Abstract
Coronaviruses encode two classes of cysteine proteases, which have narrow substrate specificities and either a chymotrypsin- or papain-like fold. These enzymes mediate the processing of the two precursor polyproteins of the viral replicase and are also thought to modulate host cell functions to facilitate infection. The papain-like protease 1 (PL1pro) domain is present in nonstructural protein 3 (nsp3) of alphacoronaviruses and subgroup 2a betacoronaviruses. It participates in the proteolytic processing of the N-terminal region of the replicase polyproteins in a manner that varies among different coronaviruses and remains poorly understood. Here we report the first structural and biochemical characterization of a purified coronavirus PL1pro domain, that of transmissible gastroenteritis virus (TGEV). Its tertiary structure is compared with that of severe acute respiratory syndrome (SARS) coronavirus PL2pro, a downstream paralog that is conserved in the nsp3's of all coronaviruses. We identify both conserved and unique structural features likely controlling the interaction of PL1pro with cofactors and substrates, including the tentative mapping of substrate pocket residues. The purified recombinant TGEV PL1pro was shown to cleave a peptide mimicking the cognate nsp2|nsp3 cleavage site. Like its PL2pro paralogs from several coronaviruses, TGEV PL1pro was also found to have deubiquitinating activity in an in vitro cleavage assay, implicating it in counteracting ubiquitin-regulated host cell pathways, likely including innate immune responses. In combination with the prior characterization of PL2pro from other alphacoronaviruses, e.g., human coronaviruses 229E and NL63, our results unequivocally establish that these viruses employ two PLpros with overlapping specificities toward both viral and cellular substrates.
Coronaviruses (CoVs) are enveloped, positive-stranded RNA viruses with a large genome of 26 to 31 kb. They belong to the order Nidovirales and include important pathogens of humans and other animals (22, 66). The coronavirus nonstructural proteins (nsp's) are encoded within the first two open reading frames (ORFs) (ORF1a and ORF1b), which comprise approximately the 5′-proximal two-thirds of the viral genome. These two ORFs are translated into two large polyproteins, pp1a and pp1ab, with the expression of the latter involving a ribosomal frameshift mechanism (8). The autoproteolytic processing of pp1a and pp1ab gives rise to a total of 15 or 16 mature nsp's. These proteins assemble into the viral replicase/transcriptase complex (RTC), which is associated with an intricate network of modified endoplasmic reticulum (ER) membranes and supports the synthesis of genomic RNA (replication) and subgenomic mRNAs (transcription) (36, 55, 61, 62).
The recently established Coronavirinae subfamily of the family Coronaviridae is subdivided into the genera Alphacoronavirus, Betacoronavirus, and Gammacoronavirus, which include phylogenetically compact genogroups (25, 38) whose numbers are growing rapidly (45, 67). The genus Alphacoronavirus (formerly known as CoV group 1) includes subgroups 1a and 1b, which are prototyped by human coronavirus 229E (HCoV-229E) and HCoV-NL63, as well as the newly established species Alphacoronavirus 1 (including porcine transmissible gastroenteritis virus [TGEV]), respectively. The genus Betacoronavirus (formerly CoV group 2) includes several subgroups, with the most prominent (subgroups 2a and 2b) being prototyped by the species Murine coronavirus (including mouse hepatitis virus [MHV]) and Severe acute respiratory syndrome-related coronavirus, respectively. The genus Gammacoronavirus includes all avian coronaviruses identified until 2009.
Coronaviruses encode two types of cysteine proteases that have narrow substrate specificities: a chymotrypsin-like main protease (Mpro, contained in nsp5) and one or two papain-like proteases (PL1pro and PL2pro, residing in nsp3) (70). Their main function is pp1a and pp1ab processing. Both PLpro domains reside in the N-terminal half of nsp3 and cleave the nsp1|nsp2, nsp2|nsp3, and nsp3|nsp4 sites, while all sites downstream of nsp4 are processed by the nsp5 main protease. Coronaviruses have evolved different ways to process the N-terminal part of pp1a/pp1ab, with no less than four functional patterns being recognized (69). The nsp3 subunits of alphacoronaviruses and subgroup 2a betacoronaviruses contain two active PLpro domains. The processing of the N-terminal part of pp1a/pp1ab in the species Alphacoronavirus 1 is poorly characterized. TGEV PL1pro processes the nsp2|nsp3 site (56) and may be involved in the processing of nsp1|nsp2 and nsp3|nsp4 cleavage sites, whose precise positions remain to be identified. For the alphacoronavirus HCoV-229E, it was shown previously that its papain-like proteases have overlapping specificities toward multiple sites in the replicase polyproteins and that PL1pro is dispensable for virus replication (69). In the case of MHV, PL1pro is responsible for the processing of the nsp1|nsp2 and nsp2|nsp3 cleavage sites, while PL2pro is dedicated to the nsp3|nsp4 cleavage site (26). Severe acute respiratory syndrome CoV (SARS-CoV) nsp3 contains only the PL2pro domain (often called PLpro), which cleaves all three sites in the region of nsp1 to nsp4 (27, 28). Until now, SARS-CoV PL2pro was the only structurally characterized representative of the coronaviral PLpro enzymes (58). Gammacoronaviruses encode an active PL2pro (44) but also carry an inactive remnant of PL1pro (71). Moreover, their replicase lacks an nsp1 moiety, and therefore, PL2pro cleaves only the nsp2|nsp3 and nsp3|nsp4 sites.
Apart from being responsible for replicase polyprotein processing, PL2pro domains possess an additional but related enzymatic activity: in HCoV-NL63 (11), MHV (68), and SARS-CoV (5, 12, 42), they have been shown to be deubiquitinating enzymes (DUBs). Like ubiquitinating enzymes, DUBs are key players in modulating the innate immune response that ultimately leads to interferon (IFN) secretion and the upregulation of interferon-stimulated genes (ISGs) to induce an antiviral state in the host cell. A variety of signaling events in this cascade is mediated by ubiquitination and can thus potentially be countered by the expression of viral DUBs. It was proposed previously that the CoV PL2pro DUB activity may directly affect the transcription factor interferon regulatory factor 3 (IRF3) (68). However, the DUB activity of SARS-CoV PL2pro appears to target all proteins in the cell, including IRF3, suggesting that its activity may not be restricted to IRF3 alone (21). In addition, the PL2pro domains of SARS-CoV and HCoV-NL63 are also able to remove ISG15, a ubiquitin-like protein whose expression and activity are tightly regulated by the innate immune response (12, 43).
Here we report the first structure of a coronaviral PL1pro domain, which was determined for the alphacoronavirus TGEV. In in vitro cleavage assays, we also characterized the purified protease enzymatically, using substrates representing both viral and cellular targets. We show that TGEV PL1pro possesses DUB activity and displays a slight preference for Lys48-linked polyubiquitin chains over Lys63-linked ones.
MATERIALS AND METHODS
TGEV PL1pro expression and purification.
The sequence encoding the TGEV PL1pro domain (residues 1062 to 1319 of the polyprotein pp1a from TGEV strain Purdue 46-MAD; GenBank accession number AJ271965.2 [http://www.ncbi.nlm.nih.gov/GenBank/index.html]) (6) was delineated with the assistance of Viralis software (24), using an updated version of a previously reported multiple-sequence alignment of coronavirus nsp3 (71). The domain was PCR amplified from a full-length PUR46-MAD cDNA clone (kindly provided by Luis Enjuanes, Madrid, Spain) and cloned into the pETM-11 vector (EMBL Hamburg). Protein expression was performed with Escherichia coli strain Rosetta (DE3)pLysS cells (Novagen). Cultures were grown in LB medium at 37°C until the optical density (A600) reached 0.8, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and left shaking overnight at 15°C. Cells were collected by centrifugation at 4,000 × g (30 min at 4°C) and frozen at −20°C. The bacterial pellet from a 1-liter culture was resuspended in 20 ml of lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole, and 1 mM MgCl2). Cells were lysed by sonication and centrifuged at 38,000 × g for 50 min at 4°C. The supernatant was filtered through a 0.45-μm-pore-size membrane (Sartorius Stedim Biotech), loaded onto 5 ml of Ni-nitrilotriacetic acid (NTA) beads (Qiagen), preequilibrated with lysis buffer, and incubated for 30 min at 4°C. Beads were then washed with high-salt buffer (50 mM Tris [pH 8.0], 500 mM NaCl), and the protein was eluted with a solution containing 50 mM Tris (pH 8.0), 150 mM NaCl, and 500 mM imidazole. The fractions collected were analyzed by SDS-PAGE, and the buffer of the protein sample was exchanged for buffer A (50 mM Tris [pH 8.0], 150 mM NaCl, 5% glycerol) using PD-10 columns (GE Healthcare). The recombinant protein was incubated overnight with the His6-tagged main protease from tobacco etch potyvirus (TEV) (TEV protease; EMBL Hamburg) at 4°C (at a 50:1 ratio). The sample was then loaded onto Ni-NTA beads equilibrated with buffer A. The flowthrough was collected, and the beads were washed with 20 ml of buffer A. The fractions collected were analyzed by SDS-PAGE. The cleaved protein sample was loaded onto a Superdex 75 gel filtration column (GE Healthcare) equilibrated with buffer A, and the purity of the sample was checked by SDS-PAGE. The protein solution was concentrated to 8.5 mg/ml by using a 30-kDa-molecular-mass-cutoff centrifuge concentrator (Vivaspin; Vivascience), and the concentration was determined with a NanoDrop spectrophotometer (Thermo Scientific). The protein sample was stored at 4°C for up to 1 week or flash-frozen in the presence of 20% glycerol and kept at −80°C for up to 6 months.
Protein crystallization.
The first crystallization trials were carried out by using the sitting-drop vapor diffusion method with 96-well plates (Greiner) at 19°C at the EMBL Hamburg High-Throughput Crystallization Facility (47). Crystals were obtained from a solution containing 0.2 M (NH4)2SO4, 0.1 M MES (morpholineethanesulfonic acid) (pH 6.5), and 30% polyethylene glycol (PEG) 5000. Further optimization of the crystallization conditions was performed manually with 24-well plates (Qiagen) by using the hanging-drop vapor diffusion method at 19°C. Crystals were obtained from a 2-μl drop of 8.5 mg/ml protein in a solution containing 0.1 to 0.18 M (NH4)2SO4, 0.1 M MES (pH 6.5), and 30% PEG 5000.
Data collection and processing.
A crystal from the crystallization drop was loop mounted, directly transferred into liquid nitrogen, and stored. The single-wavelength X-ray diffraction data were collected from the single crystal at 100 K with the help of an SC3 sample changer on the ESRF ID23-2 beamline equipped with a MAR225 charge-coupled-device (CCD) detector. The crystal-to-detector distance was maintained at 280.3 mm, and an oscillation range of 1° was used. Two hundred images were collected to a maximum resolution of 2.5 Å.
A second crystal was soaked in reservoir solution containing 1 mM KAu(CN)2 for 10 min and quickly transferred into a nitrogen gas stream at 100 K. The single-wavelength X-ray diffraction data were collected from a single crystal at 100 K on EMBL beamline X13 at DESY by using a MAR165 CCD detector. Data were measured with a crystal-to-detector distance of 250 mm using an oscillation range of 1°. Three hundred sixty images were collected to a maximum resolution of 2.6 Å.
For both data sets the recorded images were processed with XDS (35), and the reflection intensities were processed with COMBAT and scaled with SCALA (19) from the CCP4 program suite (13). Data collection statistics are shown in Table 1.
TABLE 1.
Data collection statistics
| Parameterb | Value for data set |
|
|---|---|---|
| Native | Derivative | |
| X-ray source | ID 23-2 | X13 |
| Space group | P41212 | P41212 |
| Unit cell parameters (Å) | a = b = 62.5, c = 199.4 | a = b = 61.1, c = 192.1 |
| Wavelength (Å) | 0.873 | 0.801 |
| Resolution range (Å) | 50.0-2.5 | 50.0-2.6 |
| Highest resolution shell | 2.65-2.50 | 2.76-2.61 |
| Mosaicity | 0.18 | 0.26 |
| Mean I/σ(I) | 9.7 (2.0)a | 38.3 (10.6)a |
| Rfac (linear) (%) | 16.0 (77.8)a | 6.3 (27.3)a |
| Redundancy | 8.3 | 15.2 |
| Rmeas (%) | 17.0 (83.0)a | 6.5 (28.4)a |
| No. of observations | 215456 | 321582 |
| No. of unique reflections | 25792 | 21221 |
| Completeness (%) | 99.2 (95.1)a | 99.4 (96.5)a |
Numbers given in parentheses are from the highest resolution shell.
Rfac = [ΣhklΣi|Ii(hkl) − <I(hkl)>1)/ΣhklΣIi(hkl)], where Ii(hkl) is the intensity of the ith measurement of reflection (hkl) and <I(hkl)> is the average intensity. Rmeas = (Σhkl[1/N − 1]1/2Σi|Ii(hkl) − <I(hkl)>1)/ΣhklΣIi(hkl), where Ii(hkl) is the intensity of the ith measurement of reflection (hkl), <I(hkl)> is the average intensity, and N is the number of measurements (or redundancy).
Structure determination.
The structure was solved by using the SAD protocol of Auto-Rickshaw, the automated crystal structure determination platform (50). A single heavy-atom position was found by using the program SHELXD (60) and refined with the program BP3 (51, 52). The initial phases were improved by using density modification with the program DM (14), and a partial model was produced by using the programs HELICAP (46) and BUCCANEER (15). The resulting model, containing 169 residues, was used as an input for the MRSAD protocol of Auto-Rickshaw. CNS (9, 10) refinement and heavy-atom refinement with MLPHARE (49) were performed. The resultant phases were used to continue model building by using the program ARP/wARP (53), resulting in the placement of 173 residues in the electron density. A second run of the MRSAD protocol of Auto-Rickshaw led to a model containing 195 residues. Further model building was performed manually by using the COOT graphics program (18). Refinement was carried out by using the program REFMAC5 (48). The stereochemistry of the model was evaluated with the program MOLPROBITY (16).
In vitro assay of the TGEV PL1pro enzymatic activity.
An assay to determine the cleavage of a fluorescent resonance energy transfer (FRET) peptide substrate, DABCYL-MYNKMGGGDKTVSF(E-EDANS)-amide (where DABCYL stands for 4-{[4-(dimethyloamino)phenyl]azo}benzoic acid and EDANS stands for 5-[(2-aminoethyl)amino]naphthalene-1-sulfonic acid; Alta Bioscience), was carried out by using different substrate concentrations (10 to 50 μM) and 0.5 μM purified TGEV PL1pro. Assays were performed with a 96-well microplate (Greiner Bio-One) using a Safire fluorimeter (Tecan). The reaction was carried out in the presence of a solution containing 50 mM Tris (pH 8.0), 150 mM NaCl, 5% glycerol, and 0.5 mM Tris(2-carboxyethyl)phosphine at 30°C. The rate of substrate hydrolysis was determined by monitoring the fluorescence as a function of time (excitation λ, 330 nm; emission λ, 490 nm). Initial velocities were calculated from the linear part of the curves. Since no saturation was observed in the plot of initial velocities versus substrate concentrations, data points were fitted to the equation v/[E]tot = kapp[S], where v is the initial velocity (μm/min), [E]tot is the enzyme concentration (μM), kapp is the pseudo-first-order rate constant (assuming [S] ≪ Km) [1/(μM·min)], and [S] is the substrate concentration (μM). For this equation, the kapp approximates kcat/Km.
TGEV PL1pro deubiquitination assays.
The deubiquitinating activity of purified TGEV PL1pro was assessed by using the fluorogenic substrate ubiquitin-7-amino-4-methylcoumarin (AMC) (Ub-AMC) (Enzo Life Sciences). The reaction was carried out in the presence of a solution containing 50 mM Tris (pH 8.0), 150 mM NaCl, 5% glycerol, and 0.2 to 0.8% dimethyl sulfoxide (DMSO) at 26°C. Assays were performed with a 96-well microplate (Greiner Bio-One) using a Safire fluorimeter (Tecan). Reaction mixtures contained 0.5 μM protein and different concentrations of Ub-AMC (0.2 to 1 μM). The fluorescence of the released AMC was monitored continuously (excitation λ, 380 nm; emission λ, 460 nm). The initial velocities were plotted against the Ub-AMC concentration and fitted to the equation presented above to determine the pseudo-first-order rate constant. The standard curve was generated by measuring the fluorescence of AMC dissolved in the reaction buffer at different concentrations (range, 0.05 to 0.5 μM).
As an alternative substrate we employed Lys48- and Lys63-linked polyubiquitin chains (Boston Biochem), of which 2.5 μg was incubated in a 10-μl volume of buffer B (50 mM Tris [pH 7.5], 5 mM MgCl2, 2 mM dithiothreitol [DTT]) and 2-fold serial dilutions of purified TGEV PL1pro (final enzyme concentrations, 50, 25, 12.5, and 6.25 ng/μl) or isopeptidase T (final concentration, 50 ng/μl) as a positive control. The reaction mixtures were incubated for 2 h at 37°C and subsequently mixed with Laemmli sample buffer and loaded onto a 15% SDS-PAGE gel. Protein bands were visualized by using Coomassie brilliant blue staining, and gels were scanned by using a GS-800 calibrated densitometer (Bio-Rad).
Protein structure accession number.
Atomic coordinates and structure amplitudes reported here have been deposited in the Protein Data Bank (PDB) under accession number 3MP2.
RESULTS
Protein production, crystallization, and structural studies.
To clone the TGEV PL1pro domain into bacterial expression vector pETM-11, a sequence encoding a 258-amino-acid domain of nps3 (pp1a residues 1062 to 1319, hereafter numbered residues 1 to 258) was PCR amplified. The expression product contained an additional 28 amino acids at the N terminus, which included a hexahistidine tag and a TEV main protease cleavage site. TEV protease treatment removes most of the tag, leaving 4 of the additional N-terminal residues. The N-terminally modified TGEV PL1pro domain was expressed in E. coli cells, yielding up to 30 mg per liter of cell culture, and purified with two metal ion affinity chromatography steps (before and after TEV protease cleavage), followed by a size-exclusion chromatography step. The purified protein was found to be monomeric in solution and was crystallized overnight as described in Materials and Methods. Molecular replacement with SARS-CoV PL2pro structure coordinates (PDB accession number 2FE8) as a search model was unsuccessful. Therefore, the structure was solved ab initio with the SAD technique using anomalous scattering from a classical gold derivative soak. The TGEV PL1pro crystals belonged to the space group P41212 with the following unit cell dimensions: a = b = 62.5, c = 199.4 Å. There was 1 protein molecule per asymmetric unit. The structure was determined to a resolution of 2.5 Å and refined to a final R value of 17.3% (Rfree = 22.3%). Refinement statistics are shown in Table 2. Residues 10 to 220 could be placed into the electron density. The Ramachandran plot shows 95.2% of the residues in the preferred region and 4.8% of the residues in the allowed region. The refined model also contained 1 zinc ion and 173 solvent molecules.
TABLE 2.
Refinement statistics
| Parametera | Value |
|---|---|
| Resolution range (Å) | 43.1-2.5 |
| No. of working reflections/no. of free reflections | 13,743/718 |
| No. of protein residues | 211 |
| No. of waters | 173 |
| No. of zinc ions | 1 |
| Rwork/Rfree (%) | 17.3/22.3 |
| Avg B (Å2) | 37.4 |
| RMS deviations from ideal values | |
| Bond lengths (Å) | 0.022 |
| Bond angles (°) | 1.9 |
Rwork = Σhkl|Fo − Fc|/ΣhklFo, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree was calculated as described above for Rwork but from a randomly selected subset of the data (5%), which were excluded from the refinement.
Overall structure of TGEV PL1pro.
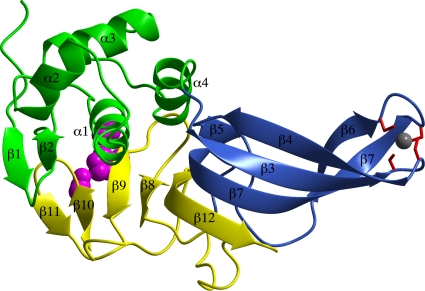
The structure of TGEV PL1pro resembles a right hand with three distinct regions, the palm, thumb, and finger (Fig. 1). The thumb region is formed by a two-stranded β sheet (β1 and β2) and four α helices (α1 to α4). The finger region is formed by a short-strand β5 and an antiparallel, twisted β sheet with the strand order β4-β3-β7-β6. In the tip region of the finger domain, four cysteine residues, Cys103, Cys105, Cys131, and Cys134 (forming the motif CXC-X25-CXXC), chelate the zinc ion. This zinc-binding domain belongs to the zinc ribbon fold (37), as was predicted previously by protein modeling (31). The presence of the zinc in the crystal of TGEV PL1pro was confirmed by an energy scan over the zinc absorption edge. The finger and palm regions are connected by a long, 17-amino-acid loop that contains a β-turn. Strands β8 to β12 make up the palm region. Together with finger strands β5 and β7, they form a seven-stranded β sheet with the strand order β5-β7-β12-β8-β9-β10-β11. With the exception of β5, the strands are antiparallel. The C-terminal part of the twisted strand β12 also participates in the formation of the antiparallel sheet β4-β3-β7-β12 present in the finger region. The strand and helix numberings used here are necessarily different from those of SARS-CoV PLpro (58), first because the structure reported here does not have the N-terminal ubiquitin-like domain and second because subtle structural differences (described below) exist.
FIG. 1.
Ribbon representation of the TGEV PL1pro structure. The thumb region is shown in green, the fingers are shown in blue, and the palm is shown in yellow. Residues Cys32, His183, and Asp196 form the catalytic triad and are shown as magenta spheres. Cysteine residues Cys103, Cys105, Cys131, and Cys134, which are coordinating the zinc ion (a gray sphere), are shown as red cylinders. The figure was created with the molecular graphics program ccp4 mg (54).
TGEV PL1pro includes the spatially proximal residues Cys32, His183, and Asp196, which are implicated in catalysis; they will be referred to as the catalytic triad, although Asp196 may play only a stabilizing role for the other two residues (see Discussion). Cys32 is positioned at the N terminus of α1 in the thumb region. Both the histidine and the aspartic acid residue are located in the palm region. His183 is positioned at the N terminus of β9, and Asp196 is situated at the C terminus of β10. The distance between Cys32 and His183 is 3.8 Å, and that between His183 and Asp196 is 3.3 Å.
The TGEV PL1pro structure was compared with all structures in the PDB by using the DALI server (http://ekhidna.biocenter.helsinki.fi/dali_server/). It is very similar to that of SARS-CoV PL2pro (Z score of 18.4); to cellular DUBs (human ubiquitin-specific proteases [USPs] 2, 7, 8, 14, and 21 and yeast Ubp6, Z scores of 12.6 to 11.2); as well as to the foot-and-mouth disease virus leader protease (Lpro) (Z score of 10.9). PL2pro and the USPs, but not Lpro, also include a Zn ribbon between the two domains that give rise to the papain-like fold.
Structural comparison of TGEV PL1pro and SARS-CoV PL2pro.
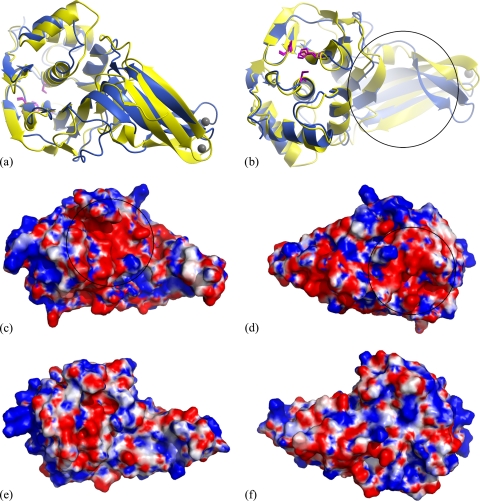
The crystal structure of TGEV PL1pro is similar to that reported previously for SARS-CoV PL2pro (58), with a 3.1-Å root mean square (RMS) deviation for 202 superimposed Cα atoms. The overall superposition of the structures is excellent in the thumb and palm regions, whereas the β strands of the fingers do not superimpose as well, even though the structural organization of the zinc-binding site in the fingertip region (Fig. 2a and b) is similar. The residues of the proposed catalytic triad are very well aligned. Interestingly, there are several regions with significant structural differences. SARS-CoV PL2pro has a relatively evenly distributed electrostatic surface potential with no apparent clusters of charged residues (Fig. 2e and f), whereas TGEV PL1pro has a more negatively charged surface with two distinguishable patches (Fig. 2c and d). The side of TGEV PL1pro opposite of the active site (Fig. 2d) has a lobe of negative charge, which mainly includes residues from loops Lβ1-β2, Lβ9-β10, and Lα1-α2 and helix α1. These are parts of the structure that differ significantly from that of the corresponding region in SARS-CoV PL2pro. A second large region of negative charge in PL1pro includes the active-site groove and the surrounding region, which is responsible for substrate binding and specificity (Fig. 2c). It includes residues from loop Lα3-α4, helix α4, the N-terminal part of region II (see below), as well as residues Tyr175, Tyr184, and Thr209.
FIG. 2.
Comparison of the TGEV PL1pro and SARS-CoV PL2pro structures. (a and b) Superposition of the TGEV PL1pro and SARS-CoV PL2pro structures shown in two orientations (rotated 180° around the horizontal axis). The TGEV PL1pro structure is shown in light blue, and the SARS-CoV PL2pro structure is shown in yellow. The catalytic triad residues are shown as magenta sticks, and zinc ions are shown as gray spheres. The black circle indicates the region where the part of the substrate upstream of the recognition site is predicted to bind. (c to f) Electrostatic surface potential of TGEV PL1pro (c and d) and SARS-CoV PL2pro (e and f). The electrostatic surface was calculated with the APBS plug-in for PyMOL (1, 40) and is rendered in blue and red to illustrate positive and negative electrostatic potentials, respectively, in the spectrum ranging from +4 kBT/e to −4 kBT/e. Two regions of the TGEV PL1pro surface with negative charge are labeled with black circles.
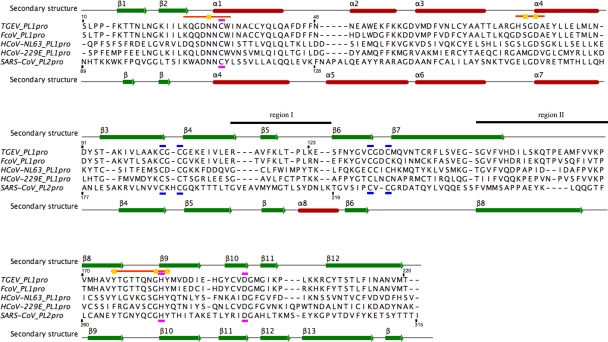
There are two regions, named regions I and II, that differ most substantially in terms of secondary structure in the two proteases. In TGEV PL1pro, region I lies between strands β4 and β6 and comprises a part of the Zn ribbon (Fig. 3). It consists of 10 amino acids (residues 115 to 124) and contains the short strand β5 (residues 116 to 118), which is the outermost strand of the structure's main, seven-stranded, β sheet. The corresponding region in the SARS-CoV PL2pro structure, located between β5 and β6, contains 17 amino acids (residues 203 to 219) and includes an α-turn, a short β strand (residues 207 to 209), and α8 (residues 213 to 219). Region II of the TGEV PL1pro structure consists of 23 residues, residues 147 to 169. It includes the C-terminal part of β7 and loop Lβ7-β8, which contains a β-turn. Its counterpart in the SARS-CoV PL2pro structure (residues 240 to 259) consists of 20 residues and includes loop Lβ7-β8, strand β8 (residues 242 to 255), and loop Lβ8-β9. In the TGEV PL1pro structure the C-terminal parts of regions I and II are in proximity to each other (the distance between the Cα's of Val166 and Lys123 is 5.1 Å) and interact through two main-chain hydrogen bonds (Fig. 4a). In contrast, in the SARS-CoV PL2pro structure the C-terminal parts of regions I and II are far apart (the distance between Cα's of Gln256 and Thr219 is 20.5 Å) and do not interact.
FIG. 3.
Multiple-sequence alignment of coronaviral R-group papain-like proteases. The alignment is based on structures of TGEV PL1pro and SARS-CoV PL2pro (PDB accession number 2FE8; molecule A). The secondary structure of TGEV PL1pro is shown on the top of the alignment, and that of SARS-CoV PL2pro is shown on the bottom. Regions I and II are labeled. Cysteine residues chelating the zinc ion are indicated by dashed blue lines, and the catalytic triad residues are indicated by dashed magenta lines. Orange lines indicate binding-site signature motifs, while yellow squares highlight USP-like binding-site signature residues (based on data from reference 63). Accession numbers (and sequence ranges [based on the numbering of the polyprotein pp1a]) of papain-like proteases are as follows: FCoV PL1pro, NC_007025.1 (residues 1018 to 1228); HCoV-NL63 PL1pro, NC_005831.2 (residues 1039 to 1250); HCoV-229E PL1pro, AF304460.1 (residues 1032 to 1246). The structure-based alignment was generated with the Dali server (http://ekhidna.biocenter.helsinki.fi/dali_lite/start) using the DaliLite pairwise option (29). Using Clustal 2.0.12 (39) in the ClustalX environment and the sequences-to-profile mode, the structural alignment was extended to include sequences of three additional papain-like proteases. The resulting alignment was visualized with Jalview (65).
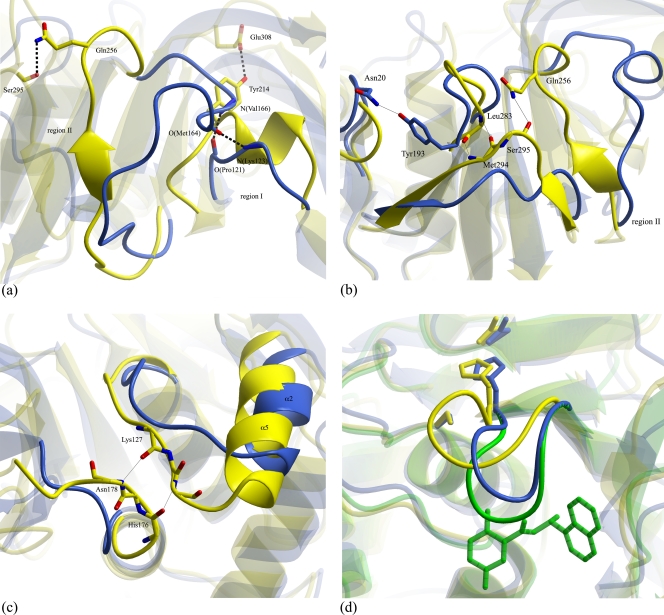
FIG. 4.
Structural differences between TGEV PL1pro and SARS-CoV PL2pro. The TGEV PL1pro structure is shown in light blue, the SARS-CoV PL2pro structure is shown in yellow, the SARS-CoV structure in an inhibitor-bound form is shown in green, oxygen atoms are shown in red, and nitrogen atoms are shown in blue. (a) Differences in C-terminal parts of regions I and II. Hydrogen bonds formed between O(Met164)-N(Lys123), N(Val166)-O(Pro121), Oη(Tyr214)-Oɛ2(Glu308), and Nɛ2(Gln256)-O(Ser295) are shown with dashed lines. (b) Region neighboring region II. Hydrogen bonds formed between Oη(Tyr193)-Oδ(Asn20), Nɛ2(Gln256)-O(Ser295), N(Met294)-O(Leu283), and O(Met294)-N(Leu283) are shown with thin black lines. (c) Structural differences in the region between α1 (SARS-CoV α4) and α2 (SARS-CoV α5). The main chain of residues 127 to 129 and 176 to 178 is shown. Hydrogen bonds formed between O(Lys127)-N(Asn178) and N(Asn129)-O(His176) are shown with thin black lines. (d) Highly mobile loop (residues 177 to 182). The inhibitor GRL0617 is shown as green sticks. Catalytic triad residues Cys34 (SARS-CoV Cys112), His183 (SARS-CoV His273), and Asp196 (SARS-CoV Asp287) are shown as sticks. The figure was created with the molecular graphics program ccp4 mg (54).
Loop Lβ12-β13 in the SARS-CoV PL2pro structure interacts with region II (as described above) and with loop Lβ10-β11 through three hydrogen bonds (Fig. 4b). In the TGEV PL1pro structure, strand β11 is shorter than the corresponding strand β12 in SARS-CoV, and loop Lβ11-β12 (equivalent to SARS-CoV loop Lβ12-β13) is 4 residues longer. This results in a different orientation of this part of the structure. In the SARS-CoV PL2pro structure, helix α5 consists of 15 residues, and loop Lα4-α5 interacts with loop Lα7-β4 through two hydrogen bonds (Fig. 4c). In contrast, in the TGEV PL1pro structure, helix α2 consists of only 9 residues, and loops Lα1-α2 (SARS-CoV loop Lα4-α5) and Lα4-β3 (SARS-CoV loop Lα7-β4) do not interact. It is notable that this difference between the SARS-CoV and TGEV structures maps to a region that is distant from the active site, including the substrate-binding region.
SARS-CoV PL2pro loop Lβ9-β10 (267-GNYQCG-272) was previously described to be a highly mobile loop, which is differently positioned in the three monomers of the asymmetric unit (58). This loop corresponds to so-called loop BL2 in the USP proteins. Loop BL2 (427-GRSSSSG-433), together with loop BL1 (not present in coronaviral PLpros), was proposed previously to modulate the DUB activity of USP14 (33). Two glycine residues flanking this loop are conserved in PLpros (and several other USPs, including USP2, USP7, USP8, and USP14) and might be responsible for the high flexibility of this region. It was also shown previously that in the SARS-CoV PL2pro structure, this loop significantly changes conformation upon the binding of the inhibitor GRL0617 (57). Interestingly, the corresponding TGEV PL1pro loop, Lβ8-β9 (177-GTTQNG-182), has a conformation similar to that of the inhibitor-bound form of SARS-CoV PL2pro (Fig. 4d).
Substrate specificity of PL1pro.
The cleavage specificity of SARS-CoV PL2pro is restricted to amino acids at substrate positions P4 to P1 (5, 27, 64). Based on the three sites within the polyprotein pp1a/pp1ab that are cleaved, it was concluded that LXGG↓ is the consensus sequence recognized by this protease (64). TGEV PL1pro cleaves at the nsp2|nsp3 site (56), whose P4-P1 positions are occupied by the KMGG peptide, resembling the LXGG peptide recognized by SARS-CoV PL2pro. This similarity enables modeling to locate substrate-binding residues in the TGEV PL1pro structure by superimposing it on SARS-CoV PL2pro bound to a ubiquitin aldehyde (C-terminal sequence LRGG), as examined by previously reported docking studies (58).
It was shown previously that the oxyanion hole of SARS-CoV PL2pro is stabilized by Tyr107. The replacement of this residue by alanine abolished the protease activity (58). In TGEV PL1pro the spatially equivalent position is occupied by Gln27 (3.2 Å away from Asp196 and 3.6 Å away from His183). The highly conserved Asn110 of SARS-CoV PL2pro (Asn30 in TGEV PL1pro) was also proposed to contribute to the stabilization of the oxyanion hole (58). The region between Thr103 and Asn110 in SARS-CoV PL2pro (region between Ile23 and Asn30 in TGEV PL1pro) limits access to the narrow active site. The hydrogen bond between Oδ2(Asp109) and Nɛ1(Trp94) was proposed to prevent the collapse of this region into the active site. In TGEV PL1pro there is no interaction between the corresponding residues, Asp29 (SARS-CoV PL2pro Asp109) and Phe14 (SARS-CoV PL2pro Tyr94). However, Asp29 forms a hydrogen bond with Arg75 [Oγ1(Asp29)-Nɛ(Arg75)], which may keep the region between Ile23 and Asn30 in the same orientation as that in the case of SARS-CoV PL2pro.
SARS-CoV PL2pro requires glycine residues in the P1 and P2 positions of its substrate. Two tyrosine residues (Tyr113 and Tyr274) as well as Asn110 and Leu163 of the substrate pocket may be partially responsible for this strict requirement. In TGEV PL1pro, the substrate of which also has glycines in the P1 and P2 positions, Trp33 and Tyr184 are spatially equivalent to SARS-CoV PL2pro Tyr113 and Tyr274, respectively. The TGEV PL1pro S1 subsite is also formed by Asn30 and Ser78 (Asn110 and Leu163 in SARS-CoV). One of the main determinants of substrate specificity at the P1 and P2 positions in papain-like proteases seems to be the above-described highly mobile loop (TGEV PL1pro loop Lβ8-β9). Interestingly, in the case of SARS-CoV PL2pro, docking studies did not reveal interactions between the substrate and this loop.
In the proposed model of the SARS-CoV PL2pro (58), the S3 and S5 subsites are occupied by arginine residues. It was shown that the backbone of the substrate's P3 residue is stabilized by an interaction with Tyr256, while P5 is stabilized by Asp165. Residues Tyr175 and Asp80 of TGEV PL1pro may stabilize the backbone of the Met and Arg residues in the substrate P3 and P5 positions. SARS-CoV PL2pro Glu168, which was proposed to interact with the side chains of the P3 and P5 arginines, is replaced by Tyr83 in TGEV PL1pro, the side chain of which is facing away from the substrate-binding site.
The P4 position is occupied by very different residues, Leu and Lys, in substrates of SARS-CoV PL2pro and TGEV PL1pro, respectively. Accordingly, their S4 subsites are formed by different residues. In the SARS-CoV PL2pro structure, S4 resides in a hydrophobic cleft created by residues Pro249, Thr302, and Tyr265. In the TGEV PL1pro structure, Tyr175 and Thr209 form the spatially equivalent site, while residue Pro249 is replaced by Ile155. The Ile155 Cα atom is 3 Å away from the corresponding proline's Cα. The displacement of this part of the TGEV PL1pro structure may create enough space to accommodate the side chain of a lysine at the substrate's P4 position.
Coronaviral papain-like proteases have narrow substrate specificities, which seem to be modulated by subtle changes in the specificity pockets. In order to fully understand the TGEV PL1pro substrate recognition mechanism, the nsp1|nsp2 and nsp3|nsp4 sites in the TGEV replicase polyproteins need to be identified.
Peptide-based in vitro cleavage assay.
We extended the characterization of purified TGEV PL1pro to determine its proteolytic activity toward peptides mimicking viral and cellular substrates. First, the enzyme was used to determine the kinetic parameters of the cleavage of a synthetic substrate, which consisted of 14 amino acids encompassing the nsp2|nsp3 cleavage site, including P4-P1 residues KMGG, which determine cleavage specificity. DABCYL and EDANS moieties were fused to the N and C termini of the peptide, respectively, to allow the detection of peptide hydrolysis by using a FRET-based assay. Upon the cleavage of the DABCYL-MYNKMGGGDKTVSF(E-EDANS)-amide substrate, the fluorescence intensity (measured at 490 nm) increased, because the released EDANS was no longer quenched by the DABCYL group.
Because no saturation was observed in the plot of initial velocities versus substrate concentrations, the pseudo-first-order rate constant, kapp, which approximates kcat/Km, was determined. TGEV PL1pro hydrolyzes the short synthetic DABCYL-MYNKMGGGDKTVSF(E-EDANS)-amide substrate with a kapp rate constant of 1.81 ± 0.03 min−1 mM−1. This value is very similar to the SARS-CoV PL2pro rate constants obtained for the hydrolysis of the substrate Z-LRGG-AMC (3.6 min−1 mM−1) (42) and (E-EDANS)RELNGGAPI(K-DABCYL)S (24.4 min−1 mM−1) (5).
In vitro deubiquitinating activity of TGEV PL1pro.
The P4-P1 KMGG specificity sequence of the TGEV PL1pro nsp2|nsp3 cleavage site resembles the C-terminal ubiquitin sequence LRGG that is recognized by a variety of viral and cellular DUBs. It was previously shown that SARS-CoV PL2pro, which recognizes the LXGG consensus sequence, possesses DUB activity (5, 42, 43). Based on these observations and structure modeling, PL1pro of alphacoronaviruses was predicted to also possess DUB activity (63), which we probed using different substrates in two types of in vitro assays.
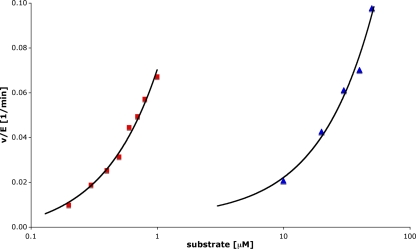
DUB activity was first tested in a reaction using the fluorescent substrate ubiquitin-AMC (Ub-AMC). The change of fluorescence intensity was measured over time at different substrate concentrations. TGEV PL1pro was found to cleave the bond between ubiquitin and AMC, resulting in the release of the fluorescent dye and an increase of the fluorescence intensity. Since the initial rate of Ub-AMC hydrolysis increased linearly with the substrate concentration, the data were fitted to the same equation as that used for analyzing the data from the peptide cleavage assay (see above). The pseudo-first-order rate constant for TGEV PL1pro was determined to be 74 ± 1 min−1 mM−1, significantly lower than the rate constants previously determined for SARS-CoV PL2pro: 4,480 min−1 mM−1 (60×) (5), 1,188 min−1 mM−1 (16×) (43), and 786 min−1 mM−1 (10×) (42). However, this value is higher than that of USP7 (kcat/Km = 13 min−1 mM−1) (32). TGEV PL1pro, SARS-CoV PL2pro, and the USP7 have low affinities for the Ub-AMC substrate but quite high turnover rates. Interestingly, the preference of TGEV PL1pro for the Ub-AMC substrate is much higher than for the FRET peptide, as seen from a 40-times-higher kapp (Fig. 5).
FIG. 5.
TGEV PL1pro in vitro activity assays. The hydrolysis of ubiquitin-AMC (squares) and cleavage of DABCYL-MYNKMGGGDKTVSF(E-EDANS)-amide (triangles) were measured at different substrate concentrations (0.2, 0.3, 0.4, 0.5, 0.6. 0.7, 0.8, and 1 μM in the case of the Ub-AMC and 10, 20, 30, 40, and 50 μM in the case of the FRET peptide). The TGEV PL1pro concentration was 0.5 μM. For both experiments the initial velocities were calculated and data were fit to the equation (see text), which allowed the determination of the kapp value. For clarity, a logarithmic scale was used for the horizontal axis.
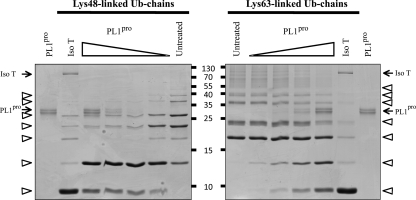
To confirm the results obtained with the fluorimetric assay and examine whether TGEV PL1pro can hydrolyze isopeptide bonds, an in vitro dose-range experiment with commercially available polyubiquitin substrates was performed. Lys63- and Lys48-linked polyubiquitin chains were incubated with different amounts of purified TGEV PL1pro. The samples were analyzed on an SDS-PAGE gel and visualized with Coomassie staining (Fig. 6). The results clearly show that TGEV PL1pro can process both types of polyubiquitin chains but seems to have a slight preference for Lys48-linked ones. In conclusion, both in vitro assays convincingly demonstrate that TGEV PL1pro is a multifunctional protease that possesses DUB activity.
FIG. 6.
Dose-range experiment with Lys48- and Lys63-linked polyubiquitin chains. Isopeptidase T served as a positive control. Untreated samples contain undigested Lys48- or Lys63-linked polyubiquitin chains. Two-fold serial dilutions of TGEV PL1pro were used in this experiment (the highest concentration was 50 ng/μl, also used for running the control reaction). Polyubiquitin chains are indicated with an arrow, and the lowest band corresponds to single, unchained ubiquitin molecules.
DISCUSSION
In this paper we report the first structure of a coronavirus PL1pro domain, one of the three nonstructural proteases of the alphacoronavirus TGEV. Historically, PL1pro was the first coronaviral papain-like protease to be experimentally identified (17) and to have its coding sequence mapped to the genome (MHV PL1pro) (2, 3). Subsequently, its sequence served as a template for the discovery of the paralogous PL2pro domain by bioinformatics (23). The structure of TGEV PL1pro resembles a right hand, with three distinct regions, the palm, thumb, and fingers. It contains a zinc-binding domain, with four cysteine residues chelating the zinc ion, and a catalytic triad that is formed by residues Cys32, His183, and Asp196.
This overall structural organization and the assignment of functional residues were previously proposed by using a combination of protein homology modeling and the biochemical characterization of HCoV-229E PL1pro (31). Site-directed mutagenesis (30, 31) revealed that the replacement of any of the Zn-coordinating residues or the catalytic Cys and His is detrimental for proteolytic activity toward the nsp1|nsp2 site. In contrast, the protease tolerated a range of replacements of the putative catalytic Asp residue (31). It was proposed, by analogy with cellular PLpros, that this Asp residue might stabilize the catalytic His through hydrogen bonding rather than play a catalytic role per se. In the TGEV PL1pro structure, the proximity of the catalytic Asp and His residues is compatible with either model, indicating that further study is required to assess the precise role of the Asp residue in PLpro catalysis.
The crystal structure of TGEV PL1pro also offers structural details not previously available. A comparison with the structure of the paralogous SARS-CoV PL2pro is instructive (Fig. 3 and 4). While they are very similar in many parts, there are a few regions with significant structural differences of potential functional significance. The most striking difference concerns a ubiquitin-like domain that is N-terminally fused with SARS-CoV PL2pro and conserved in the PL2pro domains of all coronaviruses but not in the PL1pro domains. Regions I and II are considerably dissimilar in the TGEV PL1pro and the SARS-CoV PL2pro structures. The distance between the main-chain carbons of the sequentially equivalent C-terminal parts of region II residues, Val166 (TGEV) and Gln256 (SARS-CoV), is 14.8 Å. Except for their central parts, those two regions are located far from the active site and, therefore, are not likely to be involved in substrate specificity (substrate regions S1 to S4). Based on the comparison with human USP structures, it is most likely that this part of the structure (Fig. 2b) is responsible for binding the distal part of the substrate. The observed structural difference between TGEV PL1pro and SARS-CoV PL2pro may be related to differences in the substrates that these proteases recognize (the C termini of nsp1, nsp2, and nsp3 and yet-to-be-identified host proteins) or to the properties of cofactors that may modulate the activities of these proteases.
The variable region, neighboring region II, is present in the part of the structure that is lying opposite to the active site and is, most likely, not involved in interactions with the substrate. It might compensate for the huge conformational changes in regions I and II and, therefore, maintain the conserved core of the structure that contains the active site (helix α1 and strands β9 and β10). Another very striking difference between SARS-CoV PL2pro and TGEV PL1pro occurs in the highly mobile loop, which in TGEV PL1pro adopts a fold similar to that in inhibitor-bound SARS-CoV PL2pro (Fig. 4d). We have shown that our recombinant TGEV PL1pro possesses enzymatic activity in solution. The significant variation, 3 residues out of 4, in the amino acid sequence of this loop of SARS-CoV PL2pro and TGEV PL1pro may be responsible for the difference in substrate specificity.
During their replicative cycle, many viruses interact with the host cell's ubiquitin and ubiquitin-like pathways, which can lead to, e.g., a modulation of the cell cycle by DNA viruses, viral interference with the innate and adaptive immune responses, and promoting the entry and exit of viral particles (34). Mechanistically speaking, most viruses hijack the cellular ubiquitin system, but some encode their own ubiquitin ligases and DUBs. Deubiquitinating activity has been demonstrated for the adenovirus protease adenain, herpesvirus UL36USP, SARS-CoV PL2pro, arterivirus nsp2, and bunyavirus L proteins (20, 34). Herpesviral DUBs represent a new family of deubiquitinating enzymes and might be involved in the entry, assembly, and release of virus particles (41, 59). The adenovirus protease adenain is responsible for a general decrease in the pool of ubiquitinated proteins, mainly in the nucleus, of the infected cell (4). However, cognate cellular substrates of adenain have not been identified (34). SARS-CoV PL2pro, which was proposed to be involved in counteracting the host's innate immune response, was shown previously to possess DUB activity in vitro (5, 42, 43). MHV PL2pro was shown to bind to IRF3, deubiquitinate it, and thereby prevent its nuclear translocation, which strongly inhibits IFN-β-controlled reporter gene activity (68). Here, we show that TGEV PL1pro also has DUB activity in vitro. The determined pseudo-first-order rate constant is between that of SARS-CoV PL2pro and that of USP7. Interestingly, TGEV PL1pro is capable of hydrolyzing the isopeptide bonds in both Lys48- and Lys63-linked polyubiquitin chains, which are both relevant in the regulation of innate immunity and proinflammatory signaling (7). TGEV PL1pro seems to have a preference for the Lys48-linked polyubiquitin chains, which may allow it to rescue viral proteins from the proteasomal degradation pathway. Additionally, host cells use the ubiquitin-proteasome system to fight viral infection by presenting viral antigens to T cells through the use of the major histocompatibility complex (MHC) class I pathway. TGEV PL1pro, through interference with the proteasomal degradation machinery, may alter this host immune response.
It was previously proposed that alphacoronavirus PL1pros, which contain a so-called USP-like binding site, are likely to exhibit DUB activity (63). Here, we provide the first experimental support for this hypothesis by documenting the in vitro DUB activity of purified TGEV PL1pro. The PL1pro and PL2pro domains of another virus of the genus Alphacoronavirus, HCoV-229E, are known to have overlapping substrate specificities and can both cleave the nsp1|nsp2 and nsp2|nsp3 junctions (69). It was speculated that other alphacoronaviruses, including TGEV, may share this property, which could also include an overlapping specificity for ubiquitinated substrates. However, in a recent study, Clementz et al. (12) concluded that PL1pro of HCoV-NL63, unlike its PL2pro paralog, does not exhibit general DUB activity. In the light of the structural considerations outlined above and the convincing DUB activity now observed for TGEV PL1pro, we suggest that this conclusion may have been premature and that the HCoV-NL63 PL1pro DUB activity may have gone unnoticed for technical reasons. First, the transient expression approach that was used by Clementz et al. is not suitable to reveal DUB activity toward one or a few specific ubiquitinated substrates. Second, and more important, our in vitro cleavage assay uses a purified recombinant enzyme and an “unambiguous” substrate, resulting in a more straightforward and sensitive analysis of DUB activity. We therefore believe that a more rigorous biochemical characterization of NL63 PL1pro—of which an active recombinant form is yet to be purified—will likely reveal that DUB activity is indeed common to all alphacoronavirus PL1pro domains.
The identification of the full range of natural substrates of viral enzymes remains an ongoing challenge that may not have been met for any enzyme to date, given the enormous size and diversity of the cellular proteome. Once identified, any substrate can be exploited in the context of the development of strategies to control coronavirus infections. The coronaviral papain-like proteases, including PL1pro, are no exception in this respect, and their more detailed characterization will yield important insights into their role in the viral replicative cycle as well as their potential as a target for antiviral drug development. The TGEV PL1pro structure and proteolytic activities toward virus and cellular substrates in vitro reported here will facilitate this quest.
Acknowledgments
We thank Linda Boomaars-van der Zanden (LUMC) for excellent technical assistance; Igor Sidorov and Dmitry Samborskiy (LUMC) and Alexander Kravchenko (Moscow State University, Russia) for administration of Viralis; and Luis Enjuanes (Madrid, Spain) for providing a TGEV full-length cDNA clone.
This work was supported by the European project VIZIER (Comparative Structural Genomics of Viral Enzymes Involved in Replication), funded by the 6th Framework Programme of the European Commission under reference number LSHG-CT-2004-511960.
Footnotes
Published ahead of print on 28 July 2010.
REFERENCES
- 1.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. C., C. K. Shieh, L. H. Soe, M. F. Chang, D. M. Vannier, and M. M. Lai. 1989. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 63:3693-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, S. C., K. Yokomori, S. Dong, R. Carlisle, A. E. Gorbalenya, E. V. Koonin, and M. M. Lai. 1993. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 67:6056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakirev, M. Y., M. Jaquinod, A. L. Haas, and J. Chroboczek. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 76:6323-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and E. W. Sayers. 2009. GenBank. Nucleic Acids Res. 37:D26-D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhoj, V. G., and Z. J. Chen. 2009. Ubiquitylation in innate and adaptive immunity. Nature 458:430-437. [DOI] [PubMed] [Google Scholar]
- 8.Brierley, I. 1995. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 76(Pt. 8):1885-1892. [DOI] [PubMed] [Google Scholar]
- 9.Brunger, A. T. 2007. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2:2728-2733. [DOI] [PubMed] [Google Scholar]
- 10.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Z., Y. Wang, K. Ratia, A. D. Mesecar, K. D. Wilkinson, and S. C. Baker. 2007. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 81:6007-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clementz, M. A., Z. Chen, B. S. Banach, Y. Wang, L. Sun, K. Ratia, Y. M. Baez-Santos, J. Wang, J. Takayama, A. K. Ghosh, K. Li, A. D. Mesecar, and S. C. Baker. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 14.Cowtan, K. 1994. DM: an automated procedure for phase improvement by density modification. CCP4 ESF-EACBM Newsl. Protein Crystallogr. 31:34-38. [Google Scholar]
- 15.Cowtan, K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62:1002-1011. [DOI] [PubMed] [Google Scholar]
- 16.Davis, I. W., A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denison, M. R., and S. Perlman. 1986. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J. Virol. 60:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 19.Evans, P. R. 1993. Data collection and processing, p. 114-122. In Proceedings of the CCP4 Study Weekend. CLRC Daresbury Laboratory, Warrington, United Kingdom.
- 20.Frias-Staheli, N., N. V. Giannakopoulos, M. Kikkert, S. L. Taylor, A. Bridgen, J. Paragas, J. A. Richt, R. R. Rowland, C. S. Schmaljohn, D. J. Lenschow, E. J. Snijder, A. Garcia-Sastre, and H. W. Virgin IV. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frieman, M., K. Ratia, R. E. Johnston, A. D. Mesecar, and R. S. Baric. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 83:6689-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorbalenya, A. E., P. Lieutaud, M. R. Harris, B. Coutard, B. Canard, G. J. Kleywegt, A. A. Kravchenko, D. V. Samborskiy, I. A. Sidorov, A. M. Leontovich, and T. A. Jones. 2010. Practical application of bioinformatics by the multidisciplinary VIZIER consortium. Antiviral Res. 87:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorbalenya, A. E., E. J. Snijder, and W. J. Spaan. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78:7863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham, R. L., and M. R. Denison. 2006. Replication of murine hepatitis virus is regulated by papain-like proteinase 1 processing of nonstructural proteins 1, 2, and 3. J. Virol. 80:11610-11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han, Y. S., G. G. Chang, C. G. Juo, H. J. Lee, S. H. Yeh, J. T. Hsu, and X. Chen. 2005. Papain-like protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS-CoV): expression, purification, characterization, and inhibition. Biochemistry 44:10349-10359. [DOI] [PubMed] [Google Scholar]
- 28.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasegawa, H., and L. Holm. 2009. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 19:341-348. [DOI] [PubMed] [Google Scholar]
- 30.Herold, J., A. E. Gorbalenya, V. Thiel, B. Schelle, and S. G. Siddell. 1998. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J. Virol. 72:910-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herold, J., S. G. Siddell, and A. E. Gorbalenya. 1999. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 274:14918-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu, M., P. Li, M. Li, W. Li, T. Yao, J. W. Wu, W. Gu, R. E. Cohen, and Y. Shi. 2002. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111:1041-1054. [DOI] [PubMed] [Google Scholar]
- 33.Hu, M., P. Li, L. Song, P. D. Jeffrey, T. A. Chenova, K. D. Wilkinson, R. E. Cohen, and Y. Shi. 2005. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 24:3747-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isaacson, M. K., and H. L. Ploegh. 2009. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch, W. 1988. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. Appl. Crystallogr. 21:916-924. [Google Scholar]
- 36.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishna, S. S., I. Majumdar, and N. V. Grishin. 2003. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31:532-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae, p. 1163-1185. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 39.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 40.Lerner, M. G., and H. A. Carlson. 2008. APBS plugin for PyMOL. University of Michigan, Ann Arbor, MI.
- 41.Lindner, H. A. 2007. Deubiquitination in virus infection. Virology 362:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindner, H. A., V. Lytvyn, H. Qi, P. Lachance, E. Ziomek, and R. Menard. 2007. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 466:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, D. X., I. Brierley, and T. D. Brown. 1995. Identification of a trypsin-like serine proteinase domain encoded by ORF 1a of the coronavirus IBV. Adv. Exp. Med. Biol. 380:405-411. [DOI] [PubMed] [Google Scholar]
- 45.Mihindukulasuriya, K. A., G. Wu, J. St. Leger, R. W. Nordhausen, and D. Wang. 2008. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 82:5084-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris, R. J., P. H. Zwart, S. Cohen, F. J. Fernandez, M. Kakaris, O. Kirillova, C. Vonrhein, A. Perrakis, and V. S. Lamzin. 2004. Breaking good resolutions with ARP/wARP. J. Synchrotron Radiat. 11:56-59. [DOI] [PubMed] [Google Scholar]
- 47.Mueller-Dieckmann, J. 2006. The open-access high-throughput crystallization facility at EMBL Hamburg. Acta Crystallogr. D Biol. Crystallogr. 62:1446-1452. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski, Z. 1991. Maximum likelihood refinement of heavy atom parameters, p. 80-86. In CCP4 Daresbury Study Weekend Proceedings. CLRC Daresbury Laboratory, Warrington, United Kingdom.
- 50.Panjikar, S., V. Parthasarathy, V. S. Lamzin, M. S. Weiss, and P. A. Tucker. 2005. Auto-Rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 61:449-457. [DOI] [PubMed] [Google Scholar]
- 51.Pannu, N. S., A. J. McCoy, and R. J. Read. 2003. Application of the complex multivariate normal distribution to crystallographic methods with insights into multiple isomorphous replacement phasing. Acta Crystallogr. D Biol. Crystallogr. 59:1801-1808. [DOI] [PubMed] [Google Scholar]
- 52.Pannu, N. S., and R. J. Read. 2004. The application of multivariate statistical techniques improves single-wavelength anomalous diffraction phasing. Acta Crystallogr. D Biol. Crystallogr. 60:22-27. [DOI] [PubMed] [Google Scholar]
- 53.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 54.Potterton, L., S. McNicholas, E. Krissinel, J. Gruber, K. Cowtan, P. Emsley, G. N. Murshudov, S. Cohen, A. Perrakis, and M. Noble. 2004. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 60:2288-2294. [DOI] [PubMed] [Google Scholar]
- 55.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78:9977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Putics, A., A. E. Gorbalenya, and J. Ziebuhr. 2006. Identification of protease and ADP-ribose 1″-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 87:651-656. [DOI] [PubMed] [Google Scholar]
- 57.Ratia, K., S. Pegan, J. Takayama, K. Sleeman, M. Coughlin, S. Baliji, R. Chaudhuri, W. Fu, B. S. Prabhakar, M. E. Johnson, S. C. Baker, A. K. Ghosh, and A. D. Mesecar. 2008. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 105:16119-16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlieker, C., W. A. Weihofen, E. Frijns, L. M. Kattenhorn, R. Gaudet, and H. L. Ploegh. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider, T. R., and G. M. Sheldrick. 2002. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58:1772-1779. [DOI] [PubMed] [Google Scholar]
- 61.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stertz, S., M. Reichelt, M. Spiegel, T. Kuri, L. Martinez-Sobrido, A. Garcia-Sastre, F. Weber, and G. Kochs. 2007. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361:304-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sulea, T., H. A. Lindner, E. O. Purisima, and R. Menard. 2006. Binding site-based classification of coronaviral papain-like proteases. Proteins 62:760-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 65.Waterhouse, A. M., J. B. Procter, D. M. Martin, M. Clamp, and G. J. Barton. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo, P. C., S. K. Lau, C. S. Lam, K. K. Lai, Y. Huang, P. Lee, G. S. Luk, K. C. Dyrting, K. H. Chan, and K. Y. Yuen. 2009. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 83:908-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng, D., G. Chen, B. Guo, G. Cheng, and H. Tang. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 18:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ziebuhr, J., B. Schelle, N. Karl, E. Minskaia, S. Bayer, S. G. Siddell, A. E. Gorbalenya, and V. Thiel. 2007. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 81:3922-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 71.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]