P interaction, alanine substitution was introduced into the residues corresponding to the B-3 region in untagged N protein, as shown in Fig. 4A, and were designated sAla1 to sAla5. BHK cells were transfected with the sAla mutants and wild-type P protein, and immunoprecipitation was performed using anti-NiV P antibody. All mutants except for sAla5 have been coimmunoprecipitated with P proteins, indicating that the residues substituted in sAla5 are critical for N-P interaction (Fig. 4B). The same tendency has been observed in IFA using the same expression vectors; that is, the sAla5 mutant has lost the dotted N-P colocalization (Fig. 5). Neither precipitated when no antibody was added, suggesting that the protein precipitation was due to anti-NiV P antibody activity. These results suggest that the loss of N-P colocalization reflects the loss of N-P interaction.
FIG. 4.
Alanine scanning. (A) Schematic representation of the mutants with alanine substitution. The B-3 region was further divided into five overlapping amino acid groups, each consisting of four alanine substitutions. Since the first residue of B-3 was A, the sAla1 mutant had only three substitutions compared to the intact amino acid sequence. (B) Coimmunoprecipitation of N and P proteins. BHK-T7 cells were transfected with sAla mutants and P protein and labeled with 35S. The lysates were subjected to immunoprecipitation using anti-NiV P antibody, and the presence of N mutants in the coimmunoprecipitates was verified by SDS-PAGE followed by autoradiography. no Ab, no anti-P antibody has been added during immunoprecipitation.
FIG. 5.
IFA of N and P proteins in minigenome assay conditions. IFA performed on BHK cells transfected with sAla mutants and P expression vectors, along with the L expression vector and minigenome plasmid. Only the sAla5 mutant exhibited diffuse distribution.
Minigenome assay.
To examine the possible correlation between N-P interaction and function, we performed minigenome assays. BHK-T7 cells were transfected with the NiV minigenome plasmid encoding luciferase, the T7-driven N or sAla mutants, and P and L expression plasmids. Luciferase was expressed when the minigenome RNA was properly encapsidated by N protein and used for transcription by the transcriptase complex that consisted of P and L proteins. As shown in Fig. 6, left, sAla5 has adversely affected minigenome replication. The untagged Nmut1 protein also failed to function in minigenome assays (Fig. 6, right), suggesting that the loss of N-P interaction abolished the function of N protein in minigenome replication. These results suggest that the N-P interaction is important in the viral life cycle. However, the functional impairment of sAla1, sAla3, and sAla4 mutants was observed, implying that these regions also are involved in the proper functioning of N protein.
FIG. 6.
Minigenome assay. BHK-T7 cells that expressed T7 polymerase were transfected as described for Fig. 5, and 24 h after transfection, the cells were lysed and subjected to luciferase assay. Nmut1 also was used as the negative control. Rightmost lanes (−N) show control transfectants from which N expression plasmid was omitted. The result was expressed as the relative luciferase activity (± standard deviations) (−N = 1) of triplicate samples.
DISCUSSION
Viral infection and replication are dynamic processes. The conventional methods using nonnative samples or fixed and permeabilized cells cannot provide the whole view of the natural replication cycle in cells. In the present study, the use of fluorescent tags enabled us to observe the behavior of the viral proteins in live cells and to screen efficiently for the novel N-P binding region. Previously, Chan et al. have suggested that, in addition to the 29-aa C-terminal region that they have identified, an unidentified P-interacting site exists in the non-C-terminal region of N (7). The protein-blotting protein overlay assay used by Chan et al. is for the analysis of nonnative conditioned proteins, and it is impossible to detect conformation-dependent interactions. In this study, we have identified the additional 12-aa region, which is relatively closer to the N terminal of the protein, by observing the speckles formed by fluorescence-tagged viral proteins in live cells and alanine scanning combined with IFA. The result was confirmed by coimmunoprecipitation, and the importance of the region in the proper functioning of N protein was further demonstrated by minigenome assay.
NiV N is known to oligomerize to form a herringbone-like nucleocapsid structure by itself (33), and in line with this, our study showed that wild-type NiV N or N-ECFP protein formed speckles by itself, likely representing N-N oligomerization (Fig. 1). Since the appearance of the speckle was not changed significantly by the cotransfection of NiV P protein, the speckles formed by N alone likely represent the nucleocapsids. NiV P and P-mRFP diffused throughout the cytoplasm when expressed alone, but they merged and formed N-P speckle-like colocalization upon cotransfection with NiV N (Fig. 1). It is likely that P protein stabilizes N protein as it does in the other family members of the Mononegavirales. In vesicular stomatitis virus, P protein is known to interact with N to maintain it in a soluble form (25). In measles virus, P protein acts as a chaperone to enhance the specificity of N for viral RNA (46). During actual NiV infection, NiV P protein may bind to N protein shortly after synthesis to stabilize and sequester the proteins from the unrelated cellular RNA.
Throughout our study, untagged wild-type NiV N and N-ECFP proteins always localized in the cytoplasm of all the cells examined. The cytoplasmic localization of NiV N and P proteins is in accordance with most other paramyxoviruses that replicate in the cytoplasm, except for morbilliviruses. Although henipavirus is most closely related to morbilliviruses, whose N proteins are known to contain an nuclear localization signal (NLS) (43) and whose P proteins retain them to the cytoplasm (27), whether NiV N protein has an NLS and the potential to travel to the nucleus or whether NiV P protein retains N protein to the cytoplasm is unknown.
In the present study, the cells expressing N-ECFP and its mutants, other than NmutB-ECFP and NmutD-ECFP, exhibited N-P speckles in various sizes and shapes (Fig. 1 and 2B). The significance of such variation in N-P speckles is unknown, but since untagged N protein also shows N-P speckles in various sizes, the same phenomenon may occur in the actual viral replication cycle.
The cells expressing NmutD-ECFP showed increased diffuse N-protein distribution to an extent similar to that of both single infection and cotransfection (Fig. 2B and C). The mechanism of the loss of N speckles in cells that expressed NmutD-ECFP was not clarified in our study, but it is likely that the conformational change induced by the deletion leads to the disruption of the N-N interaction and herringbone-like structure formation. Recently, it has been suggested that aa 322 to 342 are involved in NiV N capsid assembly (33), which is included in the deleted region in NmutD-ECFP. Meanwhile, the speckle formation in NmutB-ECFP was more strongly affected when expressed with P-mRFP than in NmutD-ECFP (Fig. 3). Why the speckles disappeared in NmutB is not clarified, but a possibility is that P and the mutant N proteins interact in aberrant forms with lower affinity, and such aberrant interaction results in the resolving of the speckles owing to the chaperone activity of P protein as seen in other paramyxoviruses (12, 15).
We have identified the critical residues required for the efficient interaction of N and P proteins by IFA and coimmunoprecipitation using mutant N proteins with alanine substitution in the region lacked by NmutB-ECFP (Fig. 4 and 5). All mutants except for sAla5 formed N-P speckles and coimmunoprecipitated with anti-P antibody, suggesting that N-P speckle formation is correlated with N-P interaction. Therefore, aa 145 and 146 were considered indispensable for the N-P colocalization and interaction. We also revealed that the region identified was critical for minigenome replication (Fig. 6). The minigenome system is useful for studying the roles of individual viral proteins in replication, especially when working with high levels of virus containment, such as with NiV. So far, two minigenome systems exist for NiV (20, 30), which utilize CAT and GFP. In our study, a modified minigenome that encoded firefly luciferase was generated, which is known to be more sensitive than CAT (49) and is easy to quantify. The minigenome introduced in the cells should first be encapsidated by N-P complex in order to be replicated and translated. While sAla1 mutation had a somewhat milder effect, sAla3, sAla4, and sAla5 as well as sNmut1 almost nullified the ability of N protein to take part in the replication of the minigenome, implying the functional significance of the regions (Fig. 6). Interestingly, sAla3 and sAla4, which had no effect on N-P interaction as observed in fluorescence microscopy and coimmunoprecipitation, significantly inhibited minigenome replication. This indicates another essential role for the sAla3 and sAla4 region in the function of N. Taken together, these results suggest that the N-P interaction is mandatory for N to function properly, but the interaction alone cannot account for the function of N. In other paramyxoviruses, it is known that once N binds to viral RNA, it dissociates from P, and P in turn binds to other free N proteins or polymerase L protein to form an RNA polymerase complex (11, 12, 16, 23, 24). Thus, for N to function in viral replication and transcription, N must sequentially interact with P, genome RNA, and P-L polymerase complex. Since N protein has such a variety of interacting partners in each viral replication step, the loss of its function in sAla3 and sAla4 mutants may have resulted from a mechanism distinct from the N-P interaction.
The B-3 region is strongly conserved in Hendra virus, the other member of Henipavirus. However, amino acid sequence comparison using ClustalW has revealed that the overall sequence of the region is not common to the other paramyxoviruses (data not shown). Thus, the region is unique to the Henipavirus genus. The most conserved residues were the latter four residues (TPFV) corresponding to the Ala-5 region, which were shared by Newcastle disease virus and simian virus 5, implying the importance of the residues. Otherwise, the region contains one of the least conserved residues in nucleocapsid protein among paramyxoviruses.
This is the first study to visualize NiV N-P intracellular colocalization in live cells. N-P colocalization should be the essential step for the proper functioning of N in viral replication, since mutant N proteins without the ability to form N-P complex cannot function in minigenome assays. The method to monitor fluorescence-tagged viral proteins in live cells is useful for the analysis of viral protein interaction and function and for live cell tracking of the viral infection process (47). Further studies using such fluorescence-tagged mutant proteins and/or recombinant NiV generated by our established reverse genetics (51) should shed light on the steps of the viral replication cycle and the behavior of viral proteins in live cells.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research and by a Global COE Program Center of Education and Research for Advanced Genome-Based Medicine (for personalized medicine and the control of worldwide infectious diseases), Ministry of Education, Science, Sports and Culture (MEXT), Japan.
Footnotes
Published ahead of print on 28 July 2010.
REFERENCES
- 1.Bankamp, B., S. M. Horikami, P. D. Thompson, M. Huber, M. Billeter, and S. A. Moyer. 1996. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216:272-277. [DOI] [PubMed] [Google Scholar]
- 2.Baron, M. D., and T. Barrett. 1997. Rescue of rinderpest virus from cloned cDNA. J. Virol. 71:1265-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, S., C. Rinne, U. Hofsass, H. D. Klenk, and E. Muhlberger. 1998. Interactions of Marburg virus nucleocapsid proteins. Virology 249:406-417. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz, C. J., C. Retzler, H. E. Homann, and W. J. Neubert. 1994. The carboxy-terminal domain of Sendai virus nucleocapsid protein is involved in complex formation between phosphoprotein and nucleocapsid-like particles. Virology 204:770-776. [DOI] [PubMed] [Google Scholar]
- 5.Butler, D. 2004. Fatal fruit bat virus sparks epidemics in southern Asia. Nature 429:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chadha, M. S., J. A. Comer, L. Lowe, P. A. Rota, P. E. Rollin, W. J. Bellini, T. G. Ksiazek, and A. Mishra. 2006. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 12:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, Y. P., C. L. Koh, S. K. Lam, and L. F. Wang. 2004. Mapping of domains responsible for nucleocapsid protein-phosphoprotein interaction of henipaviruses. J. Gen. Virol. 85:1675-1684. [DOI] [PubMed] [Google Scholar]
- 8.Chenik, M., K. Chebli, Y. Gaudin, and D. Blondel. 1994. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J. Gen. Virol. 75:2889-2896. [DOI] [PubMed] [Google Scholar]
- 9.Chua, K. B. 2003. Nipah virus outbreak in Malaysia. J. Clin. Virol. 26:265-275. [DOI] [PubMed] [Google Scholar]
- 10.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 11.Curran, J., H. Homann, C. Buchholz, S. Rochat, W. Neubert, and D. Kolakofsky. 1993. The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J. Virol. 67:4358-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curran, J., J. B. Marq, and D. Kolakofsky. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, T., and A. K. Banerjee. 1992. Role of the phosphoprotein (P) in the encapsidation of presynthesized and de novo synthesized vesicular stomatitis virus RNA by the nucleocapsid protein (N) in vitro. Cell Mol. Biol. 38:17-26. [PubMed] [Google Scholar]
- 14.De, B. P., M. A. Hoffman, S. Choudhary, C. C. Huntley, and A. K. Banerjee. 2000. Role of NH(2)- and COOH-terminal domains of the P protein of human parainfluenza virus type 3 in transcription and replication. J. Virol. 74:5886-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Errington, W., and P. T. Emmerson. 1997. Assembly of recombinant Newcastle disease virus nucleocapsid protein into nucleocapsid-like structures is inhibited by the phosphoprotein. J. Gen. Virol. 78:2335-2339. [DOI] [PubMed] [Google Scholar]
- 16.Fooks, A. R., J. R. Stephenson, A. Warnes, A. B. Dowsett, B. K. Rima, and G. W. Wilkinson. 1993. Measles virus nucleocapsid protein expressed in insect cells assembles into nucleocapsid-like structures. J. Gen. Virol. 74:1439-1444. [DOI] [PubMed] [Google Scholar]
- 17.Garcia, J., B. Garcia-Barreno, A. Vivo, and J. A. Melero. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243-247. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Barreno, B., T. Delgado, and J. A. Melero. 1996. Identification of protein regions involved in the interaction of human respiratory syncytial virus phosphoprotein and nucleoprotein: significance for nucleocapsid assembly and formation of cytoplasmic inclusions. J. Virol. 70:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groseth, A., J. E. Charton, M. Sauerborn, F. Feldmann, S. M. Jones, T. Hoenen, and H. Feldmann. 2009. The Ebola virus ribonucleoprotein complex: a novel VP30-L interaction identified. Virus Res. 140:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halpin, K., B. Bankamp, B. H. Harcourt, W. J. Bellini, and P. A. Rota. 2004. Nipah virus conforms to the rule of six in a minigenome replication assay. J. Gen. Virol. 85:701-707. [DOI] [PubMed] [Google Scholar]
- 21.Halpin, K., P. L. Young, H. Field, and J. S. Mackenzie. 1999. Newly discovered viruses of flying foxes. Vet. Microbiol. 68:83-87. [DOI] [PubMed] [Google Scholar]
- 22.Harty, R. N., and P. Palese. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76:2863-2867. [DOI] [PubMed] [Google Scholar]
- 23.Homann, H. E., W. Willenbrink, C. J. Buchholz, and W. J. Neubert. 1991. Sendai virus protein-protein interactions studied by a protein-blotting protein-overlay technique: mapping of domains on NP protein required for binding to P protein. J. Virol. 65:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howard, M., and G. Wertz. 1989. Vesicular stomatitis virus RNA replication: a role for the NS protein. J. Gen. Virol. 70:2683-2694. [DOI] [PubMed] [Google Scholar]
- 26.Hsu, V. P., M. J. Hossain, U. D. Parashar, M. M. Ali, T. G. Ksiazek, I. Kuzmin, M. Niezgoda, C. Rupprecht, J. Bresee, and R. F. Breiman. 2004. Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 10:2082-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber, M., R. Cattaneo, P. Spielhofer, C. Orvell, E. Norrby, M. Messerli, J. C. Perriard, and M. A. Billeter. 1991. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology 185:299-308. [DOI] [PubMed] [Google Scholar]
- 28.Liston, P., R. Batal, C. DiFlumeri, and D. J. Briedis. 1997. Protein interaction domains of the measles virus nucleocapsid protein (NP). Arch. Virol. 142:305-321. [DOI] [PubMed] [Google Scholar]
- 29.Lu, B., R. Brazas, C. H. Ma, T. Kristoff, X. Cheng, and H. Jin. 2002. Identification of temperature-sensitive mutations in the phosphoprotein of respiratory syncytial virus that are likely involved in its interaction with the nucleoprotein. J. Virol. 76:2871-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magoffin, D. E., K. Halpin, P. A. Rota, and L. F. Wang. 2007. Effects of single amino acid substitutions at the E residue in the conserved GDNE motif of the Nipah virus polymerase (L) protein. Arch. Virol. 152:827-832. [DOI] [PubMed] [Google Scholar]
- 31.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, S. Kusagawa, H. Komada, and Y. Ito. 1999. Mapping of domains on the human parainfluenza virus type 2 nucleocapsid protein (NP) required for NP-phosphoprotein or NP-NP interaction. J. Gen. Virol. 80:2017-2022. [DOI] [PubMed] [Google Scholar]
- 32.Nishio, M., M. Tsurudome, M. Kawano, N. Watanabe, S. Ohgimoto, M. Ito, H. Komada, and Y. Ito. 1996. Interaction between nucleocapsid protein (NP) and phosphoprotein (P) of human parainfluenza virus type 2: one of the two NP binding sites on P is essential for granule formation. J. Gen. Virol. 77:2457-2463. [DOI] [PubMed] [Google Scholar]
- 33.Ong, S. T., K. Yusoff, C. L. Kho, J. O. Abdullah, and W. S. Tan. 2009. Mutagenesis of the nucleocapsid protein of Nipah virus involved in capsid assembly. J. Gen. Virol. 90:392-397. [DOI] [PubMed] [Google Scholar]
- 34.Parashar, U. D., L. M. Sunn, F. Ong, A. W. Mounts, M. T. Arif, T. G. Ksiazek, M. A. Kamaluddin, A. N. Mustafa, H. Kaur, L. M. Ding, G. Othman, H. M. Radzi, P. T. Kitsutani, P. C. Stockton, J. Arokiasamy, H. E. Gary, Jr., and L. J. Anderson. 2000. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998-1999 outbreak of severe encephalitis in Malaysia. J. Infect. Dis. 181:1755-1759. [DOI] [PubMed] [Google Scholar]
- 35.Paton, N. I., Y. S. Leo, S. R. Zaki, A. P. Auchus, K. E. Lee, A. E. Ling, S. K. Chew, B. Ang, P. E. Rollin, T. Umapathi, I. Sng, C. C. Lee, E. Lim, and T. G. Ksiazek. 1999. Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet 354:1253-1256. [DOI] [PubMed] [Google Scholar]
- 36.Peluso, R. W., and S. A. Moyer. 1988. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology 162:369-376. [DOI] [PubMed] [Google Scholar]
- 37.Precious, B., D. F. Young, A. Bermingham, R. Fearns, M. Ryan, and R. E. Randall. 1995. Inducible expression of the P, V, and NP genes of the paramyxovirus simian virus 5 in cell lines and an examination of NP-P and NP-V interactions. J. Virol. 69:8001-8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randall, R. E., and A. Bermingham. 1996. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology 224:121-129. [DOI] [PubMed] [Google Scholar]
- 39.Ryan, K. W., and D. W. Kingsbury. 1988. Carboxyl-terminal region of Sendai virus P protein is required for binding to viral nucleocapsids. Virology 167:106-112. [DOI] [PubMed] [Google Scholar]
- 40.Ryan, K. W., E. M. Morgan, and A. Portner. 1991. Two noncontiguous regions of Sendai virus P protein combine to form a single nucleocapsid binding domain. Virology 180:126-134. [DOI] [PubMed] [Google Scholar]
- 41.Ryan, K. W., and A. Portner. 1990. Separate domains of Sendai virus P protein are required for binding to viral nucleocapsids. Virology 174:515-521. [DOI] [PubMed] [Google Scholar]
- 42.Ryan, K. W., A. Portner, and K. G. Murti. 1993. Antibodies to paramyxovirus nucleoproteins define regions important for immunogenicity and nucleocapsid assembly. Virology 193:376-384. [DOI] [PubMed] [Google Scholar]
- 43.Sato, H., M. Masuda, R. Miura, M. Yoneda, and C. Kai. 2006. Morbillivirus nucleoprotein possesses a novel nuclear localization signal and a CRM1-independent nuclear export signal. Virology 352:121-130. [DOI] [PubMed] [Google Scholar]
- 44.Shaji, D., and M. S. Shaila. 1999. Domains of Rinderpest virus phosphoprotein involved in interaction with itself and the nucleocapsid protein. Virology 258:415-424. [DOI] [PubMed] [Google Scholar]
- 45.Slack, M. S., and A. J. Easton. 1998. Characterization of the interaction of the human respiratory syncytial virus phosphoprotein and nucleocapsid protein using the two-hybrid system. Virus Res. 55:167-176. [DOI] [PubMed] [Google Scholar]
- 46.Spehner, D., R. Drillien, and P. M. Howley. 1997. The assembly of the measles virus nucleoprotein into nucleocapsid-like particles is modulated by the phosphoprotein. Virology 232:260-268. [DOI] [PubMed] [Google Scholar]
- 47.Sugimoto, K., M. Uema, H. Sagara, M. Tanaka, T. Sata, Y. Hashimoto, and Y. Kawaguchi. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82:5198-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuckis, J., S. Smallwood, J. A. Feller, and S. A. Moyer. 2002. The C-terminal 88 amino acids of the Sendai virus P protein have multiple functions separable by mutation. J. Virol. 76:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, T. M., J. E. Burlein, S. Ogden, L. J. Kricka, and J. A. Kant. 1989. Advantages of firefly luciferase as a reporter gene: application to the interleukin-2 gene promoter. Anal. Biochem. 176:28-32. [DOI] [PubMed] [Google Scholar]
- 50.Yang, J., D. C. Hooper, W. H. Wunner, H. Koprowski, B. Dietzschold, and Z. F. Fu. 1998. The specificity of rabies virus RNA encapsidation by nucleoprotein. Virology 242:107-117. [DOI] [PubMed] [Google Scholar]
- 51.Yoneda, M., V. Guillaume, F. Ikeda, Y. Sakuma, H. Sato, T. F. Wild, and C. Kai. 2006. Establishment of a Nipah virus rescue system. Proc. Natl. Acad. Sci. U. S. A. 103:16508-16513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao, H., and A. K. Banerjee. 1995. Interaction between the nucleocapsid protein and the phosphoprotein of human parainfluenza virus 3. Mapping of the interacting domains using a two-hybrid system. J. Biol. Chem. 270:12485-12490. [DOI] [PubMed] [Google Scholar]