Abstract
Tandem stop mutations K26X and H27X in human immunodeficiency virus type 1 (HIV-1) vif compromise virus replication in human T-cell lines that stably express APOBEC3F (A3F) or APOBEC3G (A3G). We previously reported that partial resistance to A3G could develop in these Vif-deficient viruses through a nucleotide A200-to-T/C transversion and a vpr null mutation, but these isolates were still susceptible to restriction by A3F. Here, long-term selection experiments were done to determine how these A3G-selected isolates might evolve to spread in the presence of A3F. We found that A3F, like A3G, is capable of potent, long-term restriction that eventually selects for heritable resistance. In all 7 instances, the selected isolates had restored Vif function to cope with A3F activity. In two isolates, Vif Q26-Q27 and Y26-Q27, the resistance phenotype recapitulated in molecular clones, but when the selected vif alleles were analyzed in the context of an otherwise wild-type viral background, a different outcome emerged. Although HIV-1 clones with Vif Q26-Q27 or Y26-Q27 were fully capable of overcoming A3F, they were now susceptible to restriction by A3G. Concordant with prior studies, a lysine at position 26 proved essential for A3G neutralization. These data combine to indicate that A3F and A3G exert at least partly distinct selective pressures and that Vif function may be essential for the virus to replicate in the presence of A3F.
Human APOBEC3F (apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3F) (A3F) is a DNA cytidine deaminase that, like its prototypical relative APOBEC3G (A3G), restricts the infectivity of human immunodeficiency virus type 1 (HIV-1) in the absence of the viral accessory protein virion infectivity factor (Vif) (4, 26, 43, 51, 61). Vif is thought to permit productive HIV-1 infection by mediating the proteasomal degradation of A3G and A3F through recruitment of a ubiquitin ligase complex consisting of Elongins B and C, Cullin 5, and Rbx2 (9, 26, 31, 34, 35, 44, 49, 51, 58, 59, 61). In addition, several groups have suggested other mechanisms by which Vif may counteract APOBEC3 proteins, including directly inhibiting packaging, decreasing translation, and inhibiting deaminase activity (22, 29, 36, 42, 49).
In contrast with HIV-1, direct and indirect observations indicate that many retroviruses do not need Vif to evade restriction by cellular APOBEC3 proteins. For instance, the lentivirus equine infectious anemia virus (EIAV) lacks a vif gene despite the existence of an extensive repertoire of equine APOBEC3 proteins (6, 62), and among other types of Vif-deficient retroviruses, a number of alternative APOBEC3 resistance mechanisms have evolved. Foamy viruses use an auxiliary protein called Bet to neutralize APOBEC3 proteins (27, 40). Human T-cell leukemia virus type 1 (HTLV-1) avoids APOBEC3 encapsidation through its unique nucleocapsid protein (11). Analogously, murine leukemia virus (MLV) excludes certain APOBEC3 proteins from virions and cleaves encapsidated murine APOBEC3 (1, 13), while Mason-Pfizer monkey virus (MPMV) also avoids the encapsidation of its cognate rhesus macaque A3G (12).
Overall, it appears that most retroviruses must possess a mechanism to evade the APOBEC3 proteins of their hosts, and this requirement does not always involve Vif. Even in the case of HIV-1, the most influential study to analyze the activity of primary isolates of Vif against A3F and A3G so far demonstrated that 10/40 genetically intact vif alleles tested were defective for the neutralization of one or both restriction factors (45). In addition to intact but defective vif alleles, diverse studies suggest the occurrence of vif alleles with gross genetic lesions to be quite common, ranging from 3.4 to 31% of clones analyzed (e.g., 3.4% [23], 5.8% [60], 6.3% [45], 10.2% [57], 17.4% [47], 21.1% [56], or 31.1% [50]), and readily detectable in most studies in a sizeable fraction of patients analyzed (e.g., 0/50 [19], 6/55 [52], 2/18 [53], 3/14 [38], 2/9 [60], 2/7 [45], 4/10 [47], 7/10 [57], 3/4 [56], or 5/6 [50]). Our own analysis of 2,522 subtype B sequences from the Los Alamos National Laboratory HIV Sequence Database indicated approximately 4% of vif sequences contain one or more premature termination codons (data not shown) (http://www.hiv.lanl.gov/). One may therefore conservatively estimate that, concordant with the overall proportion of defective vif alleles estimated by Simon and colleagues (45), at least 20% of vif alleles are likely inactive against one or more APOBEC3 proteins. Beyond that, some unknown percentage will likely display suboptimal neutralizing activity relative to the “wild type” as also observed previously (30, 45). Combining these findings with the fact that HIV-1 has had decades in which to adapt to the human APOBEC3 repertoire, while its Vif-encoding simian immunodeficiency virus (SIV) ancestors may have been coexisting with primate APOBEC3 repertoires for millions of years before that (15, 54), it is important to consider the possibility that Vif function may not be the sole mechanism of resistance to APOBEC3 proteins available to HIV-1.
To address this, we have previously carried out long-term in vitro selection experiments in which we passaged HIV-1 containing tandem stop codons at positions 26 and 27 of vif in the presence of A3G. These studies yielded viral variants that could spread effectively in the presence of A3G despite retention of these tandem stop codons; rather, all isolates had acquired a pyrimidine at nucleotide 200 and a stop codon in vpr (18). While the effect of the vpr null mutation remains unknown, the substitution of A to T or C at nucleotide 200 (T/C200) functions to optimize viral translation, resulting in increased particle production and a relative decrease in A3G encapsidation to levels tolerable to the viral population (17). However, we also observed that these T/C200 vif− vpr− variants were still susceptible to restriction by A3F and by the APOBEC3 repertoire in nonpermissive CEM cells (18).
We therefore set out to select variants of these viruses that might additionally resist A3F. Given the literature on the Vif-independent evasion of APOBEC3 proteins by the other viruses noted above as well what is known of the mechanisms of human APOBEC3-mediated restriction, we hypothesized that several mechanistically distinct resistance mutations might emerge: (i) changes in nucleocapsid or viral nucleic acid structures that inhibit A3F encapsidation (e.g., see the references above and many references addressing the mechanism of A3G encapsidation as discussed in reference 8), (ii) mutations in integrase or viral nucleic acid motifs that impede the effects of APOBEC3 proteins on integration (28, 32, 33), (iii) mutations in viral nucleic acid structures or nucleocapsid that impede APOBEC3 effects on reverse transcription (5, 16, 21, 25, 55), (iv) nonspecific fitness-enhancing mutations (e.g., T/C-200 in reference 18), or (v) the restoration of functional vif alleles.
MATERIALS AND METHODS
Plasmids.
All novel constructs described were confirmed by DNA sequencing. A3F and A3G coding sequences are identical to those found in GenBank, NM_145298 and NM_021822, respectively. The pcDNA3.1-derived constructs used for the stable expression of untagged A3F and A3G have been described previously (18). pcDNA3.1-V5 and pcDNA3.1-A3F-V5 expression constructs containing a V5 epitope tag were made by digesting pcDNA3.1-3xHA or pcDNA3.1-A3F-3xHA (48) with XhoI/XbaI and ligating these plasmids with synthetic complementary oligonucleotides 5′-TC GAG GGA GTC GAG GGC GGC GGT AAG CCT ATC CCT AAC CCT CTC CTC GGT CTC GAT TCT ACG TAG T-3′ and 5′-CT AGA CTA CGT AGA ATC GAG ACC GAG GAG AGG GTT AGG GAT AGG CTT ACC GCC GCC CTC GAC TCC C-3′, which replaces the 3× HA coding sequence with one for the V5 epitope. pcDNA3.1-A3G-V5 was derived by amplifying A3G from pcDNA3.1-A3G using primers 5′-NNN GAA TTC GAG CTC GGT ACC ACC ATG AAG CCT CAC TTC AG AAA C-3′ and 5′-NNN GTC GAC TCC GTT TTC CTG ATT CTG GAG AAT-3′, digesting with KpnI/SalI and ligating the purified product into pcDNA3.1-V5 digested with KpnI/XhoI.
We codon optimized HIV-1IIIB Vif and added nucleotides for a C-terminal hemagglutinin (HA) epitope (GenScript USA, Inc.). The translated Vif open reading frame is identical to GenBank accession no. EU541617 (18). VifIIIB-HA was excised from parental vector pU57 using BamHI/SalI and subcloned into a similarly cut pVR1012-derived plasmid provided by Xiao-Fang Yu (Johns Hopkins University, Baltimore, MD). This BamHI/SalI-digested expression construct was religated to itself to create a vector control. pVR1012-VifIIIB-HA was modified by site-directed mutagenesis with Pfu polymerase (Stratagene) to create variants encoding QQ and YQ at positions 26 and 27 using primers 5′-T TGG AAG CGC CTC GTG CAG CAG CAT ATG TAC ATC TCC C-3′ and 5′-G GGA GAT GTA CAT ATG CTG CTG CAC GAG GCG CTT CCA A-3′ or primers 5′-ACT TGG AAG CGC CTC GTG TAT CAG CAT ATG TAC ATC TCC CGC-3′ and 5′-GCG GGA GAT GTA CAT ATG CTG ATA CAC GAG GCG CTT CCA AGT-3′.
Proviral constructs for wild-type HIV-1IIIB (GenBank accession no. EU541617 [18]) and a vif-deficient derivative with tandem stop codons at positions 26 and 27 were obtained from Michael Malim (King's College, London, England). Derivative molecular clones with A3G-resistant virus 2 (A3G-R2) and A3G-resistant virus 3 (A3G-R3) mutations, as well as a vif-deficient HIV-1IIIB containing a cytidine at nucleotide 200, have been reported (18) (Table 1). Substitution of a given vif allele in these backgrounds was done by site-directed mutagenesis on a pCR4-Blunt (Invitrogen) plasmid containing the PCR-amplified and cloned HIV-1IIIB vif/vpr region extending from nucleotides 3095 to 5485 (GenBank accession no. EU541617 [18], where +1 is the transcriptional start site). Of note, the A3G-R3 used here is a variant of the published sequence containing an additional nucleotide deletion at position 5285 in the vpr open reading frame downstream of the reported vpr W18X mutation (18). Mutated fragments were reintroduced into full-length molecular clones by subcloning the SwaI/SalI fragment from the shuttle vector into a similarly cut parental HIV-1IIIB proviral plasmid. A comprehensive listing of the molecular clone genotypes used in these studies is provided in Table 1.
TABLE 1.
HIV-1 molecular clone genotypes used in this study
| HIVIIIB clone | Nucleotide at position 200 | vif codons 26 and 27 | vpra | Other mutation(s)b | Reference(s) |
|---|---|---|---|---|---|
| A200 XX | A | XX | WT | N/A | Fig. 1B to E; Fig. 3A to D |
| C200 XX | C | XX | WT | N/A | Fig. 4A and B; Fig. 5A and B; Fig. 6B to D; Fig. 7 |
| A200 KH | A | KH | WT | N/A | Fig. 1B to E; Fig. 3A to D |
| C200 KH | C | KH | WT | N/A | Fig. 4A and B; Fig. 6B to D; Fig. 7 |
| C200 KH SLQ>AAA | C | KH | WT | vif codons 144 to 146 SLQ>AAA | Fig. 6; data not shown |
| T200 XX vpr− (A3G-R2) | T | XX | W54X | gag (nucleocapsid) R32K; pol (integrase) silent | Fig. 1B to E; Fig. 3A to D |
| T200 XX vpr− (A3G-R3) | T | XX | W18X | Deletion of A5285 (vpr) | Fig. 1B to E; Fig. 3A to D |
| T200 KH vpr− (A3G-R2) | T | KH | W54X | gag (nucleocapsid) R32K; pol (integrase) silent | Fig. 4; data not shown |
| T200 KH vpr− (A3G-R3) | T | KH | W18X | Deletion of A5285 (vpr) | Fig. 4; data not shown |
| T200 QQ vpr− (A3F-R7) | T | W54X | gag (nucleocapsid) R32K; pol (integrase) silent | Fig. 3A to D | |
| T200 YQ vpr− (A3F-R5) | T | YQ | W18X | Deletion of A5285 (vpr) | Fig. 3A to D |
| C200 QQ | C | WT | N/A | Fig. 4A and B; Fig. 6B to D; Fig. 7 | |
| C200 QQ SLQ>AAA | C | WT | vif codons 144 to 146 SLQ>AAA | Fig. 6; data not shown | |
| C200 YQ | C | YQ | WT | N/A | Fig. 4A and B; Fig. 6B to D |
| C200 YQ SLQ>AAA | C | YQ | WT | vif codons 144 to 146 SLQ>AAA | Fig. 6; data not shown |
| C200 KQ | C | KQ | WT | N/A | Fig. 6B to D; Fig. 7; data not shown |
| C200 QH | C | QH | WT | N/A | Fig. 6B to D |
| C200 YH | C | YH | WT | N/A | Fig. 6B to D |
| C200 RQ | C | RQ | WT | N/A | Fig. 7; data not shown |
WT, wild-type Vpr (GenBank accession no. EU541617).
N/A, not applicable.
Stable cell lines.
CEM-SS cells stably expressing A3F, A3G, or a vector control have been described previously (18). SupT1 was obtained from the AIDS Research and Reference Reagent Program (catalog no. 100). SupT11 is a single-cell subclone of SupT1 isolated by limiting dilution. APOBEC3-expressing derivatives were made by electroporating with 20 μg linearized plasmid (250 V, 950 μF; Bio-Rad), selecting with 1.0 mg/ml G418 (Mediatech), and screening clones for APOBEC3 expression by immunoblotting using anti-A3F or anti-A3G antibodies (clone 1474 or 10201 from Michael Malim or Jaisri Lingappa, respectively, obtained through the AIDS Research and Reference Reagent Program). T-cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin-streptomycin, β-mercaptoethanol, and in some cases, 0.5 mg/ml G418.
Virus stocks.
Infectious virus was produced by transfecting 5 or 10 μg of proviral molecular clone plasmids into 293T cells at approximately 70% confluence in 10-cm dishes using Trans-IT (Mirus Bio) or Fugene 6 (Roche) transfection reagents. Two or three days later, virus-containing supernatants were harvested and filtered through 0.45-μm filters. 293T cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin.
Virus titration.
The titers of viruses derived from molecular clones were determined by infecting 50,000 CEM-GFP reporter cells (14, 18). Approximately 3 days later, cells were fixed in 4% paraformaldehyde, and green fluorescent protein (GFP)-positive infected cells were quantified using flow cytometry. A Beckman-Coulter Quanta MPL instrument or a Becton-Dickinson FACSCalibur instrument was used for data collection, and FlowJo (Tree Star) or CellQuest (Becton-Dickinson) software was used for data analysis. Live cells were gated by forward versus side scatter (or by electronic volume versus side scatter when using the Quanta MPL instrument), and the proportion of GFP-positive cells in this population was quantified. These percentages were then graphed against the volume of virus stock used, and linear regression was employed to determine the percentage of cells infected at a given volume of a particular virus stock. Calculations of multiplicity of infection (MOI) on CEM-GFP cells were derived from these percentages and used as a standard by which to normalize the quantity of infectious virus used to initiate infections. Quantification of resistant virus isolates differed from this protocol due to the limited amount of passaged supernatants available. In this case, titers were calculated from single infections of 25,000 CEM-GFP cells with 150 μl of virus saved on a given day in a total volume of 250 μl.
HIV-1 spreading infection experiments.
Spreading infections were initiated by infecting 50,000 cells of a given cell line in a total volume of 1 ml in one well of a 24-well plate at a CEM-GFP MOI of 0.01 to 0.05; the MOI in a given experiment is indicated in the associated figure legend. Spreading infections were monitored by periodically using 150 μl of cell-free supernatant to infect fresh target CEM-GFP cells at a concentration of 25,000 cells per 100 μl per well of a 96-well plate. At approximately 2 to 4 days postinfection, these cells were fixed in 4% paraformaldehyde and analyzed by flow cytometry as described above. The original infected cultures were split, and their media were replenished as needed at each time point taken to prevent the overgrowth of infected cells. The accumulation of multiple cycles of infection in a spreading infection leads to some variability in the peaks of replication, and clonal variation among the cell lines used in the experiments described introduces an additional variable (see “Stable cell lines” above for cell line derivation procedures). Nevertheless, general trends of growth/no growth for each virus on each cell line are apparent in the data shown here and in many additional experiments not shown. Additional observations of import are provided in the text or associated figure legend to supplement raw infectivity curves. Except where specifically indicated, the restriction of a given virus is indicated by increasingly flat replication curves. Viruses that spread effectively, in contrast, will generally yield a sharp infectivity peak, followed by a decline as cells are killed by viral spread.
Resistant virus selection procedures.
Resistant viruses were selected by initiating spreading infections in a given cell line (e.g., CEM-SS clones stably transfected with A3F) as indicated above at an MOI of 0.05 or 0.03. Cell-free supernatants were periodically used to infect target CEM-GFP cells as described above, and these CEM-GFP cultures were visually monitored for the outgrowth of notably more infectious viruses as evidenced by an increased proportion of bright GFP-positive cells. Cultures were split and fed as needed at each monitoring point to prevent the overgrowth of infected cells on which viral spread remained restricted until the emergence of resistance. Upon outgrowth of a virus displaying enhanced infectivity, cell-free supernatants were saved at each monitoring point for subsequent confirmatory passage. Passage of resistant isolates was then initiated using virus produced in the presence of A3F, and these passages were conducted as indicated above for a typical spreading infection.
Proviral DNA sequencing.
Genomic DNA was extracted from target CEM-GFP cells infected at the end of the third passage of resistant virus isolates using Qiagen DNeasy kits. This gDNA was then used as template for the amplification of integrated proviral sequences using PCR with high-fidelity Phusion polymerase (NEB) and primers 5′-CAAGGCCAATGGACATATCA-3′ and 5′-CAAACTTGGCAATGAAAGCA-3′. PCR products were cloned into pJET (Fermentas), TOPO pCR4-Blunt or TOPO pCR-Blunt II (Invitrogen). Cloned viral fragments were recovered in Escherichia coli TOP10 or DH10B, and plasmid DNA was prepared with a Qiagen or Clontech miniprep kit. Plasmids were then sequenced using universal or virus-specific primers. Sequences were compiled and analyzed using Sequencher software (Gene Codes Corp.).
APOBEC3 immunoblotting experiments.
T-cell lines were grown to confluence, harvested, and lysed in approximately 50 μl of lysis buffer (25 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 50 μM ZnCl2, 10% glycerol, and 1% Triton X-100 lysis buffer containing 50 μM MG132 [American Peptide] and complete protease inhibitor [Roche]) per milliliter of cells. A 5× sample buffer consisting of 62.5 mM Tris (pH 6.8), 20% glycerol, 2% sodium dodecyl sulfate (SDS), 5% β-mercaptoethanol, and 0.05% bromophenol blue was added to each lysate to a final concentration of 2×, and the mixture was boiled for approximately 10 min prior to loading. Proteins were fractionated by 10% SDS-polyacrylamide gel electrophoresis (PAGE), transferred to a polyvinylidene difluoride (PVDF) membrane, blocked in 4% milk dissolved in phosphate-buffered saline (PBS) containing 0.01% Tween, incubated with rabbit anti-A3F (above), rabbit anti-A3G (above) or mouse antitubulin (Covance) antibodies, and probed with secondary horseradish peroxidase (HRP)-conjugated goat anti-rabbit or goat anti-mouse antibodies (Bio-Rad). The blots were then developed using HyGlo chemiluminescent HRP antibody detection reagent (Denville Scientific) and exposed to film. The membranes were stripped using 62.5 mM Tris (pH 6.8), 2% SDS, and 100 mM β-mercaptoethanol at 50°C and washed in PBS containing 0.01% Tween prior to sequential blocking and reprobing with each primary antibody.
Single-cycle infectivity assays.
A total of 250,000 293T cells/well were plated in a total volume of 2 ml/well in 6-well plates. Approximately 24 h later, each well was transfected using Trans-IT transfection reagent (Mirus Bio) with 200 ng of a vector control or an APOBEC3-V5 expression construct, a pVR1012 vector control, or 50, 100, or 200 ng of a given Vif-HA expression construct supplemented as necessary to 200 ng total with the pVR1012 vector and 1.6 μg of the full-length replication-competent HIV-1IIIB C200 XX provirus (Table 1). Two days after transfection, virus-containing supernatants were filtered through 0.45-μm filters and used to infect CEM-GFP cells in 96-well plates as described above. Three days after infection, these CEM-GFP cells were fixed and analyzed by flow cytometry as described above.
In addition to particle infectivity, producer cell APOBEC3 expression levels were determined. Cells were lysed in 25 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 50 μM ZnCl2, 10% glycerol, and 1% Triton X-100 lysis buffer containing 50 μM MG132 (American Peptide) and complete protease inhibitor (Roche). Immunoblots were then prepared as described above, except primary mouse anti-V5 (Invitrogen), mouse anti-HA.11 (Covance), or mouse antitubulin (Covance) antibodies were used for protein detection in conjunction with secondary HRP-conjugated goat anti-mouse (Bio-Rad) antibodies.
RESULTS
A3F selects for the restoration of Vif function.
To determine how vif-deficient HIV-1IIIB may evolve to overcome restriction by A3F, we used four starting virus templates to initiate a total of 288 selection cultures across four independent experiments. In the first two experiments, we used viruses derived from molecular clones of our previous A3G-resistant isolates to infect A3F-expressing CEM-SS cells (i.e., A3G-R2 is HIV-1IIIB T200 vif-X26X27 vpr-W54X and A3G-R3 is T200 vif-X26X27 vpr-W18X [18]; Tables 1 and 2). Within approximately 8 to 12 weeks of long-term continuous culture, a total of 7 resistant isolates out of 96 infected were obtained; representative isolates from experiments 1 and 2 are described below and in Table 2. All attempts to select resistance from the parental A200 vif-X26X27 vpr+ or a C200 derivative of this parent in these experiments and in two subsequent experiments were unsuccessful (data not shown). Immunoblotting showed that the A3F levels in stably transfected clones of CEM-SS and the naturally nonpermissive CEM and H9 cell lines are similar (Fig. 1A) (18). A recent report further indicates that A3F expression levels in CEM cells are similar to those in primary CD4+ T cells, suggesting that the selective pressure applied in these studies is comparable to that exerted by A3F in primary cells (39).
TABLE 2.
Summary of experimental results selecting A3F- or CEM-resistant HIV-1 variants
| Expt no. | Cell line | Parental HIV-1IIIB genotype | Resistant isolatea | Nucleotides for vif codons 26 and 27 | Amino acids for vif codons 26 and 27 | Frequency by sequencingb |
|---|---|---|---|---|---|---|
| A200 KH vpr+ | N/A | AAA CAC | KH | N/A | ||
| A200 XX vpr+ | N/A | TAA TAG | XX | N/A | ||
| 1 | CEM-SS F1 | T200 XX vpr W54X (A3G-R2) | A3F-R3 | CAA CAG | 11/13 | |
| T200 XX vpr W18X (A3G-R3) | A3F-R5 | TAT CAG | YQ | 12/13 | ||
| 2 | CEM-SS F2 | T200 XX vpr W54X (A3G-R2) | A3F-R7 | CAA CAG | 5/12 | |
| CAA TGG | QW | 2/12 | ||||
| 5 | CEM | C200 QQ vpr+ | CEM-R1 | AAA CAG | KQ | 5/5 |
| C200 QQ vpr+ | CEM-R2 | CGA CAG | RQ | 3/3 |
N/A, not applicable.
Frequency of base change. The number of clones with the base change to the total number of sequenced clones.
FIG. 1.
Restriction of Vif-deficient HIV-1 by APOBEC3F (A3F) selects resistant virus variants. (A) Western blot showing the expression of A3F and A3G in H9 and CEM cells and the CEM-SS-derived cell lines used in these studies. F1 and F2, A3F-expressing CEM-SS cell lines; G1 and G2, A3G-expressing CEM-SS cell lines; V1 and V2, CEM-SS transfected with a vector control. Tub, tubulin. (B to E) Growth curves in the indicated cells for the following HIV-1 isolates: Vif-proficient (A200 KH), Vif-deficient (A200 XX), A3G-resistant [T200 XX vpr− (A3G-R2) and T200 XX vpr− (A3G-R3)], and representative A3F-resistant viruses derived from parent A3G-resistant viruses [T200 YQ vpr− (A3F-R5) and T200 QQ vpr− (A3F-R7) where vif genotypes are retrospectively indicated according to subsequent sequencing (Table 2)]. The starting MOI was approximately 0.02, and similar results were obtained using the second A3F-expressing and vector control CEM-SS cell lines (data not shown). The low peaks observed for some A200 KH growth curves, particularly in panel D, are due to the high cytotoxicity of this virus, which sometimes results in low apparent titers as infected cells are rapidly killed. In addition to the notably low A200 KH CEM-SS V1 curve, a growth curve for A200 KH in another CEM-SS vector line from this experiment, CEM-SS V3, is indicated by a black arrow in panel D to visually demonstrate that the wild-type virus spreads in the absence of restrictive levels of APOBEC3 proteins. The x axis is offset from zero in all curves to permit better visualization of viruses showing little or no growth. Throughout, open symbols indicate viruses lacking Vif expression; closed symbols indicate full-length vif alleles. Similarly, broken lines are used for vpr− viruses, while solid black lines are used for vpr+ viruses. A3F+, A3F expressing; A3−, not expressing A3; GFP+, GFP positive; multi-A3+, expresses ≥5 APOBEC3 proteins.
A3F-resistant isolates were passaged sequentially two additional times in A3F-expressing CEM-SS cells. Replication curves from the third passage indicated that the resistant viruses spread with improved titer and kinetics in A3F-expressing cell lines with peaks generally occurring between 10 and 20 days postinfection as shown for 2 representative isolates (Fig. 1B). As anticipated, the putative A3F-resistant isolates maintained the ability to replicate in A3G-expressing or vector control CEM-SS clones (Fig. 1C and D). However, these viruses did not acquire the capacity to replicate in CEM cells (Fig. 1E; also see below and see Discussion). In some instances, the growth curves of the parental virus (A200 Vif-K26-H27) are relatively weak due to high cytotoxicity, but this does not affect our overall interpretations or conclusions (e.g., Fig. 1D).
To genotype the A3F-selected viruses, we amplified and cloned a region of the proviral genome encompassing vif and vpr, nucleotides 3095 to 5485 (GenBank accession no. EU541617 [18]). We obtained a minimum of 10 sequences per isolate, and analysis revealed that the A3F-resistant isolates had changed nonsense vif codons to missense, while the vpr null mutations remained intact (Table 2 and data not shown). A composite analysis of the sequences derived from three A3F-resistant isolates showed a high hypermutation load averaging 2.4 G-to-A mutations per kilobase (Fig. 2A to C). These mutations generally occurred with notable bias toward a 5′-GA-3′ dinucleotide context, which is characteristic of the deaminase activity of A3F attacking cDNA strand 5′-TC-3′ motifs (underlining indicates the base that changes by APOBEC3 action). However, substantial 5′-GG-3′ context hypermutations consistent with the action of A3G were also evident, particularly in A3F-resistant isolate 7 (Fig. 2C). These G-to-A mutations were likely the result of the low levels of endogenous A3G in CEM-SS cells (e.g., Fig. 1A, see below, and published quantifications [24, 39]). The overall hypermutation levels in these isolates was also likely influenced by the existence of an initially mixed resistant population due to the sequential accumulation of the missense vif mutations, with codon 26 mutating first and codon 27 mutating second. This order of events is suggested by the fact that the same X-to-Q missense mutation at codon 26 was detected in isolate 7 alongside two different codon 27 missense mutations, X-to-Q or X-to-W mutations. From hereon, analyses will focus exclusively on two resistance-associated vif alleles encoding missense mutations at codons 26 and 27, vif-QQ and vif-YQ (Tables 1 and 2).
FIG. 2.
Hypermutation patterns in selected A3F-resistant isolates. The frequency of each base change is given for the clones described in experiments 1 and 2 of Table 2, as is the predominance of the dinucleotide context in which G-to-A mutations occur. Similar to previous authors, we note a substantial C-to-T transition rate in the presence of A3F in addition to the expected G-to-A hypermutations (4, 20, 26). The viruses selected also contain substantial 5′-GG-3′ to 5′-AG-3′ transitions, particularly in the case of A3F-resistant isolate 7, suggesting ongoing mutation by A3G in addition to A3F.
Resistance to A3F occurs through Vif.
On the basis of our proviral DNA sequence analyses and the fact that numerous prior studies have shown that Vif can counteract A3F, we hypothesized that the resistance to A3F observed in our isolates would be conferred by the aforementioned missense mutations at vif codons 26 and 27. To test this hypothesis and to eliminate the possibility that mutations elsewhere in the viral genome might contribute to A3F resistance, we incorporated the vif-QQ and vif-YQ alleles cleanly into their parent A3G-resistant molecular clone backgrounds and infected A3F-expressing CEM-SS cells with viruses produced from these molecular clones (Table 1). These viruses—HIV-1IIIB T200 vif-QQ (or vif-YQ) vpr−—displayed robust infectivity and kinetics on cells expressing A3F, A3G, or a vector control, as expected for a virus engineered to contain both A3G and A3F resistance mutations (Fig. 3A to C). In addition, like the original A3F-resistant isolates, these molecular clones still failed to replicate in CEM cells despite their enhanced replicative capacity in the presence of A3F (Fig. 3D; see below and see Discussion).
FIG. 3.
Restoration of the vif open reading frame accounts for phenotypic resistance to A3F-mediated restriction. Spreading infections at an MOI of 0.05 were initiated in CEM-SS cells stably transfected with A3F (A) or A3G (B) or a vector control (C) as well as in nonpermissive CEM cells (D) using viruses derived from proviral molecular clones with the indicated genotypes. The mildly enhanced infectivity of A3G-resistant viruses relative to their parent A200 XX viruses in A3F-expressing CEM-SS cells is sometimes observed in experiments such as the one shown that start from a higher MOI (compare the lower MOI in Fig. 1B with the higher MOI in panel A). In contrast, A3F-selected viruses consistently display robust peaks at any MOI in the presence of A3F. The peaks of A200 KH growth are indicated by arrows in panels A and B to differentiate them from the descending T200 YQ vpr− (A3F-R5) curve and the superimposed T200 YQ vpr− (A3F-R5) peak, respectively. Similar results were obtained using proviral molecular clones corresponding to other selected isolates as well as additional CEM-SS clones stably transfected with A3F, A3G, or a vector control (data not shown).
vpr deficiency does not explain the lack of resistant virus replication in CEM cells.
Given the ability of the selected viruses to grow in the presence of A3F or A3G, it was notable that they were still unable to spread in CEM cells that express these two proteins and three additional APOBEC3 proteins (39). We therefore turned to viral genotypes in search of a possible explanation. The nucleotide at position 200 was not likely to be responsible, because the vast majority of HIV-1 isolates already have a T or C at that position (for example, see reference 18 [per the Los Alamos National Laboratory HIV Sequence Database]). The A200 in the parent HIV-1IIIB is presumably disfavored, because it causes suboptimal translation, lower levels of viral particle production, and a relatively greater degree of A3G-dependent restriction (17, 18). However, the function served by vpr inactivation in the parent A3G-resistant viruses is still unknown.
To address the possibility that an undescribed Vpr function might be required for these viruses to grow in the presence of multiple APOBEC3 proteins, we incorporated the selected vif alleles into HIV-1IIIB C200 vpr+ molecular clones (Table 1). All viruses were replication competent in vector control CEM-SS and SupT11 cell lines (data not shown). Like the Vpr-deficient vif-QQ and vif-YQ viruses in Fig. 1E and 3D, however, Vpr-proficient viruses with the vif-QQ or vif-YQ allele remained restricted by nonpermissive CEM and H9 cells (Fig. 4A and B; also data not shown; see below for additional data with these molecular clones). In contrast, the wild-type vif-KH virus spread readily in these fully nonpermissive lines in both vpr+ and vpr− backgrounds (Fig. 4A and B and data not shown). We therefore concluded that the basis for the observed replication defect in naturally nonpermissive cell lines lay in the identity of the amino acids selected by A3F—vif-QQ or vif-YQ versus the wild-type vif-KH.
FIG. 4.
The identity of Vif amino acids 26 and/or 27 rather than Vpr status is critical for the ability to replicate on naturally nonpermissive cells. Spreading infection curves are shown for viruses with wild-type (KH), A3F-selected missense (QQ and YQ), and nonsense (XX) codons at positions 26 and 27 of vif in a Vpr-proficient context. Spreading infections were carried out from a starting MOI of 0.01 on CEM and H9 cells (A and B) as well as CEM-SS and SupT11 clones transfected with a vector control (data not shown). Different alleles of vif in Vpr-deficient contexts showed the same growth patterns on CEM and H9 cells as their Vpr-proficient counterparts (Table 1 and data not shown).
Variants of Vif selected by A3F are unable to effectively degrade A3G.
To mechanistically characterize Vif-QQ and Vif-YQ, we assessed the ability of these Vif variants to neutralize A3F or A3G in single-cycle infectivity experiments at several levels of Vif expression. As predicted by the spreading infection phenotypes, the Vif variants selected by A3F were as efficient at enhancing viral infectivity and lowering intracellular A3F levels as the wild-type Vif-KH was (Fig. 5A). In contrast, the Vif variants selected by A3F did not restore infectivity in the presence of A3G, and they were markedly deficient in their ability to lower intracellular A3G levels (Fig. 5B). This clear A3G susceptibility helped to explain why viruses with these vif alleles fail to spread in CEM or H9 cells (e.g., Fig. 4) and why our original A3F-resistant isolates had significant 5′-GG-3′ context hypermutations—the original CEM-SS A3F lines have permissive but detectable levels of endogenous A3G (Fig. 1A and 2) (39). However, it remains possible that other APOBEC3 proteins expressed in CEM and H9 cells beyond A3F and A3G may account for at least part of the continued restriction of A3F-selected viruses in these cells (see Discussion) (39).
FIG. 5.
Functional Vif proteins selected by A3F are deficient in their ability to degrade A3G. A titration experiment analyzing the infectivity of particles produced by the cotransfection of constant amounts of A3F-V5 (A) or A3G-V5 (B) in the presence of increasing amounts of Vif-HA. The identities of amino acids 26 and 27 are indicated for each Vif-expressing construct. Infectivity data represent the mean plus standard error of the mean (SEM) (error bar) of three independent experiments where infectivity is determined relative to that of particles produced under the same cotransfection conditions in each experiment with a vector control in place of the APOBEC3 expression construct (not shown). Immunoblots shown are taken from one of these three experiments. While Vif-QQ and Vif-YQ are notably deficient in their ability to neutralize A3G relative to the wild-type Vif-KH, a mild effect is seen at higher levels of Vif expression, which achieves statistical significance by a paired two-tailed t test for Vif-QQ, but not Vif-YQ (panel B and data not shown). V, Vif vector and A3F-V5 or A3G-V5 expression constructs cotransfected; these conditions were tested once in each experiment but are loaded in the immunoblots and plotted in the histograms three times each for direct visual comparison with the addition of each Vif protein.
The inability of the vif alleles selected by A3F to neutralize A3G maps to the identity of amino acid 26.
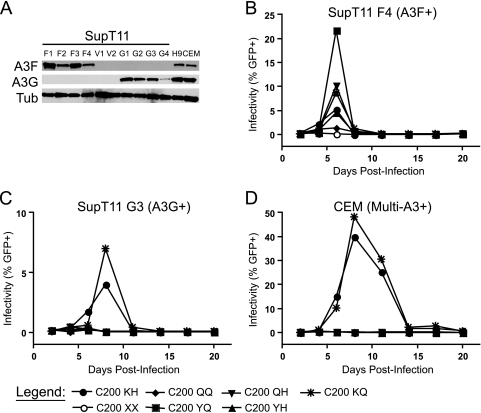
To test whether viruses with the vif alleles selected by A3F are deficient in their ability to neutralize A3G in spreading infections and, if so, to additionally determine the nature of the mutation responsible, we did a series of spreading infection experiments using a panel of full-length molecular clones with amino acid substitution mutations at vif codon 26 or 27 (i.e., wild-type HIV-1IIIB C200 viruses with the exception of vif; Table 1). Specifically, we compared viruses with K, Q, or Y at position 26 and H or Q at position 27, and we used A3F- or A3G-expressing SupT11 clones, because we recently found that this line is nearly devoid of endogenous APOBEC3 expression (39) (Fig. 6A).
FIG. 6.
A3F-selected vif alleles are nonfunctional for the neutralization of A3G but can be rescued by restoration of the wild-type K26 residue. (A) Western blots showing expression levels of A3F and A3G in the SupT11-derived cell lines used in these experiments as well as in H9 and CEM cells. Spreading infection curves from a starting MOI of 0.01 are shown for the wild type (C200 KH) and for Vif-deficient (C200 XX) and A3F-selected (C200 QQ and YQ) mutants as well as mutants completing the matrix of combinations of wild-type and selected residues at positions 26 and 27 of vif (C200 QH, YH and KQ) on SupT11 cells transfected with A3F (B) or A3G (C) as well as nonpermissive CEM cells (D). Results demonstrate that K26 is critical for the neutralization of A3G and the APOBEC3 repertoire found in CEM cells, but not A3F. Results concordant with results in panels B to D were observed using additional SupT11-derived cell lines expressing A3F or A3G as well as SupT11 clones transfected with a vector control and H9 cells (data not shown). Higher peaks than that shown in panel B are usually observed with the C200 QQ virus (e.g., Fig. 1 and 3 and data not shown); in addition to those curves and many not shown, the ability of C200 QQ to spread in the presence of A3F is indicated by the fact that it efficiently kills the culture in which it replicates (see the legend to Fig. 1).
We found that all combinations of Vif residues at positions 26 and 27 were able to spread in SupT11 cells expressing A3F and in SupT11 vector control cells (Fig. 6B and data not shown). In contrast, only viruses encoding a lysine at position 26 of Vif were able to spread effectively in cells expressing A3G or in nonpermissive CEM or H9 cell lines (Fig. 6C and D and data not shown). These data corroborate two recent Vif site-directed mutation screens, which independently found that K26 is important for the neutralization of A3G (7, 10). They are also consistent with the high conservation of K26 in sequences in the Los Alamos National Laboratory HIV Sequence Database, where 99.4% (2,507/2,522) of subtype B sequences encode lysine at position 26 (data not shown; http://www.hiv.lanl.gov/).
Given the correlation between the capacity of a particular Vif to degrade APOBEC3 proteins and the ability of viruses carrying those vif alleles to spread in the presence of APOBEC3 proteins, we further hypothesized that degradation is the predominant mechanism of Vif action at play in our spreading infection system. To test this hypothesis, we incorporated Vif BC Box mutations at positions 144 to 146 into both the wild-type and A3F-selected molecular clones. SLQ-to-AAA substitutions in this highly conserved region have been previously shown to interrupt interaction between Vif and Elongin C, thus attenuating the Vif-mediated degradation of APOBEC3 proteins (58). We observed that viruses carrying these mutations are unable to spread in SupT11 cells stably transfected with A3F or A3G as well as naturally nonpermissive CEM or H9 cells (data not shown). These data indicate that the interaction between Vif and Elongin C is required for A3F/G neutralization and, further, based on many prior studies, that degradation is an integral part of the mechanism.
Additional long-term selection experiments in nonpermissive CEM cells select for a positive charge at Vif position 26.
To determine how the vif alleles selected by A3F might evolve to become resistant to the full nonpermissive APOBEC3 repertoire in CEM cells, we passaged vif-QQ viruses in these cells until resistant isolates arose. From 48 parallel cultures, 5 isolates emerged after 8 to 12 weeks of incubation. Second-passage proviral DNA sequencing revealed that 3 isolates carried Q26R mutations, while two more isolates carried Q26K mutations; representative viruses from these selections are shown in experiment 5 of Table 2. Serial passage in CEM cells indicated that these isolates replicated with greater efficiency than the parent vif-QQ viruses, although Q26R viruses still tended to display attenuated replication kinetics (Fig. 7).
FIG. 7.
Long-term culture of A3F-resistant viruses in CEM cells selects for restoration of a positive charge at Vif residue 26. Passage of CEM-resistant viral isolates in CEM cells demonstrating their functional resistance to the nonpermissive phenotype of CEM. Phenotypic resistance of the selected alleles to the nonpermissive phenotype as encountered in CEM cells was also confirmed using viruses derived from molecular clones (Table 1 and data not shown). A summary of the sequence evolution observed in resistant isolates that were confirmed by second passage and for which sequence was available is given in experiment 5 of Table 2.
DISCUSSION
The experiments described here are the first reported selections for HIV-1 resistance to the restriction factor A3F. Our data show that A3F, like A3G, is capable of potent long-term restriction, extending to two the number of APOBEC3 proteins shown capable of selecting heritable resistance (18). The strength of A3F-mediated restriction in our spreading infection experiments combined with its ability to select resistance contrasts somewhat with the consistently weaker effects of A3F in single-cycle assays compared with A3G (3, 4, 20, 26, 45). The reason(s) for this disparity is not obvious, but the data presented here suggest that A3F may be an important part of the selective pressure that maintains Vif function in vivo.
We presume that the isolates emerging from long-term selection experiments will have taken the easiest path to resistance. Given the current literature on APOBEC3 proteins, we might have anticipated the selection of any number of Vif-independent resistance mechanisms to A3F as discussed in the introduction, but we have only ever selected for the restoration of Vif function. If Vif-independent resistance to A3F is possible, then the genetic barriers to evolving such mechanisms are apparently greater than the 1 or 2 mutational events required to restore Vif function in the alleles described here. Fitness costs to viruses acquiring these putative alternative resistance mutations may also influence the overall barrier to their development.
Aside from the central observation that A3F selects for the restoration of Vif function, we identified K26 as an important residue for the Vif-mediated degradation of A3G, but not A3F. This result is satisfying in light of recent site-directed mutagenesis studies also showing that K26 is indeed required for A3G neutralization (7, 10), and it likely explains the notable presence of G-to-A mutations consistent with A3G action in our selected viral isolates (Fig. 1A and 2). Molecularly, our characterization of the vif alleles selected by A3F is largely in agreement with these two reports in finding that viruses without the wild-type K26 lack the ability to efficiently degrade A3G, although they still continue to bind it (Fig. 5B and data not shown). Thus, our data also support the notion that K26 is part of the Vif interaction surface that contacts A3G.
In the process of characterizing the amino acid identity requirements at residue 26, we also demonstrate for the first time that the evolution of vif in response to one APOBEC3 protein (A3F) may not result in a Vif variant with activity against another APOBEC3 protein (A3G; Fig. 5 and 6). Thus, the selective pressures exerted on vif by each APOBEC3 protein are at least partially distinct. This conclusion is consistent with a growing body of mutagenesis data implying that the structural determinants of A3F and A3G binding sites within Vif can be both defined and in some instances separated genetically (reviewed in references 2 and 46).
While we have not yet succeeded in selecting Vif function as a mechanism of A3G resistance, our observations should not be extrapolated to quantify the relative abilities of A3F and A3G to select for Vif function. For instance, the vif genotype of the parental virus is highly likely to influence the outcome. Furthermore, we do not wish to imply that Vif function is unimportant for overcoming restriction by A3G. Even in vitro, the A3G-selected mutations that we have previously reported are not as robust as wild-type Vif function. They fail to complement the A3F-selected Vif alleles when replicated in naturally nonpermissive CEM and H9 cells as well as CEM-SS cells stably transfected with both A3F and A3G (Fig. 1E and 3D and data not shown), and we have also observed that they may become overwhelmed when replicating on SupT11 cells expressing higher levels of A3G but no other APOBEC3 proteins (data not shown). This is likely due to the nature of our previously characterized A3G resistance mutations, which merely tolerate the presence of A3G better than the parental virus (18). That a particular resistance mutation should be saturable by higher APOBEC3 expression levels is perhaps not all that surprising, because even Vif-proficient viruses can be overwhelmed and restricted under conditions of APOBEC3 overexpression (37, 41). Furthermore, we cannot rule out the possibility that other dominant acting APOBEC3 restriction factors in CEM and H9 cells beyond A3F and A3G may be at least in part responsible for the more potent restriction observed in these cell lines. Results of future long-term selection experiments, especially if the viral genotype and/or the cellular A3 levels differ from those described here, are likely to yield other informative outcomes.
Acknowledgments
We thank R. S. LaRue and D. Urso for technical assistance, P. Southern, N. Somia, and the reviewers for helpful feedback, M. Malim, J. Lingappa, X. F. Yu, and the AIDS Research and Reference Reagent Program for materials, and L. Mansky for sharing cell culture facilities.
This research was funded by grants from the National Institute of Allergy and Infectious Diseases (R01 AI064046), the University of Minnesota Academic Health Center and the Campbell Foundation. J.S.A. was supported in part by the University of Minnesota Medical Scientist Training Program (T32 GM008244), a Warwick Fellowship from the Minnesota Medical Foundation, and the National Institute on Drug Abuse (F30 DA026310). G.H. was supported in part by a Canadian Institutes of Health Research Predoctoral Studentship, and J.F.H. was supported in part by a National Science Foundation Predoctoral Fellowship.
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Abudu, A., A. Takaori-Kondo, T. Izumi, K. Shirakawa, M. Kobayashi, A. Sasada, K. Fukunaga, and T. Uchiyama. 2006. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr. Biol. 16:1565-1570. [DOI] [PubMed] [Google Scholar]
- 2.Albin, J. S., and R. S. Harris. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med. 12:1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, K. N., M. Verma, E. Y. Kim, S. M. Wolinsky, and M. H. Malim. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4:e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd, H. P., R. L. Tallmadge, J. L. Oaks, S. Carpenter, and B. R. Cullen. 2008. Equine infectious anemia virus resists the antiretroviral activity of equine APOBEC3 proteins through a packaging-independent mechanism. J. Virol. 82:11889-11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, G., Z. He, T. Wang, R. Xu, and X. F. Yu. 2009. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J. Virol. 83:8674-8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317-353. [DOI] [PubMed] [Google Scholar]
- 9.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 10.Dang, Y., X. Wang, T. Zhou, I. A. York, and Y. H. Zheng. 2009. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J. Virol. 83:8544-8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derse, D., S. A. Hill, G. Princler, P. Lloyd, and G. Heidecker. 2007. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. U. S. A. 104:2915-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doehle, B. P., H. P. Bogerd, H. L. Wiegand, N. Jouvenet, P. D. Bieniasz, E. Hunter, and B. R. Cullen. 2006. The betaretrovirus Mason-Pfizer monkey virus selectively excludes simian APOBEC3G from virion particles. J. Virol. 80:12102-12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doehle, B. P., A. Schafer, H. L. Wiegand, H. P. Bogerd, and B. R. Cullen. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 79:8201-8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gervaix, A., D. West, L. M. Leoni, D. D. Richman, F. Wong-Staal, and J. Corbeil. 1997. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl. Acad. Sci. U. S. A. 94:4653-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gifford, R. J., A. Katzourakis, M. Tristem, O. G. Pybus, M. Winters, and R. W. Shafer. 2008. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U. S. A. 105:20362-20367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haché, G., T. E. Abbink, B. Berkhout, and R. S. Harris. 2009. Optimal translation initiation enables Vif-deficient human immunodeficiency virus type 1 to escape restriction by APOBEC3G. J. Virol. 83:5956-5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haché, G., K. Shindo, J. S. Albin, and R. S. Harris. 2008. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr. Biol. 18:819-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassaine, G., I. Agostini, D. Candotti, G. Bessou, M. Caballero, H. Agut, B. Autran, Y. Barthalay, and R. Vigne. 2000. Characterization of human immunodeficiency virus type 1 vif gene in long-term asymptomatic individuals. Virology 276:169-180. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 282:2587-2595. [DOI] [PubMed] [Google Scholar]
- 21.Iwatani, Y., D. S. Chan, F. Wang, K. S. Maynard, W. Sugiura, A. M. Gronenborn, I. Rouzina, M. C. Williams, K. Musier-Forsyth, and J. G. Levin. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35:7096-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komoto, S., S. Tsuji, B. J. Lee, Y. Iwabu, Y. Kojima, T. Otake, K. Taniguchi, and K. Ikuta. 2005. Higher frequency of premature stop codon mutations at vpu gene of human immunodeficiency virus type 1 CRF01_AE compared with those of other subtypes. Microbes Infect. 7:139-147. [DOI] [PubMed] [Google Scholar]
- 24.Koning, F. A., E. N. Newman, E. Y. Kim, K. J. Kunstman, S. M. Wolinsky, and M. H. Malim. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, X. Y., F. Guo, L. Zhang, L. Kleiman, and S. Cen. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 282:32065-32074. [DOI] [PubMed] [Google Scholar]
- 26.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 27.Löchelt, M., F. Romen, P. Bastone, H. Muckenfuss, N. Kirchner, Y. B. Kim, U. Truyen, U. Rösler, M. Battenberg, A. Saib, E. Flory, K. Cichutek, and C. Münk. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X. F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 81:7238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mariani, R., D. Chen, B. Schröfelbauer, F. Navarro, R. König, B. Bollman, C. Münk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 30.Marin, M., S. Golem, K. M. Rose, S. L. Kozak, and D. Kabat. 2008. Human immunodeficiency virus type 1 Vif functionally interacts with diverse APOBEC3 cytidine deaminases and moves with them between cytoplasmic sites of mRNA metabolism. J. Virol. 82:987-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 32.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mbisa, J. L., W. Bu, and V. K. Pathak. 2010. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 84:5250-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehle, A., J. Goncalves, M. Santa-Marta, M. McPike, and D. Gabuzda. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 36.Mercenne, G., S. Bernacchi, D. Richer, G. Bec, S. Henriet, J. C. Paillart, and R. Marquet. 2010. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 38:633-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, G., K. J. Lei, W. Jin, T. Greenwell-Wild, and S. M. Wahl. 2006. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 203:41-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangel, H. R., D. Garzaro, A. K. Rodriguez, A. H. Ramirez, G. Ameli, C. Del Rosario Gutierrez, and F. H. Pujol. 2009. Deletion, insertion and stop codon mutations in vif genes of HIV-1 infecting slow progressor patients. J. Infect. Dev. Ctries 3:531-538. [DOI] [PubMed] [Google Scholar]
- 39.Refsland, E. W., M. D. Stenglein, K. Shindo, J. S. Albin, W. L. Brown, and R. S. Harris. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38:4274-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell, R. A., H. L. Wiegand, M. D. Moore, A. Schafer, M. O. McClure, and B. R. Cullen. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724-8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadler, H. A., M. D. Stenglein, R. S. Harris, and L. M. Mansky. 2010. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J. Virol. 84:7396-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santa-Marta, M., F. A. da Silva, A. M. Fonseca, and J. Goncalves. 2005. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J. Biol. Chem. 280:8765-8775. [DOI] [PubMed] [Google Scholar]
- 43.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 44.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 45.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, J. L., W. Bu, R. C. Burdick, and V. K. Pathak. 2009. Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends Pharmacol. Sci. 30:638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sova, P., M. van Ranst, P. Gupta, R. Balachandran, W. Chao, S. Itescu, G. McKinley, and D. J. Volsky. 1995. Conservation of an intact human immunodeficiency virus type 1 vif gene in vitro and in vivo. J. Virol. 69:2557-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837-16841. [DOI] [PubMed] [Google Scholar]
- 49.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 50.Tominaga, K., S. Kato, M. Negishi, and T. Takano. 1996. A high frequency of defective vif genes in peripheral blood mononuclear cells from HIV type 1-infected individuals. AIDS Res. Hum. Retroviruses 12:1543-1549. [DOI] [PubMed] [Google Scholar]
- 51.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wieland, U., J. Hartmann, H. Suhr, B. Salzberger, H. J. Eggers, and J. E. Kuhn. 1994. In vivo genetic variability of the HIV-1 vif gene. Virology 203:43-51. [DOI] [PubMed] [Google Scholar]
- 53.Wieland, U., A. Seelhoff, A. Hofmann, J. E. Kuhn, H. J. Eggers, P. Mugyenyi, and S. Schwander. 1997. Diversity of the vif gene of human immunodeficiency virus type 1 in Uganda. J. Gen. Virol. 78:393-400. [DOI] [PubMed] [Google Scholar]
- 54.Worobey, M., M. Gemmel, D. E. Teuwen, T. Haselkorn, K. Kunstman, M. Bunce, J. J. Muyembe, J. M. Kabongo, R. M. Kalengayi, E. Van Marck, M. T. Gilbert, and S. M. Wolinsky. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455:661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, Y., F. Guo, S. Cen, and L. Kleiman. 2007. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology 365:92-100. [DOI] [PubMed] [Google Scholar]
- 56.Yedavalli, V. R., and N. Ahmad. 2001. Low conservation of functional domains of HIV type 1 vif and vpr genes in infected mothers correlates with lack of vertical transmission. AIDS Res. Hum. Retroviruses 17:911-923. [DOI] [PubMed] [Google Scholar]
- 57.Yedavalli, V. R., C. Chappey, E. Matala, and N. Ahmad. 1998. Conservation of an intact vif gene of human immunodeficiency virus type 1 during maternal-fetal transmission. J. Virol. 72:1092-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 59.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, L., Y. Huang, H. Yuan, S. Tuttleton, and D. D. Ho. 1997. Genetic characterization of vif, vpr, and vpu sequences from long-term survivors of human immunodeficiency virus type 1 infection. Virology 228:340-349. [DOI] [PubMed] [Google Scholar]
- 61.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zielonka, J., I. G. Bravo, D. Marino, E. Conrad, M. Perkovic, M. Battenberg, K. Cichutek, and C. Münk. 2009. Restriction of equine infectious anemia virus by equine APOBEC3 cytidine deaminases. J. Virol. 83:7547-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]