Abstract
Murine gammaherpesvirus 68 (MHV-68) is closely related to Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) and provides a small-animal model with which to study the pathogenesis of gammaherpesvirus (γHV) infections. To completely explore the potential of the MHV-68 system for the investigation of γHV microRNAs (miRNAs), it would be desirable to know the number and expression patterns of all miRNAs encoded by MHV-68. By deep sequencing of small RNAs, we systematically investigated the expression profiles of MHV-68 miRNAs in both lytically and persistently infected cells. In addition to the nine known MHV-68 miRNAs, we identified six novel MHV-68 miRNA genes and analyzed the expression levels of all MHV-68 miRNAs. Furthermore, we also characterized the cellular miRNA expression signatures in MHV-68-infected versus noninfected NIH 3T3 fibroblasts and in 12-O-tetradecanoyl-phorbol-13-acetate (TPA)-treated versus nontreated S11 cells. We found that mmu-mir-15b and mmu-mir-16 are highly upregulated upon MHV-68 infection of NIH 3T3 cells, indicating a potential role for cellular miRNAs during MHV-68 infection. Our data will aid in the full exploration of the functions of γHV miRNAs.
Herpesviruses cause significant morbidity and mortality in the human population. The human gammaherpesviruses (γHV) Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) are associated with a number of tumors and lymphoproliferative disorders (37, 40).
The outcome of virus-host interactions is determined by many different factors. On the one hand, host immune responses play a pivotal role in the control of γHV infections and in pathogenesis. On the other hand, viral factors govern levels of infectivity, tropism, and immune evasion. Previous research on viral factors focused mainly on proteins encoded by viral genes. Recently, it was discovered that viruses, like the genomes of eukaryotic cells, also encode microRNAs (miRNAs). miRNAs are approximately 22-nucleotide noncoding RNAs generated from stem-loop precursors. Mature miRNAs interact directly with a member of the Argonaute (Ago) protein family to form the RNA-induced silencing complex (RISC), which silences gene expression posttranscriptionally by binding to the 3′ untranslated regions (3′ UTRs) of target mRNAs (reviewed in references 8, 18, and 31). It has been proposed that viral miRNAs participate in both lytic and latent infections and may be involved in virus-host interactions (25, 44). EBV was the first virus demonstrated to encode miRNAs (34). Shortly thereafter, other herpesviruses were found to encode miRNAs, for example, KSHV, murine gammaherpesvirus 68 (MHV-68), human cytomegalovirus, and rhesus monkey rhadinovirus (7, 32, 38, 39). The functions of most virus-encoded miRNAs are still unknown, and for the majority of viruses, only a few miRNA targets have been identified so far (21, 47). For KSHV and EBV, a systematic analysis of viral miRNA-mRNA interaction networks in latently infected cells has recently been performed by RISC immunoprecipitation assays, resulting in the identification of a large number of targets (17). Elucidation of the functions of viral miRNAs will shed light on whether they modulate pathogenesis and might also suggest new therapeutic approaches (12, 27, 33).
Due to the species specificity of the γHV, pathogenetic studies of human infections are restricted. MHV-68, a mouse γHV, allows investigation of the role of γHV miRNAs during infection in a small animal model (3, 15, 16, 20, 26, 41, 43, 46). In addition, MHV-68, unlike EBV and KSHV, readily establishes productive infections in a variety of cell lines and thus facilitates the examination of de novo lytic infections. To fully explore the potential of the MHV-68 system for the investigation of γHV miRNAs, it would be desirable to know the number and expression patterns of all miRNAs encoded by MHV-68. Pfeffer et al. (32) computationally predicted 14 miRNAs to be encoded by MHV-68 and experimentally confirmed 9 by small-RNA cloning and sequencing. In that study, the small RNAs were isolated from S11 cells, a cell line persistently infected with MHV-68 (45). Here we asked the question whether MHV-68 might encode more than the 9 miRNAs already identified. For that purpose, we analyzed the small RNAs from S11 cells, either untreated or treated with 12-O-tetradecanoyl-phorbol-13-acetate (TPA) to induce the lytic cycle, and from lytically infected NIH 3T3 fibroblasts by 454 deep sequencing. Subsequently, the expression of selected MHV-68 miRNAs was further validated by Northern blotting and stem-loop quantitative reverse transcription-PCR (RT-PCR) (10). Finally, the cellular miRNA profiles were analyzed. Our analysis identified 6 novel MHV-68 miRNAs and additionally provided the first comprehensive overview of both MHV-68 and cellular miRNA expression in infected cells. Our data will aid in the full exploration of the functions of γHV miRNAs.
MATERIALS AND METHODS
Cell culture and MHV-68 infection. (i) Cell lines.
S11 cells, a cell line persistently infected with MHV-68 (45), were kindly provided by J. P. Stewart (University of Liverpool, Liverpool, United Kingdom) and were cultured in RPMI 1640 medium (Cell Concepts, Umkirch, Germany) supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol. Ag8 cells (ATCC CRL-1580), a murine B-cell line, were cultured in RPMI 1640 medium (Cell Concepts, Umkirch, Germany) supplemented with 10% FCS, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol. NIH 3T3 cells (ATCC CRL-1658) were grown in DMEM High Glucose (Cell Concepts, Umkirch, Germany) supplemented with 10% FCS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Wild-type and Dicer−/− mouse embryonic fibroblasts (MEFs) were generated as described recently (29) and were cultured in DMEM High Glucose supplemented with 10% FCS, 10 mM HEPES, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin.
(ii) Infection of cells.
NIH 3T3 cells, MEFs, and Ag8 cells were infected with wild-type MHV-68 at a multiplicity of infection (MOI) of 1. The original stock of MHV-68 (clone G2.4) was obtained from A. Nash (University of Edinburgh, Edinburgh, United Kingdom). Working stocks of virus were prepared as previously described (1).
(iii) Lytic-cycle induction in S11 cells.
S11 cells were treated with TPA (20 ng/ml) for 48 h to induce the lytic cycle.
454 sequencing.
Small-RNA species were isolated from the cells (uninfected NIH 3T3 cells or NIH 3T3 cells 48 h after infection; S11 cells with or without TPA treatment) using the mirVana miRNA isolation kit (Applied Biosystems/Ambion, Darmstadt, Germany). The small RNAs were separated on a denaturing 12.5% polyacrylamide (PAA) gel stained with SYBR green II. After passive elution, RNAs with a length of 15 to 30 bases were concentrated by ethanol precipitation and dissolved in water. Next, RNAs were poly(A) tailed using poly(A) polymerase, and a 43-nucleotide (nt) adapter with the following sequence was ligated to the 5′ phosphate of the miRNAs: 5′-GCCTCCCTCGCGCCATCAGCTNNNNGACCTTGGCTGTCACTCA-3′ (5′-end adapter). NNNN represents a “bar code” sequence for the individual samples, as follows: ACTA for uninfected NIH 3T3 cells, AGGT for infected NIH 3T3 cells, ATCG for uninduced S11 cells, and CAAT for S11 cells induced by TPA. Next, first-strand cDNA synthesis was performed using a 3′-end oligo(dT) linker primer (61 nt), 5′-GCCTTGCCAGCCCGCTCAGACGAGACATCGCCCCGC(T)25-3′, and Moloney murine leukemia virus RNase H reverse transcriptase. The resulting cDNAs were PCR amplified in 22 cycles, using the high-fidelity Phusion polymerase (Finnzymes). Amplification products (120 to 135 bp) were confirmed by polyacrylamide gel electrophoretic (PAGE) analysis. All cDNA pools were mixed in equal amounts and were subjected to gel fractionation. The 120- to 135-bp fraction was electroeluted from 6% PAA gels. After isolation with Nucleospin extract II (Macherey and Nagel), cDNA pools were dissolved in 5 mM Tris-HCl (pH 8.5) at a concentration of 10 ng/liter and were used in single-molecule sequencing. Massively parallel sequencing was performed by 454 Life Sciences, using a Genome Sequencer 20 system.
Sequence analysis.
Base calling and quality trimming of sequence chromatograms were done by the publicly available Phred software (19). The sublibraries were sorted according to bar codes, and the adapter sequences and poly(A) tails were removed from the sequences. The remaining sequences were analyzed later. Sequences were annotated according to miRBase, version 14.0 (23), a tRNA database (9), a noncoding RNA database (24), and the MHV-68 genome sequence (NC_001826.2) from GenBank. The retrieved MHV-68 sequences other than the known miRNAs were then searched for putative novel miRNA sequences. Genomic regions containing inserts with 100-nt flanks were retrieved from GenBank in order to calculate RNA secondary structures by mFold (50). Only regions that folded into hairpins and contained an insert in one of the hairpin arms were considered putative novel miRNA sequences. The relative abundance of cellular miRNAs was calculated as the percentage of individual miRNA read numbers among total cellular miRNA read numbers from each library. The heat maps of the expression profiles were generated using the Mayday platform (1a).
Ago complex immunoprecipitation and RNA extraction.
Ago complex immunoprecipitation was performed as described previously (2). Briefly, S11 cells in 10-cm-diameter plates were lysed and centrifuged. For immunoprecipitation, 10 μg/ml of a purified monoclonal anti-mouse Ago2 (anti-mAgo2) antibody (6F4) or a commercial anti-bromodeoxyuridine (anti-BrDU) antibody (Abcam, Cambridge, United Kingdom) was mixed with cell lysates under constant rotation at 4°C for 3 h. Protein G-Sepharose (GE Healthcare) beads were added to the lysates, and the rotation continued for 1 h. After rotation, the beads were washed five times with cold RIPA-300 buffer. Finally, the beads were washed once with cold phosphate-buffered saline (PBS). The beads were treated with proteinase K under shaking at 65°C for 20 min, and coprecipitated RNA was extracted using phenol-chloroform and was subsequently precipitated from the aqueous phase by using at least 3 volumes of ethanol.
Northern blotting.
For Northern blotting, total RNAs were isolated from S11 cells, NIH 3T3, cells and Dicer−/− and Dicer+/+ MEFs by using the TRI reagent (Sigma, Taufkirchen, Germany) according to the recommendations of the manufacturer. The RNAs were separated by 15% denaturing RNA PAGE and were transferred to a nylon membrane (GE Healthcare) by semidry electroblotting. Membranes were cross-linked by incubation with a 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) chemical cross-link for 1 h at 50°C (28), prehybridized for 1 h, and hybridized overnight at 45°C with [32P]ATP-labeled probes complementary to the desired miRNAs. The probes were as follows: mghv-mir-M1-1 (5′-AAAGGAAGTACGGCCATTTCTA), mghv-mir-M1-8 (5′-GACCAAACCCCCAGTGAGTGCT), mghv-mir-M1-10 (5′-AAAGAACCTTCCGTGTAATCA), mghv-mir-M1-12 (5′-GCGTCGGGACCCGGGA), mghv-mir-M1-13 (5′-AAAGGGGTAGGACTCCCACACCAAA), mghv-mir-M1-14 (5′-AAAGAAGAGCTCACATGAGATA), mmu-mir-15b (5′-TGTAAACCATGATGTGCTGCTA), mmu-mir-16 (5′-CGCCAATATTTACGTGCTGCTA), mmu-mir-21 (5′-TCAACATCAGTCTGATAAGCTA), and tRNA-Met (5′-TGGTAGCAGAGGATGGTTTCGATCCATCGACCTCTG).
RT and qPCR.
Real-time quantification of miRNAs was performed by stem-loop quantitative RT-PCR (qRT-PCR) as described by Chen et al. (10). Total RNA was isolated using the miRNeasy kit (Qiagen, Hilden, Germany) according to the recommendations of the manufacturer. One microgram of RNA was reverse transcribed using Superscript III reverse transcriptase (Invitrogen). qPCR was performed using Power SYBR green master mix (Applied Biosystems) on the 7300 platform (Applied Biosystems). As controls, we used 5.8S rRNA and the cellular miRNA mmu-miR-191 (30). The universal primer used was Universal.rev (5′-GTGCAGGGTCCGAGGT). The specific primers used (SLP, stem-loop RT primer; qP, qPCR primer) were as follows: SLP_mghv-miR-1 (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAAAGGA), qP_mghv-miR-1.for (5′-CGGCTAGAAATGGCCGTACT), SLP_mghv-miR-13 (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAAAGAA), qP_mghv-miR-13.for (5′-CGCGTATCTCATGTGAGCTC), SLP_rRNA-5.8S-K (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAGCGAC), qP_rRNA-5.8S-K.for (5′-GCCCGCCTGTCTGAGC), SLP_mmu-miR-191-K 5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCAGCTG), and qP_mmu-miR-191-K.for (5′-CGGCAACGGAATCCCAAAAG).
To analyze the expression of the cellular gene BRCA-1, RNA was isolated 48 h after infection from infected and uninfected NIH 3T3 cells using the miRNeasy kit (Qiagen, Hilden, Germany) and was reverse transcribed using Superscript III reverse transcriptase (Invitrogen). After a 1:5 dilution, 5 μl of the cDNA was amplified by qPCR using Power SYBR green master mix (Applied Biosystems) on the 7300 platform (Applied Biosystems). As a housekeeping gene, we used the 18S RNA. The specific primers used were as follows: mBRCA-1-primer1-for (5′-TTGTGAGCGTTTGAATGAGG), mBRCA-1-primer1-rev (5′-CTGTCCTTCAAGGTGGCATT), mBRCA-1-primer2-for (5′-CAAGGCGAGAGCTAGAAGGA), mBRCA-1-primer2-rev (5′-AATGTGGGCTGGCTCTTTAG), 18S-for (5′-CGGCTACCACATCCAAGGAA), and 18S-rev (5′-GCTGGAATTACCGCGGCT).
RESULTS
Sequencing of small RNAs from MHV-68-infected cells.
In order to validate the expression of known MHV-68 miRNAs and to identify novel MHV-68 miRNAs, we infected NIH 3T3 cells with MHV-68, which results in lytic replication (Fig. 1, sample NIH 3T3+). Uninfected NIH 3T3 cells served as negative controls (sample NIH 3T3−). In addition, we analyzed S11 cells, a cell line persistently infected with MHV-68. S11 cells either were left untreated (sample S11−) or were treated with TPA to induce the lytic cycle (sample S11+). Small RNAs were extracted and sequenced using 454 sequencing technologies (GEO accession no. GSE22938). We obtained 59,922 reads from NIH 3T3−, 54,135 reads from NIH 3T3+, 43,035 reads from S11−, and 60,704 reads from S11+ after bar code sorting. After removal of the adapter sequences, more than 99% of the remaining reads were longer than 15 nt and were taken for subsequent analysis (Fig. 1A).
FIG. 1.
Comprehensive analysis of small-RNA deep-sequencing libraries constructed from NIH 3T3 and S11 cells. (A) Table of sequence distribution after annotation according to the MHV-68 genome and the mouse genome. NIH 3T3−, uninfected NIH 3T3 cells; NIH 3T3+, MHV-68-infected NIH 3T3 cells (48 h after infection); S11−, untreated S11 cells; S11+, S11 cells treated with TPA (20 ng/ml for 48 h). (B) Schemes showing the abundances of MHV-68 and mouse sequences in NIH 3T3 and S11 small-RNA deep-sequencing libraries.
After thorough analysis of the reads against both the MHV-68 and the mouse genome, we found that the majority of reads were miRNA sequences, while a minor portion was derived from degradation products of other noncoding RNAs (Fig. 1A and B), indicating the good quality of the small-RNA libraries. Accordingly, we found no MHV-68 sequences in the NIH 3T3− sample. Nearly 10% of the reads in both the S11− and S11+ libraries were MHV-68 sequences, i.e., much more than the 0.5% in MHV-68-infected NIH 3T3 cells (Fig. 1B).
Identification of novel MHV-68 miRNAs.
Analysis of the pool of MHV-68 sequences from the various libraries showed that a considerable number of reads were not matched to known mature MHV-68 miRNAs in miRBase, version 14.0. However, among these, about half of the sequences matched known miRNA hairpins and were thus characterized as novel star miRNAs (Fig. 1A and Table 1). The remaining MHV-68 sequences were analyzed using the secondary-structure prediction program mFold by extracting the flanking sequences from the genome. In this way, we identified six novel MHV-68 miRNA genes (termed mghv-mir-M1-10 to mghv-mir-M1-15), which give rise to 11 mature miRNAs in total (Table 2). mghv-mir-M1-11 is a perfectly matched hairpin but has only very low read numbers from both arms. The other five hairpins show common imperfect miRNA folding structures and are biased in generating the mature miRNAs from the 3′ arm (Tables 1 and 2). Alignment of the deep-sequencing reads to the MHV-68 miRNA hairpins revealed more variation in the 3′ than in the 5′ ends of the reads, a pattern that is commonly observed in the analysis of miRNA deep-sequencing data (see Fig. S1 in the supplemental material).
TABLE 1.
microRNA read numbers of known and novel precursors from MHV-68 samples
| Namea | Sequence | No. of reads in the libraries |
||
|---|---|---|---|---|
| NIH 3T3+ | S11− | S11+ | ||
| mghv-mir-M1-1 | UAGAAAUGGCCGUACUUCCUUU | 35 | 1,168 | 1,572 |
| mghv-mir-M1-1* | AGGAAGUGGGUCCAACUU | 0 | 2 | 0 |
| mghv-mir-M1-2 | CAGACCCCCUCUCCCCCUCUUU | 1 | 36 | 71 |
| mghv-mir-M1-2-5p | AGAGGGGGAGUGUGUGGUCUGUb | 19 | 149 | 225 |
| mghv-mir-M1-3 | GAGGUGAGCAGGAGUUGCGCUU | 7 | 29 | 42 |
| mghv-mir-M1-3* | AGCGAACCUCUGCUCACUGCCC | 0 | 9 | 15 |
| mghv-mir-M1-4 | UCGAGGAGCACGUGUUAUUCUA | 0 | 23 | 29 |
| mghv-mir-M1-4* | AGAUAGCAUGUGCCGUCCUCUUU | 0 | 6 | 7 |
| mghv-mir-M1-5 | AGAGUUGAGAUCGGGUCGUCUC | 8 | 144 | 275 |
| mghv-mir-M1-5* | AGGCAAACCCGAGCUCCUCCUU | 29 | 28 | 84 |
| mghv-mir-M1-6 | UGAAACUGUGUGAGGUGGUUUU | 1 | 38 | 101 |
| mghv-mir-M1-6* | CAACCACCUCCCACAAUUUCAGb | 0 | 22 | 28 |
| mghv-mir-M1-7-5p | AAAGGUGGAGGUGCGGUAACCU | 11 | 140 | 231 |
| mghv-mir-M1-7-3p | GAUAUCGCGCCCACCUUUAUU | 12 | 306 | 510 |
| mghv-mir-M1-8 | AGCACUCACUGGGGGUUUGGUC | 61 | 679 | 977 |
| mghv-mir-M1-8* | UGACCAACCCUAAGUGAGUUUU | 7 | 154 | 177 |
| mghv-mir-M1-9 | UCACAUUUGCCUGGACCUUUUU | 58 | 674 | 617 |
| mghv-mir-M1-10 | UGAUUACACGGAAGGUUCUUUU | 5 | 62 | 90 |
| mghv-mir-M1-10* | UUAAGAACCCUCAGUGCAAUCb | 1 | 20 | 12 |
| mghv-mir-M1-11-5p | AGCUGUCAGGGGUUACAUGb | 0 | 0 | 1 |
| mghv-mir-M1-11-3p | UGUAACCCCUGACAGCUGUC | 0 | 1 | 0 |
| mghv-mir-M1-12 | UUUGGUGUGGGAGUCCUACCCCUUU | 4 | 76 | 103 |
| mghv-mir-M1-12* | AAGGGUACUCUCAUCACCAAUGUb | 0 | 1 | 2 |
| mghv-mir-M1-13 | UAUCUCAUGUGAGCUCUUCUUU | 8 | 254 | 276 |
| mghv-mir-M1-13* | UGGGAAGAGUCUGUUGAGUGGC | 0 | 1 | 4 |
| mghv-mir-M1-14 | UGCUACAGCGUGCAGAACGUUU | 2 | 74 | 78 |
| mghv-mir-M1-14* | CCCGUUCUGGAUGCUGUGGGACb | 3 | 7 | 8 |
| mghv-mir-M1-15 | AGCUACCCGCGUGGCCGGAGUGUUU | 0 | 0 | 2 |
miRNAs and star miRNAs of a precursor are defined according to the relative abundances of the reads of both arms. Names in regular type designate the known miRNAs; those in boldface are the novel star miRNAs of known precursors; and those in italics are the novel miRNAs of novel precursors.
Due to the characteristics of 454 sequencing, the putative miRNA sequences might extend to the additional “A” at the end.
TABLE 2.
Novel candidate microRNAs of novel precursors identified from MHV-68 samples
| Novel candidate miRNAa | Stem-loop structure of putative miRNA precursorb | ΔG (kcal/mol)b | Genomic locationc |
|---|---|---|---|
| UUCAC GUCUG | |||
| mghv-mir-M1-10* | AAGAACCUCGUGAAUCACUU C | −21.5 | 262-282 |
| mghv-mir-M1-10 | UUCUUGGAGCACUUAGUGAG A | −21.5 | 301-322 |
| UUAGA GUUU | |||
| AG | |||
| mghv-mir-M1-11-5P | GACAGCUGUCAGGGGUUACAUGAG A | −44.9 | 762-780 |
| mghv-mir-M1-11-3P | CUGUCGACAGUCCCCAAUGUACUU A | −44.9 | 792-811 |
| C | |||
| GUAAU AAAUUAAU | |||
| mghv-mir-M1-12* | GGGU---ACUCUCACACCAAUGU A | −21.5 | 1722-1744 |
| mghv-mir-M1-12 | CCCAUGAGGGU-GUGGUUUCA U | −21.5 | 1764-1788 |
| UUUUCUCC AACGAUG | |||
| UUGU C- UA | |||
| mghv-mir-M1-13* | GGGAAGAG-UCUGAG-UGGC GCG G | −19.6 | 3709-3770 |
| mghv-mir-M1-13 | UUCUUCUCAGACUCAUCG UGU G | −19.6 | 3787-3808 |
| UGUGUU AA U | |||
| GCCCGG CUGA | |||
| mghv-mir-M1-14* | CGUUCUGAUGCUGUGGACA A | −28.7 | 5105-5126 |
| mghv-mir-M1-14 | GCAAGACUGCGACAUCUGU A | −28.7 | 5140-5161 |
| UUUUGG CGUU | |||
| CU -C-- CC- UU | |||
| mghv-mir-M1-15 | AGACCCGGUGGGGAGGUU | −15.7 | 5462-5486 |
| UC UGGGC CACC UCUC CA A | |||
| UC CUUC CAA C G |
Names of the putative new miRNA sequences are given according to the submission to miRBase (http://www.mirbase.org). The miRNAs and star miRNAs of a precursor are defined according to the relative abundances of the reads of both arms (see Table 1).
RNA secondary structures were predicted, and free energy was calculated, using mFold, version 3.2. The miRNA sequences are underlined. Due to the characteristics of 454 sequencing, the putative miRNA sequences might extend to the additional “A” at the end. The actual size of the stem-loop has not been determined experimentally.
Positions in the MHV-68 genome (GenBank accession number NC_0018262).
Furthermore, we investigated whether the known and newly identified MHV-68 miRNAs have seed homology to miRNAs from murine and other herpesviruses. Both KSHV and Marek's disease virus (MDV) have been reported to encode functional orthologs of the oncogenic mir-155 (22, 42, 48). We found that mghv-mir-M1-4 has the same seed sequence as mir-151-5p, which has been revealed to be an important oncogenic factor during tumor invasion and metastasis (14) (see Fig. S2 in the supplemental material). On the other hand, mghv-mir-M1-9 and mghv-mir-M1-14 have seed homology with KSHV-miR-K12-3 and MDV1-mir-M31, respectively. Binding sites for KSHV-miR-K12-3 have been identified within the 3′ UTR of the transcription factor C/EBPβ, a regulator of the transcriptional activation of interleukin-6 (IL-6) and IL-10 (35). This analysis suggests that miRNAs from different viruses or even from the mouse may regulate similar target mRNAs.
Genomic organization of the novel MHV-68 miRNAs.
Interestingly, like the nine MHV-68 miRNAs already known, all six novel miRNA genes are situated within the first 6 kbp at the left end of the MHV-68 genome and are located close to the eight viral tRNAs (vtRNAs), except for mghv-mir-M1-11 (Fig. 2 A). The novel MHV-68 miRNAs are positioned either immediately following the known miRNAs (Fig. 2B and F) or between the vtRNAs and known miRNAs (Fig. 2G and I). The vtRNA-miRNA-miRNA structure is common for six of the eight vtRNAs. Following vtRNA3 and following vtRNA7, there is only one miRNA hairpin structure; the other stretch was not able to form a hairpin, according to mFold (Fig. 2D and H), and we found no corresponding reads from the libraries. Furthermore, these specifically structured ∼200-nt RNA sequences all start with the conserved A/B box promoter for RNA polymerase III in the vtRNAs and end with a poly(U) sequence, which serves as the terminus for RNA polymerase III, implying that the vtRNA-miRNA-miRNA sequences are transcribed as a long primary transcript and are then processed further into mature miRNAs.
FIG. 2.
MHV-68 miRNAs are located directly after the vtRNA sequences. (A) Genomic locations of MHV-68 miRNAs, modified from reference 32. The first 6 kbp of the MHV-68 genome are shown. Filled triangles represent the 8 vtRNA sequences. M1 to M9 (in red) are the known MHV-68 miRNA precursors. M10 to M15 (in blue) are the 6 novel MHV-68 miRNA genes identified in this paper. ORF, open reading frame; TR, terminal repeat. (B to I) RNA folding structures of 8 vtRNA sequences and their subsequent miRNAs. Sequences are color coded as follows: green, the predicted anticodons of the vtRNA sequences (5); red, the known MHV-68 miRNAs; yellow, the novel star miRNAs of known precursors; blue, the novel miRNAs of novel MHV-68 miRNA precursors. The GenBank accession number of the MHV-68 genome and the genomic locations of the sequences are given at the left of each sequence.
Validation of the expression of the novel MHV-68 miRNAs.
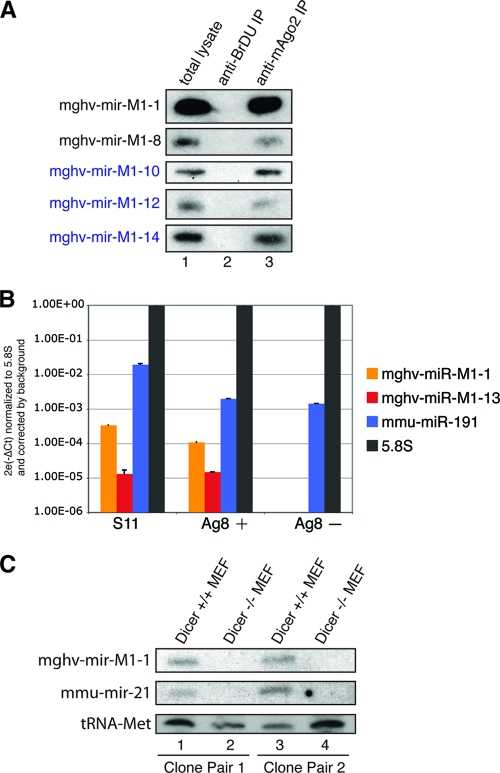
To further validate the existence of the novel MHV-68 miRNAs, we performed Northern blotting with RNAs isolated from S11 cells. RNAs were extracted either from total-cell lysates or from lysates after pulldown with a monoclonal anti-mouse Ago2 antibody. As a negative control, pulldowns were performed using an isotype-matched control antibody (anti-BrDU). Probes against the abundant miRNAs mghv-mir-M1-10, mghv-mir-M1-12, and mghv-mir-M1-14 were easily detectable not only in total-cell lysates but also in the anti-mAgo2-immunoprecipitated (IP) samples (Fig. 3A), indicating that the novel MHV-68 miRNAs, like cellular miRNAs and the MHV-68 miRNAs already known, associate efficiently with functional RISCs. Only a very weak signal could be detected from the probe against mghv-mir-M1-13 (data not shown). However, by using a more-sensitive stem-loop qRT-PCR, we were able to demonstrate the expression of mghv-mir-M1-13 (Fig. 3B) both in S11 cells and in infected Ag8 cells 48 h after infection with MHV-68. No MHV-68 miRNAs were detectable in uninfected Ag8 cells, while the cellular miRNA mmu-miR-191 was detected in all three samples.
FIG. 3.
Validation of novel MHV-68 miRNAs. (A) Northern blot validation of novel MHV-68 miRNAs. RNAs were extracted from S11 total-cell lysates (lane 1), a control IP with anti-BrDU (lane 2), and an IP with anti-mAgo2 (monoclonal antibody 6F4) (lane 3) and were blotted onto a nylon membrane. Probes complementary to known or novel MHV-68 miRNAs were labeled and used for detection. Novel MHV-68 miRNAs are highlighted in blue. (B) Stem-loop qRT-PCR validation of the novel MHV-68 miRNA mghv-mir-M1-13. RNAs were extracted from total-cell lysates of S11 cells and from infected (+) or uninfected (−) Ag8 cells. The latter served as a negative control. The samples were subjected to stem-loop RT-qPCR as described in Materials and Methods. Data shown are means ± standard deviations for triplicate determinations. As positive controls, the expression of mghv-mir-M1-1 and that of the cellular miRNA mmu-miR-191 are also shown. (C) Generation of MHV-68 miRNAs in MHV-68-infected Dicer−/− MEFs. Two independent clones of Dicer−/− MEFs (lanes 2 and 4) were infected with MHV-68 in parallel with wild-type controls (lanes 1 and 3). Total RNAs were extracted and blotted onto a nylon membrane. Probes complementary to mghv-mir-M1-1 and mmu-mir-21 were labeled and used for detection. Probing against tRNAMet served as a loading control.
Since MHV-68 miRNAs derive from unusual precursors, we investigated whether the MHV-68 miRNAs employ the same miRNA-processing machinery as cellular miRNAs. To this end, we infected Dicer-deficient mouse embryonic fibroblasts (Dicer−/− MEFs) with MHV-68 and monitored miRNA generation. Two independently derived Dicer−/− MEF clones were tested. Compared to wild-type MEFs (Dicer+/+), Dicer−/− MEFs were not able to produce either mature cellular miRNAs or MHV-68 miRNAs (Fig. 3C). Thus, MHV-68 miRNAs are processed by a Dicer-dependent pathway.
Cellular miRNA profiling in NIH 3T3 and S11 cells.
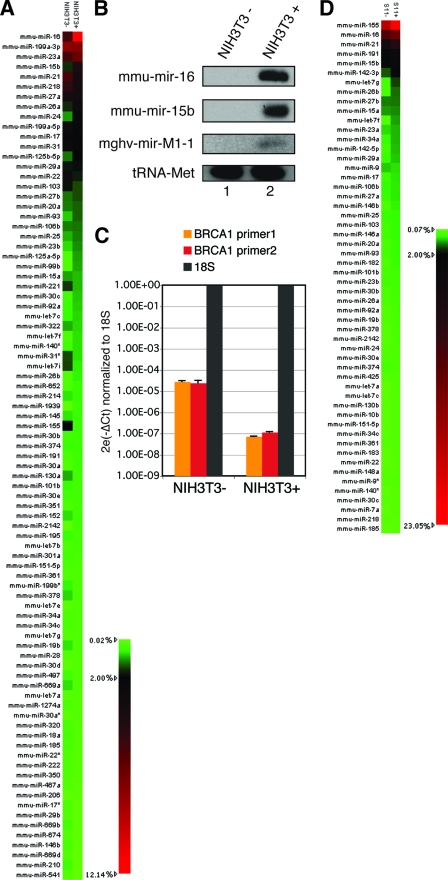
Apart from the MHV-68 sequences, the majority of reads in the sequencing libraries were murine miRNA sequences. Annotating these sequences by miRBase, version 14.0, we found a significant number of reads derived from the other arm of known murine miRNA hairpins. Altogether, 70 novel murine star miRNAs were identified and registered (see Table S1 in the supplemental material). The read numbers of annotated murine miRNAs in the libraries suggested unique miRNA expression signatures of NIH 3T3 and S11 cells (see Tables S2 and S3 in the supplemental material). NIH 3T3− and NIH 3T3+ cells showed distinct patterns of cellular miRNA expression levels, suggesting that miRNAs may play a role in the cellular response to MHV-68 infection. Among the abundantly expressed miRNAs, mmu-mir-16 and mmu-mir-15b, which belong to the same miRNA family, were found to be highly upregulated after MHV-68 infection (Fig. 4 A). This upregulation was further confirmed by Northern blotting (Fig. 4B). As expected, mghv-mir-M1-1 was present only in NIH 3T3+ cells, and tRNAMet served as loading control. The cellular miRNA expression profiles in TPA-induced versus noninduced S11 cells were also analyzed. Only subtle changes were observed after TPA induction (Fig. 4D).
FIG. 4.
(A and D) Cellular miRNA profiling of uninfected or MHV-68-infected NIH 3T3 cells and of untreated or TPA-treated S11 cells. Cellular miRNA levels were calculated and compared in the four deep-sequencing libraries. (A) Heat map depicting MHV-68-infected versus noninfected NIH 3T3 cells. (D) Heat map depicting TPA-treated versus untreated S11 cells. Only miRNAs that were more than 0.2% abundant in at least one library are shown. (B) Northern blot validation of cellular miRNAs upregulated after infection of NIH 3T3 cells. Total RNAs were extracted from uninfected (lane 1) or MHV-68-infected (lane 2) NIH 3T3 cells. RNAs were blotted onto a nylon membrane and were detected by labeled probes complementary to mmu-mir-16, mmu-mir-15b, or mghv-mir-M1-1. Probing against tRNAMet served as a loading control. (C) Downregulation of BRCA-1 in MHV-68-infected NIH 3T3 cells. To analyze the expression of BRCA-1, RNAs were isolated from uninfected and MHV-68-infected NIH 3T3 cells 48 h after infection and were subjected to qRT-PCR using primers specific for BRCA-1 and 18S RNA. Data shown are means ± standard deviations for triplicate determinations.
We have reported previously that hsa-mir-15a and hsa-mir-16 are upregulated in EBV-positive nasopharyngeal carcinoma. We found that the tumor suppressor gene BRCA-1 is one of the targets that is regulated by the miR-15/16 family (49). Therefore, we wondered whether the upregulation of mmu-mir-16 and mmu-mir-15b after MHV-68 infection might also lead to the downregulation of BRCA-1 in the MHV-68 model system. As shown in Fig. 4C, BRCA-1 expression in MHV-68-infected NIH 3T3 cells was indeed lower than that in uninfected cells, suggesting that miR-15/16 upregulation and the subsequent BRCA-1 downregulation might be common features of herpesvirus infection.
DISCUSSION
MHV-68 is an important small animal model with which to study the pathogenesis of γHV infections. By deep sequencing of small RNA, we systematically investigated the MHV-68 miRNA expression profiles in both lytically and persistently infected cells. We identified six novel MHV-68 miRNA genes and analyzed the expression levels of all MHV-68 miRNAs. Furthermore, we also characterized the cellular miRNA expression signatures in MHV-68-infected versus noninfected NIH 3T3 fibroblasts and in TPA-treated versus nontreated S11 cells. We found that mmu-mir-15b and mmu-mir-16 are highly upregulated upon MHV-68 infection of NIH 3T3 cells, indicating a potential role of cellular miRNAs during MHV-68 infection.
MHV-68 miRNAs were first identified by traditional small-RNA cloning and sequencing (32). Nine MHV-68 miRNA genes were characterized and experimentally validated, and five more hairpins were predicted. With the more-sensitive deep-sequencing approach, we were able to confirm the existence of all previously described MHV-68 miRNAs and almost all of the star miRNAs, which have not been reported before. Moreover, we identified, for the first time, the five predicted miRNA genes, together with one unpredicted miRNA gene. Notably, all MHV-68 miRNAs, except for mghv-mir-M1-11, reside immediately following the unique vtRNA sequences in the MHV-68 genome. A transcript containing mghv-mir-M1-11 might be generated by read-through transcription beyond the T clusters at which RNA polymerase III usually terminates transcription (6).
Almost all MHV-68 miRNAs were detectable in the two infected cell lines we have sequenced (Table 1). Overall, the persistently infected S11 cells showed much higher MHV-68 miRNA expression levels than the lytically infected NIH 3T3 cells. mghv-mir-M1-1, mghv-mir-M1-8, and mghv-mir-M1-9 were the three most abundant miRNAs in both cell lines. mghv-mir-M1-5* was more abundant than mghv-mir-M1-5 in infected NIH 3T3 cells, while in S11 cells, mghv-mir-M1-5 was expressed at a higher level. Such different MHV-68 miRNA expression patterns might be suggestive of distinct functions during lytic and persistent/latent infections. By Northern blotting, we could demonstrate the expression of three (mghv-mir-M1-10, -12, and -14) of the six novel MHV-68 miRNAs. Although mghv-mir-M1-13 was found with a comparable read number in the deep-sequencing data sets, it was hardly detectable by Northern blotting. This might be due to its low GC content. However, we were able to demonstrate the expression of mghv-mir-M1-13 by a more-sensitive stem-loop qRT-PCR. This is consistent with a recent report demonstrating that reverse ligation-mediated RT-PCR (RLM-RT-PCR) is able to detect mature MHV-68 miRNAs with at least 100-fold higher sensitivity than Northern blotting (13). We did not attempt to detect the expression of mghv-mir-M1-11 and -15 by either Northern blotting or stem-loop qRT-PCR because of their extremely low read numbers (1 and 2 reads, respectively) in the deep sequencing.
The MHV-68 miRNAs are among the minority of miRNAs that are under the regulation of RNA polymerase III. It has been shown recently that the generation of the MHV-68 miRNAs is dependent on the A/B boxes in the vtRNA sequences and on the presence of tRNase Z and Dicer, but not on Drosha (4, 13). By infection of Dicer-deficient MEFs, we confirmed that MHV-68 miRNAs are processed by a Dicer-dependent pathway. Importantly, while Bogerd et al. (4) showed Dicer dependency by small interfering RNA (siRNA)-mediated knockdown of Dicer in human (HeLa) cells transfected with an MHV-68 miRNA expression plasmid, we demonstrated it by infection of authentic host cells (Dicer−/− and Dicer+/+ MEFs) with MHV-68.
The vtRNAs were identified and characterized more than 10 years ago and were shown not to be aminoacylated (5). However, their function still remains unknown. We speculate that the vtRNA sequences remained as remnants during evolution and now serve as promoter sequences for the generation of the MHV-68 miRNAs. How the differential expression of the MHV-68 miRNAs is regulated, and the significance of the existence of the vtRNA-miRNA-miRNA structures, will be interesting to investigate.
The MHV-68 miRNAs were shown to be efficiently associated with the RISC by immunoprecipitation using a monoclonal anti-mouse Ago2 antibody, implying that they are functional. However, the functions of the MHV-68 miRNAs still remain unknown. By matching the seed sequences of the MHV-68 miRNAs, we found that they are predicted to target both viral and cellular proteins (data not shown). The abundance of the MHV-68 miRNAs might also imply that they could function by occupying the cellular RISC machinery, thus interfering with the normal cellular miRNA pathways. The attenuated but nonlethal phenotype of an MHV-68 mutant virus lacking the first 9.5 kbp of the genome, including all vtRNAs and miRNAs, indicated that the vtRNAs and miRNAs are not absolutely essential for lytic replication or for the establishment and maintenance of latency (11).
Apart from the MHV-68 miRNAs, the deep-sequencing data also allowed us to analyze the expression signature of the cellular miRNAs in uninfected versus infected NIH 3T3 cells. mmu-mir-15b and mmu-mir-16 were shown to be upregulated after infection, and this observation was validated by Northern blotting. Interestingly, in our previous study (49), hsa-mir-15a and hsa-mir-16 were found to be upregulated in EBV-infected nasopharyngeal carcinoma samples compared to their expression in healthy control tissues, and the tumor suppressor BRCA-1 was validated as one of the target genes of hsa-mir-15a and hsa-mir-16. Here we could also show that upregulation of mmu-mir-16 and mmu-mir-15b correlated with lower BRCA-1 expression in infected NIH 3T3 cells than in uninfected cells. Notably, other authors reported that NIH 3T3 cells, expressing an RNA transcript that is antisense to the BRCA-1 mRNA and that thus inhibited the expression of BRCA-1 protein, showed accelerated and anchorage-independent growth and tumorigenicity in nude mice (36).
Taken together, we have performed deep sequencing of small RNAs expressed in lytically infected NIH 3T3 cells and in persistently infected S11 cells in order to more completely define the miRNA coding potential of MHV-68. Our analysis identified 6 novel MHV-68 miRNAs and additionally provided the first comprehensive overview of both MHV-68 and cellular miRNA expression in infected cells. Our data will aid in the full exploration of the functions of γHV miRNAs.
Supplementary Material
Acknowledgments
This work was supported by grants from the BMBF (NGFNplus) (FKZ PIM-01GS0802-3 to H.A. and FKZ PIM-01GS0804-5 to G.M.) and from the Deutsche Forschungsgemeinschaft (DFG) and the Deutsche Krebshilfe to G.M.
We are grateful to B. Steer for expert technical assistance and to S. Grömminger for helpful discussions. We also thank B. Adler for critical reading of the manuscript.
Footnotes
Published ahead of print on 28 July 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Battke, F., S. Symons, and K. Nieselt. 2010. Mayday—intergrative analytics for expression data. BMC Bioinformatics 11:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beitzinger, M., L. Peters, J. Y. Zhu, E. Kremmer, and G. Meister. 2007. Identification of human microRNA targets from isolated Argonaute protein complexes. RNA Biol. 4:76-84. [DOI] [PubMed] [Google Scholar]
- 3.Blackman, M. A., and E. Flano. 2002. Persistent γ-herpesvirus infections: what can we learn from an experimental mouse model? J. Exp. Med. 195:F29-F32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., H. W. Karnowski, X. Cai, J. Shin, M. Pohlers, and B. R. Cullen. 2010. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol. Cell 37:135-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowden, R. J., J. P. Simas, A. J. Davis, and S. Efstathiou. 1997. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J. Gen. Virol. 78:1675-1687. [DOI] [PubMed] [Google Scholar]
- 6.Braglia, P., R. Percudani, and G. Dieci. 2005. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 280:19551-19562. [DOI] [PubMed] [Google Scholar]
- 7.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carthew, R. W., and E. J. Sontheimer. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, P. P., and T. M. Lowe. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37:D93-D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C., D. A. Ridzon, A. J. Broomer, Z. Zhou, D. H. Lee, J. T. Nguyen, M. Barbisin, N. L. Xu, V. R. Mahuvakar, M. R. Andersen, K. Q. Lao, K. J. Livak, and K. J. Guegler. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clambey, E. T., H. W. Virgin IV, and S. H. Speck. 2002. Characterization of a spontaneous 9.5-kilobase-deletion mutant of murine gammaherpesvirus 68 reveals tissue-specific genetic requirements for latency. J. Virol. 76:6532-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen, B. R. 2006. Viruses and microRNAs. Nat. Genet. 38(Suppl.):S25-S30. [DOI] [PubMed] [Google Scholar]
- 13.Diebel, K. W., A. L. Smith, and L. F. Van Dyk. 2010. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA 16:170-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding, J., S. Huang, S. Wu, Y. Zhao, L. Liang, M. Yan, C. Ge, J. Yao, T. Chen, D. Wan, H. Wang, J. Gu, M. Yao, J. Li, H. Tu, and X. He. 2010. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat. Cell Biol. 12:390-399. [DOI] [PubMed] [Google Scholar]
- 15.Doherty, P. C., J. P. Christensen, G. T. Belz, P. G. Stevenson, and M. Y. Sangster. 2001. Dissecting the host response to a gamma-herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doherty, P. C., R. A. Tripp, A.-M. Hamilton-Easton, R. D. Cardin, D. L. Woodland, and M. A. Blackman. 1997. Tuning into immunological dissonance: an experimental model for infectious mononucleosis. Curr. Opin. Immunol. 9:477-483. [DOI] [PubMed] [Google Scholar]
- 17.Dölken, L., G. Malterer, F. Erhard, S. Kothe, C. C. Friedel, G. Suffert, L. Marcinowski, N. Motsch, S. Barth, M. Beitzinger, D. Lieber, S. M. Bailer, R. Hoffmann, Z. Ruzsics, E. Kremmer, S. Pfeffer, R. Zimmer, U. H. Koszinowski, F. Grasser, G. Meister, and J. Haas. 2010. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe 7:324-334. [DOI] [PubMed] [Google Scholar]
- 18.Eulalio, A., E. Huntzinger, and E. Izaurralde. 2008. Getting to the root of miRNA-mediated gene silencing. Cell 132:9-14. [DOI] [PubMed] [Google Scholar]
- 19.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 20.Flaño, E., D. L. Woodland, and M. A. Blackman. 2002. A mouse model for infectious mononucleosis. Immunol. Res. 25:201-217. [DOI] [PubMed] [Google Scholar]
- 21.Gottwein, E., and B. R. Cullen. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3:375-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottwein, E., N. Mukherjee, C. Sachse, C. Frenzel, W. H. Majoros, J. T. Chi, R. Braich, M. Manoharan, J. Soutschek, U. Ohler, and B. R. Cullen. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths-Jones, S. 2004. The microRNA Registry. Nucleic Acids Res. 32:D109-D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, C., B. Bai, G. Skogerbo, L. Cai, W. Deng, Y. Zhang, D. Bu, Y. Zhao, and R. Chen. 2005. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 33:D112-D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair, V., and M. Zavolan. 2006. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 14:169-175. [DOI] [PubMed] [Google Scholar]
- 26.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine γ-herpesvirus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neilson, J. R., and P. A. Sharp. 2005. Herpesviruses throw a curve ball: new insights into microRNA biogenesis and evolution. Nat. Methods 2:252-254. [DOI] [PubMed] [Google Scholar]
- 28.Pall, G. S., and A. J. Hamilton. 2008. Improved Northern blot method for enhanced detection of small RNA. Nat. Protoc. 3:1077-1084. [DOI] [PubMed] [Google Scholar]
- 29.Parameswaran, P., E. Sklan, C. Wilkins, T. Burgon, M. A. Samuel, R. Lu, K. M. Ansel, V. Heissmeyer, S. Einav, W. Jackson, T. Doukas, S. Paranjape, C. Polacek, F. B. dos Santos, R. Jalili, F. Babrzadeh, B. Gharizadeh, D. Grimm, M. Kay, S. Koike, P. Sarnow, M. Ronaghi, S. W. Ding, E. Harris, M. Chow, M. S. Diamond, K. Kirkegaard, J. S. Glenn, and A. Z. Fire. 2010. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6:e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peltier, H. J., and G. J. Latham. 2008. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 14:844-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters, L., and G. Meister. 2007. Argonaute proteins: mediators of RNA silencing. Mol. Cell 26:611-623. [DOI] [PubMed] [Google Scholar]
- 32.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grässer, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 33.Pfeffer, S., and O. Voinnet. 2006. Viruses, microRNAs and cancer. Oncogene 25:6211-6219. [DOI] [PubMed] [Google Scholar]
- 34.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 35.Qin, Z., P. Kearney, K. Plaisance, and C. H. Parsons. 2010. Pivotal advance: Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J. Leukoc. Biol. 87:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao, V. N., N. Shao, M. Ahmad, and E. S. Reddy. 1996. Antisense RNA to the putative tumor suppressor gene BRCA1 transforms mouse fibroblasts. Oncogene 12:523-528. [PubMed] [Google Scholar]
- 37.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 38.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schäfer, A., X. Cai, J. P. Bilello, R. C. Desrosiers, and B. R. Cullen. 2007. Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology 364:21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulz, T. F. 1998. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 79:1573-1591. [DOI] [PubMed] [Google Scholar]
- 41.Simas, J. P., and S. Efstathiou. 1998. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 6:276-282. [DOI] [PubMed] [Google Scholar]
- 42.Skalsky, R. L., M. A. Samols, K. B. Plaisance, I. W. Boss, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836-12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speck, S. H., and H. W. Virgin IV. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2:403-409. [DOI] [PubMed] [Google Scholar]
- 44.Sullivan, C. S., and D. Ganem. 2005. MicroRNAs and viral infection. Mol. Cell 20:3-7. [DOI] [PubMed] [Google Scholar]
- 45.Usherwood, E. J., J. P. Stewart, and A. A. Nash. 1996. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J. Virol. 70:6516-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Virgin, H. W., IV, and S. H. Speck. 1999. Unraveling immunity to γ-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 47.Whitby, D. 2009. Searching for targets of viral microRNAs. Nat. Genet. 41:7-8. [DOI] [PubMed] [Google Scholar]
- 48.Zhao, Y., Y. Yao, H. Xu, L. Lambeth, L. P. Smith, L. Kgosana, X. Wang, and V. Nair. 2009. A functional microRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J. Virol. 83:489-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu, J. Y., T. Pfuhl, N. Motsch, S. Barth, J. Nicholls, F. Grasser, and G. Meister. 2009. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 83:3333-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.