Abstract
Vaccinia virus (VACV) is the prototypic orthopoxvirus and was the live vaccine used to eradicate smallpox. In addition, VACV is a possible vector for recombinant vaccines. Despite these reasons for study, the roles of many VACV genes are unknown, and some fundamental aspects, such as the total size of immune responses, remain poorly characterized. VACV gene A47L is of interest because it is highly transcribed, has no sequence similarity to any nonpoxvirus gene, and contains a larger-than-expected number of CD8+ T cell epitopes. Here it is shown that A47L is not required for growth in vitro and does not contribute to virulence in mice. However, we confirmed that this one protein primes CD8+ T cells to three different epitopes in C57BL/6 mice. In the process, one of these epitopes was redefined and shown to be the most dominant in A47 and one of the more highly ranked in VACV as a whole. The relatively high immunogenicity of this epitope led to a reevaluation of the total CD8+ T cell response to VACV. By the use of two methods, the true size of the response was found to be around double previous estimates and at its peak is on the order of 60% of all CD8+ T cells. We speculate that more CD8+ T cell epitopes remain to be mapped for VACV and that underestimation of responses is unlikely to be unique to VACV, so there would be merit in revisiting this issue for other viruses.
Poxviruses compose a family of large, double-stranded DNA viruses, with members that infect a wide range of insect and vertebrate hosts. In addition to the essential complement of genes required for basic viral replication that cluster toward the middle of the linear genome (29, 30), each poxvirus has numerous genes that enhance survival within and, presumably, transmission between hosts (22, 32, 35). Vaccinia virus (VACV) is the member of this family that is best known and studied, and it is estimated that between a third and a half of its ∼200-kb genome encodes those functions (42). In many cases, insight into the possible functions of these genes can be gained by looking for sequence similarity with genes encoding mammalian proteins of known functions (e.g., 2). In other cases, the similarities have been less directly useful, but careful characterization of the genes has revealed functions and often provided substantial new insights into virus-host relationships (see, e.g., reference 36). Finally, the sequences of a few genes and proteins have no similarity to any nonpoxvirus sequences, and these are intriguing, as their origin is entirely obscure. The subject of this paper, namely, VACV gene A47L, falls into this last class.
A47L is located toward the distal right end of the VACV genome and has been assigned a putative size of 252 amino acids (aa) in VACV strain WR in the Poxvirus Bioinformatic Resource (www.poxvirus.org). The gene is highly conserved among most of the sequenced orthopoxvirus genomes, with the exception that fragmented and truncated versions are encoded by all strains of camelpox and monkeypox viruses, respectively. The gene is absent outside the orthopoxvirus family, with the possible exception that gene 229 of fowlpox virus (strain Iowa) is distantly related. This high level of conservation within the orthopoxviruses but lack of conservation in other poxviruses suggests an important role but one that is not related to basic replicative functions. Promoter predictions suggest an early-late expression profile, with recent experimental microarray data confirming an extremely high transcription level at 30 min to 2 h postinfection that remains at substantial levels up to 24 h postinfection (5). In fact, in that analysis by Assarsson et al., A47L was found to be one of the earliest and most highly expressed of VACV genes.
Also of interest, and possibly related to its high level of expression, A47L was found to be one of a few genes in VACV that contained an unexpectedly high number of CD8+ T cell epitopes (27). Six CD8+ T cell epitopes, restricted by five different major histocompatibility complex (MHC) class I allomorphs, have been identified in A47; of those, three were previously reported to generate responses in the C57BL/6 mouse (20, 21, 27, 28, 37, 38). The term “immunoprevalent” has been used to describe such proteins that have many epitopes and that can be presented in the context of multiple MHC restriction elements (27). Most of these epitopes were defined using synthetic peptides in in vitro assays, and definitive evidence that the T cells stimulated in each case were primed by A47 in vivo is lacking (46). This was of particular concern where relatively high levels of peptide were required in the in vitro assays, as was the case for one of the epitopes found in C57BL/6 mice (20).
The initial aims of this work were to examine the role of A47L in the biology of VACV by deletional analysis and to test whether all three published CD8+ T cell epitopes for A47 in C57BL/6 mice prime CD8+ T cells in vivo. As a result of our investigations, a published CD8+ T cell epitope was redefined and the relatively large response to this peptide led us to use a new method to redefine the total size of the CD8+ T cell response to VACV.
MATERIALS AND METHODS
Viruses and cell lines.
Immortalized cell lines, including BHK-21, DC2.4 (33), 293, and BS-C-1, were maintained in Dulbecco's modified Eagle medium (DMEM; Invitrogen) with glutamine and 10% or 2% fetal bovine serum (FBS) (D10 and D2, respectively). VACV strains WR (Western Reserve NIH-TC) and WR-NP-S-GFP (4, 25) (called WR-SIIN here) were grown in BHK-21 cells and purified by centrifugation through a 36% sucrose cushion and then subjected to titration in BS-C-1 cells; standard methods were used for all of these procedures. VACV strain MVA was grown in BHK-21 cells, and its titer was determined in the same cells by the use of the immunostaining method for titrations (35). Viruses used were kind gifts from Bernard Moss or Jon Yewdell and Jack Bennink, NIH.
Generation of VACVs with the A47L open reading frame deleted.
Recombinant VACVs lacking A47L were engineered using a transient dominant method (12) in which unstable intermediates were enriched and identified through the use of a fusion protein between enhanced green fluorescent protein (GFP) and Bsd (which confers resistance to blasticidin). This fusion protein was driven by the VACV p7.5 promoter from outside the multiple cloning site of a transfer plasmid based on pCR-Blunt II (Invitrogen) and named p7.5Gb (D. C. Tscharke and Yik Chun Wong, unpublished data). Sequences flanking A47L were amplified by PCR using the following primers: for the left flank, CAGTGTGCTGGAATTTGTAAAGGATATGATATGTTGTGAT and AAGGTGATTGGAATGGGAT; for the right flank, CATTCCAATCACCTTGACCTAATCGTCTC GGATGA and GATATCTGCAGAATTCCCTGTTCTAGTTGTTCTTGTAT (underlined sequences match sequences on both sides of a unique EcoRI restriction enzyme site of p7.5Gb; boldface sequences represent an area of identity between the 3′ end of the left flank and the 5′ end of the right flank). These areas of identity allowed the flanks to be joined and inserted into EcoRI-digested vector in a three-way Infusion (Clontech) reaction to create the transfer vector p7.5GbΔA47L. This vector was transfected into 293 cells infected 1 h previously with VACV WR, and progeny were harvested after 3 days. Intermediate (GFP-positive [GFP+]) recombinants were isolated through three rounds of plaque purification. These purifications were performed with BS-C-1 cells grown in phenol red-free D2 with 10 μg/ml blasticidin (Invivogen) and 0.4% medium viscosity sodium carboxymethyl cellulose (CMC; Sigma-Aldrich catalog no. C4888). A further three or four rounds of plaque selection for GFP− viruses were then done in the absence of blasticidin to obtain A47L deletion viruses and a matched wild-type control. Deletion and wild-type viruses were identified and their purity was confirmed using PCR, and the area around the deletion was verified by DNA sequencing. The process starting from infection and transfection was performed twice in parallel experiments to make two independently generated A47L deletion viruses, which were named WR-delA47L-1 and WR-delA47L-2.
Synthetic peptides.
Peptides were purchased from Mimotopes (Clayton, Victoria, Australia) or Genscript Corp. (Piscataway, NJ). Master stocks (10 mg/ml in 100% dimethyl sulfoxide [DMSO]) were stored at −70°C, and peptides were diluted to required concentrations in serum-free DMEM before use. Peptides from the A47 protein are listed in Table 1; other peptide sequences used were as follows: B820, TSYKFESV; A8189, ITYRFYLI; A23297, IGMFNLTFI; K36, YSLPNAGDVI; A3270, KSYNYMLL; L253, VIYIFTVRL; A870, IHYLFRCV; A4288, YAPVSPIVI; A1947, VSLDYINTM (all from VACV [21, 38]); and OVA257, SIINFEKL (from chicken egg ovalbumin).
TABLE 1.
A47 peptides used
| Namea | Sequence | MHC | Rankb |
|---|---|---|---|
| A4793-101 | TMMINPFMI | H-2Db | |
| A47138-146 | AAFEFINSL | H-2Kb | |
| A47171-179 (MVA-029C)c | YAHINALEY | H-2Db | 2 |
| A47171-180 | YAHINALEYI | H-2Db | 1 |
| A47171-181 | YAHINALEYII | H-2Db | 4 |
| A47172-180 | AHINALEYI | H-2Db | 5 |
| A47172-181 | AHINALEYII | H-2Db | 3 |
| A47169-179 | LLYAHINALEY | H-2Db | 6 |
Numbers in subscripts represent the amino acid positions in A47 from VACV WR (or VACVWR-173).
Rank in H-2Db binding predictions of peptides tested using A47 (aa 168 to 187).
Name of peptide used in Mathew et al. (20). That paper based its amino acid positions for peptides on the sequence of A47 in MVA; those positions differ from the positions for VACV WR as used here.
Mice and infections.
Specific-pathogen-free, female C57BL/6 and BALB/c mice (all more than 8 weeks of age) were obtained from the Animal Resource Centre (Perth, Australia). Mice were housed and experiments were done in compliance with ethical requirements under an approval from the Australian National University (ANU) Animal Ethics and Experimentation Committee. For pathogenesis experiments using the dermal model, BALB/c mice were anesthetized by inhalation of isofluorane (4% in medical-grade oxygen at 0.8 liters/min) and intradermally (i.d.) injected with 1 × 104 PFU in 10 μl of phosphate-buffered saline (PBS) in the left ear pinnae. The sizes of the lesions that developed at the injection site were estimated daily as previously described (39, 40). For intranasal (i.n.) infections, BALB/c and C57BL/6 mice were anesthetized as described above and infected with 1 × 104 and 2 × 104 PFU of virus, respectively, in a total of 20 μl of PBS (10 μl per naris). Mice were monitored and scored daily for signs of illness and weight. Mice that lost more than 30% of starting body weight and that had an undiminished illness score were euthanized. For immunization of mice, C57BL/6 mice were injected intraperitoneally (i.p.) with 1 × 106 PFU of virus in 200 μl of PBS or i.d. as described above except with 1 × 106 PFU.
Stimulations and ICS of IFN-γ.
Mice were euthanized 7 days after infection, and spleens were taken for analysis of CD8+ T cell responses by intracellular staining (ICS) as described previously (38). Briefly, splenocytes were plated at 1 × 106 cells/well into round-bottom 96-well plates. Unless otherwise stated, synthetic peptides were added to achieve a final concentration of 10−7 M and plates were incubated at 37°C and 5% CO2. In some experiments, DC2.4 cells (33) infected with VACV for 5 h were used instead of peptides as stimulators. After 1 h, 5 μg/ml brefeldin A (Sigma) was added and plates were incubated for another 3 h. Plates were spun at 4°C, medium was removed, and cells were resuspended in 50 μl of anti-CD8-phycoerythrin (anti-CD8-PE) (clone 53-6.7; BioLegend) diluted 1:150. After 30 min of incubation on ice, cells were washed, resuspended in 50 μl of 1% paraformaldehyde, and incubated at room temperature for 20 min before another two washes and staining with 50 μl of anti-gamma interferon (IFN-γ)-allophycocyanin (anti-IFN-γ-APC) (clone XMG1.2; BioLegend) (diluted 1:200) overnight in PBS with 0.5% saponin (Sigma) at 4°C. Cells were washed three times before acquisition was performed using an LSR II flow cytometer (BD Biosciences). Analysis was done using Flowjo software (Tree Star Inc.). Events were gated for live lymphocytes on forward scatter (FSC) against side scatter (SSC) followed by CD8+ T cells against IFN-γ. Background levels (determined using irrelevant peptides or no peptide) were usually on the order of 0.1 to 0.2% of CD8+ events and in most experiments were subtracted from the values presented for test samples.
Granzyme B (GzmB), CD62L, and DimerX staining.
DimerX reagents (BD Biosciences) represent a variant of peptide-MHC tetramer technology and were used as previously published (14). Briefly, H-2Kb:Ig fusion protein was loaded overnight at 37°C with a 40 M excess of B820 or OVA257 peptide. Peptide-loaded dimers were then incubated for 1 h at room temperature with α-mouse IgG1-PE (clone A85-1; BD Biosciences). Single-cell suspensions of splenocytes were labeled with DimerX reagent, anti-CD62L-fluorescein isothiocyanate (anti-CD62L-FITC) (clone 53-6.7; BioLegend), and anti-CD8-APC-Cy7 (BD Biosciences). After several washes, cells were fixed in 1% paraformaldehyde, washed again, and stained with anti-human GzmB-APC (clone GB12; Invitrogen [supplied by Caltag]) in PBS with 0.5% saponin (Sigma). In some cases, staining was done as described above, but the Dimer-X reagent was omitted and a PE conjugate of anti-CD8+ was used instead of an APC-Cy7 conjugate. After staining, cells were washed twice before acquisition was performed using an LSR II cytometer (BD Biosciences). Analysis was done using Flowjo software (Tree Star Inc.) with gating for lymphocytes on FSC against SSC followed by CD8+ events or CD8+ and DimerX+ events. Final plots displayed GzmB against CD62L, with gates drawn based on the populations seen with naïve and VACV-infected mice.
Purification and transfer of OT-I CD8+ T cells.
Spleens and lymph nodes were collected from OT-I mice (17), and single-cell suspensions were made; the percentage of CD8+ T cells that recognized OVA257 in these mice was >90%, as determined by DimerX staining. OT-I CD8+ T cells were then purified using a magnetic separation protocol based on negative selection (CD8α+ T cell isolation kit II; Miltenyi Biotec). A total of 5 × 107 or 1 × 105 CD8+ OT-I cells were transferred into C57BL/6 mice by intravenous injection 1 day prior to infection with VACV WR or VACV-WR-SIIN, respectively.
Statistical analysis.
Statistical comparisons were done with the aid of Prism software (version 5.01; GraphPad) using the nonparametric Mann-Whitney U test. To calculate the error for the differences in the numbers of CD8+ T cells in infected and naïve mice, we took the square root of the sum of squares of the standard errors of the means (SEMs) associated with the average numbers of CD8+ T cells in these two groups of mice.
RESULTS
A47L is not required for the growth of VACV in culture.
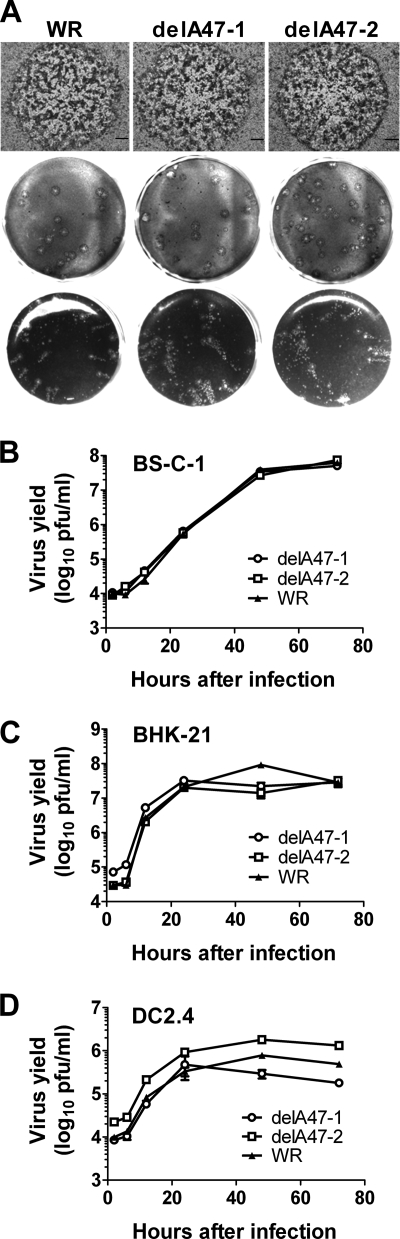
A47L homologues are not found in poxvirus genera outside the orthopoxviruses, and the gene is located toward the right end of the VACV genome, outside the central region where most genes encoding basic replicative functions are located (42). Therefore, we reasoned that it was likely that A47L would not be required for growth of VACV in vitro. To test this hypothesis, we set out to generate VACVs with all but the last three amino acids of the A47L open reading frame deleted. The method used was a recently developed variant of transient-dominant selection that uses a combination of fluorescence (enhanced green fluorescent protein [eGFP]) screening and antibiotic resistance (specifically, blasticidin resistance) to choose the unstable intermediates (Tscharke and Wong, unpublished). Two independent deletions of A47L were made, each representing progeny from independent transfection-infection experiments, to control for possible second-site mutations that might be inadvertently chosen through the plaque purification process. The isolation of these viruses demonstrates that A47L is indeed dispensable for growth in culture. To determine whether the loss of A47L had a more subtle effect on virus growth in vitro, we compared plaque size and morphology characteristics after infection of monolayers of BS-C-1 cells with the two deletion viruses and a wild-type control (Fig. 1 A). Growth kinetics for these three viruses resulting from a low multiplicity of infection were likewise determined by the use of two fibroblastoid cell lines (BS-C-1 and BHK-21) and a mouse dendritic cell line (DC2.4); no consistent differences were found (Fig. 1B to D). From these experiments, we concluded that A47L plays no role in basic replication of the virus in culture.
FIG. 1.
Deletion of A47L does not affect VACV growth in vitro. VACV-delA47-1, VACV-delA47-2, and a matched wild-type (WT) strain were grown in BS-C-1 (A and B), BHK-21 (C), or DC2.4 (D). (A) Plaque size and morphology, from experiments performed with (top two rows) and without (bottom row) CMC in the growth medium to inhibit the spread of virus from plaques. Bars shown in the top row represent 300 μm. (B to D) Virus growth and spread in monolayers of the cells as seen after infection at a multiplicity of infection (MOI) of 0.01 PFU/cell.
A47L does not affect VACV pathogenesis in mice.
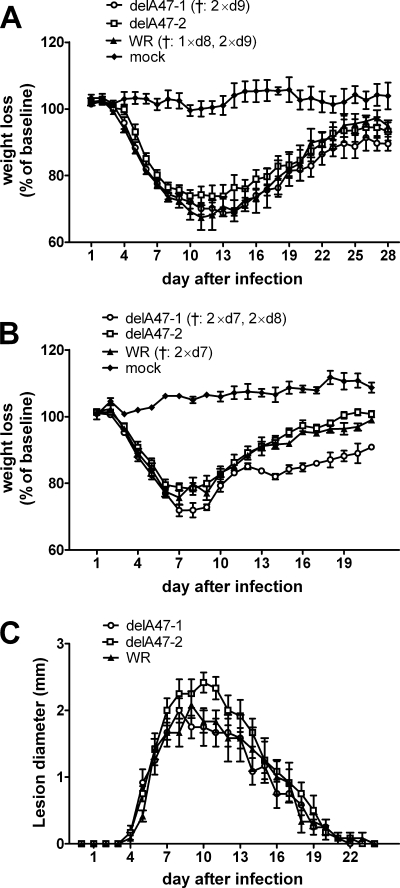
VACV genes that do not play roles in replication in vitro are generally assumed to be important for pathogenesis. With this in mind, we studied the pathogenesis of the two WR-delA47L viruses in mice. We first used intranasal models in BALB/c and also C57BL/6 mice, monitoring body weight as a sign of illness, and subsequently used the intradermal model in BALB/c mice, measuring the lesion size at the site of injection (Fig. 2). Across these three models, no significant differences in pathogenesis were seen at any time. These data suggest that, whatever the role played by A47L may be, it cannot be revealed in such mouse models.
FIG. 2.
Deletion of A47L does not affect VACV pathogenesis in vivo. Groups of 6 BALB/c mice (A and C) or 5 C57BL/6 mice (B) were infected with VACV-delA47-1, VACV-delA47-2, or a matched wild-type (WT) strain using an i.n. (A and B) or i.d. (C) model. (A and B) Mice were weighed daily, and averages and SEMs of percentages of weight loss are shown. †, numbers of mice that were euthanized for humane reasons on the day (d) indicated. (C) Averages and SEMs of estimated lesion diameters on infected ears are shown.
All CD8+ T cell epitopes mapped in A47L prime CD8+ T cells in vivo.
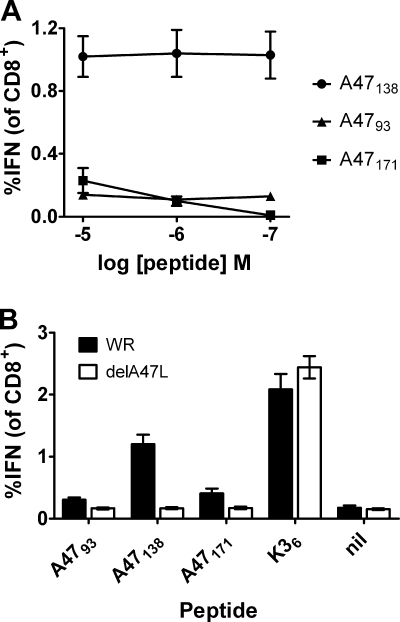
A47L was also of interest because it has been reported to contain many CD8+ T cell epitopes, but in each case these have been identified using synthetic peptides and in vitro assays (20, 21, 27, 28, 37, 38). Having made the delA47L viruses, we had an opportunity to formally test the proposition that the T cells detected by some of these peptides in vitro are primed in vivo by the A47 protein. The C57BL/6 model was used because three CD8+ T cell epitopes in A47L have been reported for this strain: A4793-101 (TMMINPFMI) (21); A47138-146 (AAFEFINSL) (38); and A47171-179 (YAHINALEY) (named MVA-029C in reference 20). Before going forward with these experiments, it was important to ensure that we could detect responses to the three published peptides. Each peptide was tested at three concentrations (from 1 × 10−5 M to 1 × 10−7 M) for its ability to stimulate CD8+ T cells from the spleens of mice that had been infected with VACV WR 7 days earlier, as seen by production of intracellular IFN-γ (Fig. 3 A). As shown previously (38), A47138 stimulated similar percentages of CD8+ T cells at all three peptide concentrations, as did A4793, albeit at a lower level. In contrast, while A47171 stimulated CD8+ T cells at 1 × 105 M, fewer were stimulated at 1 × 10−6 M; at 1 × 10−7 M, the response was no longer detectable, echoing previous findings that this peptide requires high concentrations for such responses (20). It is very unlikely that a peptide that requires the use of such unnaturally high concentrations in assays such as this is generated in vivo and is able to prime CD8+ T cells. It was then postulated that this peptide acted as a mimic or mimotope that is able to stimulate in vitro T cell populations primed by another peptide in vivo (7, 46). We next tested whether generation of responses to all three peptides during VACV infection required an intact A47L gene. This was done by comparing responses to the peptides in the same assay 7 days after infection with WR and WR-delA47L-1 (Fig. 3B). We used the higher concentration (1 × 10−5 M) for A47171 to ensure detection but used the standard concentration (1 × 10−7 M) for the other peptides. For each peptide, including A47171, the percentage of CD8+ T cells stimulated was significantly (P < 0.02) higher for splenocytes from WR-infected mice than for those from WR-delA47-1-infected mice. Further, splenocytes from WR-delA47L-1-infected mice did not show responses significantly higher than the background level (all P > 0.5), unlike those from WR-infected mice (all P < 0.02). Finally, all mice showed strong responses to two peptides not derived from A47, namely, K36 (Fig. 3B) and B820 (not shown), demonstrating that WR-delA47L-1 was not inherently less immunogenic for CD8+ T cells. Taken together, these data show that, for all three epitopes mapped to A47L, the T cell populations revealed in vitro were primed by A47 in vivo.
FIG. 3.
Responses to three CD8+ T cell epitopes in A47. Groups of C57BL/6 mice were infected i.p. with 106 PFU of VACV; 7 days later, peptide-specific CD8+ T cell responses were measured using ICS for IFN-γ. Average percentages (and SEMs) of CD8+ T cells that produced IFN-γ in ex vivo stimulations with the indicated peptides are shown. (A) CD8+ T cell responses that were detected using the peptides shown at the concentrations given on the x axis. (B) CD8+ T cell responses to the peptides shown on the x axis in mice infected with VACV WR or VACV-delA47-1. A4793, A47138, and K36 were used at 1 × 10−7 M; A47171 was used at 1 × 10−5 M. nil, a control where no peptide was used. All data are from two independent experiments, each performed with groups of three mice.
A redefined CD8+ T cell epitope in A47L stimulates strong responses.
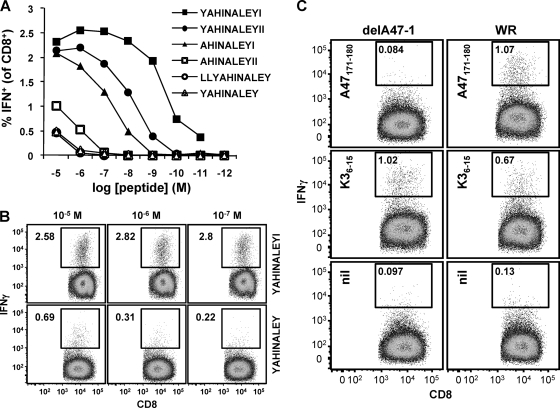
Having demonstrated that A47171 was very unlikely to be a mimotope of a peptide derived from a protein other than A47, the reason that such high concentrations of this peptide were required in assays remained unclear. Another possible explanation was that the peptide reported did not represent the minimal optimal sequence. An algorithm (ANN [24], accessed via the Immune Epitope Database and Analysis Resource [IEDB; www.iedb.org]) was then used to predict possible H-2Db-binding peptides in a 20-amino-acid sequence (aa 168 to 187) based on the originally defined peptide. The list of possible binding peptides was reduced by excluding peptides that did not contain a core sequence (aa 172 to 179), as defined using data from Mathew et al. (20) and unpublished data (D. C. Tscharke), or that had been previously used (20). The remaining six peptides and their rank order as determined from the binding prediction are listed in Table 1. These peptides were then used over a range of concentrations to stimulate splenocytes from a mouse that had been infected with VACV 7 days earlier in our standard ICS assay (Fig. 4 A). The peptide ranked first in the predictions, A47171-180 (YAHINALEYI), performed best, as it was able to stimulate IFN-γ production at concentrations as low as 1 × 10−10 M. In contrast, the original peptide (A47171-179 YAHINALEY), although ranked second in the predictions, performed as poorly as the peptides ranked last. A comparison of the best-performing peptide to the originally defined epitope over the three highest concentrations is shown in Fig. 4B. We also tested versions of A47171-180 that were phosphorylated at either of the two tyrosines, but both were slightly less able to stimulate IFN-γ from CD8+ T cells than the unphosphorylated peptide at low concentrations (not shown). Therefore, we concluded that the optimal minimum epitope in this region was unphosphorylated A47171-180. To ensure that this new peptide also stimulated T cell populations primed by A47 in vivo, we used this peptide in ICS assays of splenocytes from WR- and WR-delA47L-1-infected mice (Fig. 4C). As expected, responses to the redefined A47171-180 peptide were not seen in mice infected with WR-delA47L-1.
FIG. 4.
Redefining a CD8+ T cell epitope in A47. C57BL/6 mice were infected i.p. with 106 PFU VACV; 7 days later, splenocytes were used in ICS assays. (A and B) Testing synthetic peptides for antigenic potency. Peptides at the indicated concentrations were used as stimulators for ICS (IFN-γ) assays. (A) Data are expressed as percentages of CD8+ T cells responding to each peptide concentration and are representative of 2 or more experiments. (B) Flow cytometry plots from the experiment described above, showing a direct comparison between the optimal peptide (YAHINALEYI [A47171-180]) and originally defined peptide (YAHINALEY [A47171-179]) at the 3 highest concentrations. (C) The optimal peptide stimulated responses by splenocytes only for those mice that were infected with a VACV with an intact A47L gene. Data represent flow cytometry plots of ICS performed using splenocytes from representative mice that had been infected with VACV WR or VACV-delA47-1 7 days earlier. The peptide used for stimulation in the assay is shown in each plot. Data are representative of the results seen with three mice.
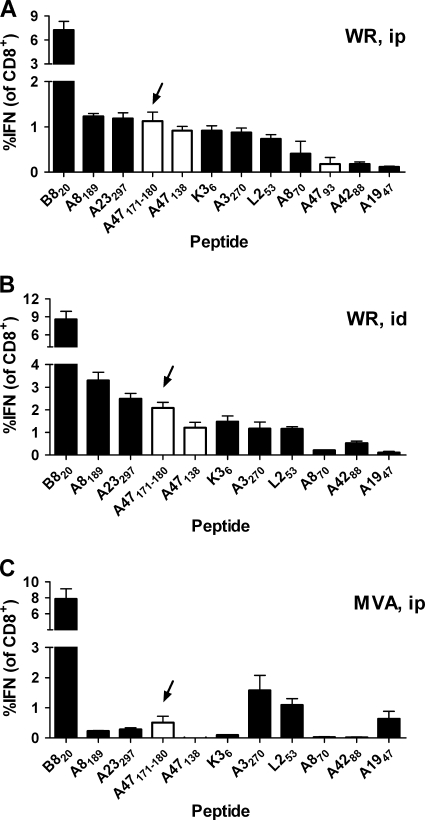
To put this redefined peptide epitope into the context of a more complete immunodominance hierarchy, we used this peptide with a panel of other known VACV epitopes, including the six most dominant in C57BL/6 mice. We did these experiments for mice infected with WR by i.p. and i.d. routes and for VACV strain MVA administered by the i.p. route (Fig. 5). For mice infected with VACV WR either i.p. or i.d., the newly defined A47171-180 was ranked fourth among the peptides; for mice immunized with MVA, it was ranked fifth. Notably, this peptide was in all cases ranked higher than the other A47-derived peptides, making it the most dominant peptide from this protein (results for A4793 are not shown for i.d. or MVA experiments, as it was not detectable above background levels).
FIG. 5.
The position of A47171-180 in the VACV immunodominance hierarchy. Groups of 3 C57BL/6 mice were infected with 106 PFU VACV WR i.p. (A) or i.d. (B) or with VACV MVA i.p. (C); 7 days later, splenocytes were used in ICS assays. Average percentages (and SEMs) of CD8+ T cells that produced IFN-γ in ex vivo stimulations with the indicated peptides are shown. Open bars show data for peptides from A47; the newly defined A47171-180 ([YAHINALEYI]) peptide is indicated with arrows. A4793 was not used for i.d. or MVA experiments, because preliminary data indicated that responses to this peptide were always at background levels after these immunizations. Results were confirmed by repeated experiments.
Taken together, the data in this section redefine a CD8+ T cell epitope in A47 that is the most dominant H-2b-restricted peptide from this protein and is among the more dominant epitopes in VACV for mice of this MHC haplotype.
Reevaluating the total size of the anti-VACV CD8+ T cell response.
It has been previously shown that, in C57BL/6 mice, the vast majority (indeed, almost 95%) of the CD8+ T cell response to VACV can be accounted for by the epitopes mapped to date (21). We were therefore interested to discover that the redefined A47171-180 peptide stimulated a relatively strong response for between 1% and 2% of CD8+ T cells in the spleen 7 days after infection. In the context of the current best estimates of a total anti-VACV CD8+ T cell response of between 25% and 30% of CD8+ T cells in the spleen (21), this new peptide would account for nearly 5% of this total. This suggests that either (i) with this new peptide defined, all peptides that generate substantial responses have now been identified (95 plus 5 is 100%) or (ii) the total anti-VACV response and, possibly, the responses to mapped peptides have been underestimated by current assays. It seems that the second scenario is more likely, because current measurements of the total CD8+ T cell response require restimulation of splenocytes in vitro by virus-infected cells, followed by ICS for IFN-γ. We reasoned that an infected cell found in the 3-to-4-h window of a standard stimulation used prior to ICS for IFN-γ is unlikely to be able to present all possible peptides than can be generated during infection. Here, we took two independent approaches to estimate the total CD8+ T cell response to VACV; importantly, neither of the methods relies on identification of known epitopes or on cells that present virus peptides in vitro.
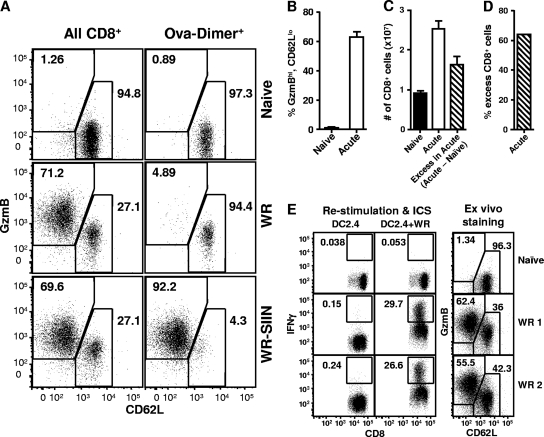
First, we took advantage of increased GzmB and decreased CD62L expression as a marker of activated CD8+ T cells. Unlike IFN-γ production, these markers can be detected directly ex vivo without the requirement for restimulation. GzmB has previously been used alone (13), but we found that use of this marker alone was not able to fully discriminate between activated and other CD8+ T cells (not shown). However, GzmB and CD62L used together effectively separated CD8+ T cells taken directly from the spleens of VACV WR-infected mice into two discrete populations on flow cytometry plots (Fig. 6 A). We defined these populations as GzmBhi-CD62Llo and GzmBlo-CD62Lhi, representing activated and naïve CD8+ T cells, respectively. In naïve mice, almost all CD8+ T cells (an average of 95%) are in the GzmBlo-CD62Lhi population, with 1 or 2% being GzmBhi-CD62Llo. In contrast, on day 7 after infection with VACV, an average of 63% of splenic CD8+ T cells were GzmBhi-CD62Llo and less than 40% were in the nonactivated population (Fig. 6A and B). To ensure that these markers are not expressed on non-virus-specific cells, perhaps as a result of bystander activation, OT-I T cells (specific for OVA257, a non-VACV peptide) (17) were transferred into mice 1 day prior to infection with VACV to provide a population of non-VACV-specific cells that could be tracked. After 7 days, splenocytes were stained for the OT-I cells (by the use of OVA257-loaded DimerX in addition to the other markers). The vast majority (94%) of the CD8+-DimerX+ events remained in the GzmBlo-CD62Lhi gate. This was in contrast to the results seen with the bulk of CD8+ T cells from the same mouse, of which 71% were in the GzmBhi-CD62Llo (activated) population. Finally, we demonstrated that OT-I CD8+ T cells can move into the GzmBhi-CD62Llo population (if activated) by infecting mice previously injected with OT-I CD8+ T cells with a VACV that presents OVA257 (WR-SIIN). In this case, 92% of the CD8+-DimerX+ events were in the GzmBhi-CD62Llo population (Fig. 6A). This result mirrors that found when mice were infected with VACV WR and a DimerX reagent loaded with the B820 peptide (an endogenous VACV epitope) was used (not shown). Therefore, by this method we estimated that, at 7 days after infection with VACV, 63% of CD8+ T cells were responding to the virus.
FIG. 6.
Reevaluation of the total CD8+ T cell response to VACV. (A) C57BL/6 mice were administered OT-I CD8+ T cells by i.v. injection and then left uninfected (top plots) or were infected with 106 PFU VACV WR (middle plots) or VACV-WR-SIIN (bottom plots). After another 7 days, spleens were taken and analyzed for surface CD8, CD62L, OVA257-DimerX, and intracellular GzmB. GzmB against CD62L plots are shown for CD8+ (left plots) and for CD8+ and OVA257-DimerX+ (right plots). Data are representative of the results obtained with 3 to 6 mice analyzed in 2 experiments. (B) Mice were infected with 106 PFU VACV (Acute) or left uninfected (Naïve); after 7 days, spleens were taken and analyzed for surface CD8, CD62L, and intracellular GzmB. The percentages of CD8+ events representing the GzmBhi-CD62Llo population are shown. Data are representative of the results obtained with 5 mice in 2 experiments. (C) Total numbers of CD8+ cells in spleens were measured by flow cytometry for 5 naïve (filled bar) and 30 acutely infected (day 7; open bar) C57BL/6 mice over multiple experiments; data representing averages and SEMs of the results are shown. Using these values, the difference between the acutely infected and naïve (acute − naïve = excess in acute [striped bar]) mice was calculated. (D) The percentage of excess (acute − naïve) CD8+ cells in VACV-infected mice (shown as a percentage of all CD8+ cells in acutely infected mice) was calculated using the data shown in panel C. (E) A direct comparison between ICS for IFN-γ after restimulation of splenocytes with VACV-infected cells (left 2 columns) and staining of the same splenocytes (without restimulation) for CD8, CD62L, and GzmB (right column). For restimulations, uninfected cells (DC2.4) were used as a control for assays using VACV-infected cells (DC2.4+WR). Splenocytes were taken from uninfected (Naïve) or infected (WR 1 and WR 2) mice.
Second, we used a simple method based on the increase in the total number of CD8+ T cells in acutely infected mice compared with the level seen with naïve mice. This method assumes (i) that the fraction of CD8+ T cells that are VACV specific in a naïve mouse is not significant (<1/1,000) (32), (ii) that bystander proliferation of non-virus-specific cells is negligible (22), (iii) that there is no substantial reduction in the numbers of naïve CD8+ T cells in acutely infected mice, and (iv) that the majority of CD8α+ cells in the spleen are conventional TCRα/β T cells (all of which are reasonable assumptions). According to this method, the number of VACV-specific CD8+ T cells in infected mice represents the difference between the total numbers of CD8+ T cells in acutely infected and naïve mice. In other words, it represents the number of “excess” CD8+ T cells observed in mice at the peak of acute infection compared to the number seen in naïve mice. The number of excess CD8+ T cells in acutely infected mice was calculated using data obtained with 30 infected and 5 naïve mice (Fig. 6C). Finally, to allow direct comparison with the data obtained using the GzmB-CD62L method described above, the number of “excess” CD8+ T cells in acutely infected mice was expressed as a percentage of the total number of CD8+ T cells in these mice. That percentage was calculated as follows: number of excess CD8+ T cells/total number of CD8+ T cells in acutely infected mice × 100%. The value thus calculated was 64% (Fig. 6D), which is in very close agreement with the value obtained with the GzmB-CD62L staining method.
Taken together (compare Fig. 6B and D), the results obtained by these two independent methods suggest that approximately 60% of all CD8+ T cells in the spleen of mice acutely infected with VACV WR are involved in the antiviral response. This is roughly double the previous best estimate of around 30% (15, 21, 38, 41).
Finally, we did a direct comparison of restimulation of splenocytes with VACV-infected cells followed by ICS to direct ex vivo staining for CD62L and GzmB as methods for estimating the size of the CD8+ T cell response to VACV in individual mice (Fig. 6E). Both methods have low background levels in naïve mice, and, for the two VACV-infected mice for which results are shown, the ex vivo method defined the size of the anti-viral CD8+ T cell response as being over twice the estimate provided by restimulation and ICS. These data demonstrate that, not only as an overall result for groups of mice, but also for individual mice, the method previously considered to be best led to underestimation of the size of the VACV-specific CD8+ T cell response by 2-fold.
DISCUSSION
VACV pathogenesis and immunity have been studied in animal models since the late 1930s (10, 23), but many unknowns remain. Indeed, the roles of many VACV genes in pathogenesis have yet to be explored or explained. In other cases, genes have been characterized and tested in more than a single model of pathogenesis, but characterization of their roles remains elusive (3, 6, 31, 39). From the data shown here, A47L fits into the latter class of VACV genes. That A47L was dispensable for growth was not surprising, given that it is not conserved beyond the orthopoxviruses and is fragmented (and most likely not expressed) in monkeypox virus. However, the level of conservation seen with most other orthopoxviruses led us to expect that a pathogenesis phenotype might be found for an A47L knockout. In contrast, although we investigated two strains of mice and two models of infection, no role was found for A47L. Our conclusion here is not that A47L has no role but rather that its role is dispensable or redundant for VACV pathogenesis in mice. There are no amino acid changes from WR in A47 that are not found in at least one other orthopoxvirus strain; therefore, it seems unlikely that A47L is simply dysfunctional in this strain. However, it remains possible that A47 works in concert with other virus functions; one or more of these might have been disrupted by the multiple artificial passages of VACV WR, thus nullifying the action of A47 in this strain. Alternatively, A47 may have a role in transmission of virus that cannot be captured in simple models of pathogenesis. Further, VACV persists in a natural reservoir and it is reasonable to suggest that it and some of the other orthopoxviruses, most notably cowpox virus, may have niches that involve multiple hosts (1, 9). In this scenario, not all virus genes would necessarily be required to aid growth in and/or transmission from all host species. We note that A47L is very highly conserved across VACV and cowpox and ectromelia viruses. Given the difficulty and expense of investigations using larger-animal models, mouse models of A47L deletions in cowpox and, especially, ectromelia virus are likely to have the best chance of revealing a function. Finally, the lack of a pathogenesis phenotype for the A47L deletion viruses shows that loss of individual CD8+ T cell epitopes (or, as in the case of C57BL/6 mice, loss of three epitopes) does not compromise control of infection. This is not surprising, as many more targets for CD8+ T cells remain; indeed, there is much redundancy in the control of VACV across the immune response as a whole (44).
Despite the fact that no role for A47 in VACV pathogenesis emerged from this study, the protein remains of interest from an immunological point of view because it contains so many CD8+ T cell epitopes. We confirm here that all the T cell populations thought to recognize peptides derived from A47 are indeed primed by A47 in vivo. To our knowledge, this is the first such published demonstration for any of the CD8+ T cell epitopes of VACV, although we have done similar experiments to validate the dominant B820 epitope (Tscharke, unpublished). Why A47L (or, indeed, the other immunoprevalent antigens) should encode so many epitopes is not clear, but the report of an early and high level of mRNA expression in a recent genome-wide analysis of VACV transcription is consistent with a good presentation of A47-derived peptides on infected cells (5). Further recent studies suggested that direct presentation (i.e., presentation from infected dendritic cells) is important for priming CD8+ T cell responses to VACV (16, 26, 45).
The extension of our finding of in vivo priming by A47 was the redefinition of a CD8+ T cell epitope based on an amino acid sequence previously shown to detect a T cell population only when used at a high concentration. Why this optimal peptide was not found previously in a large study based on prediction is not clear. The minimal peptide as mapped here was predicted to be a good binder in the previous study, and at least one other form of the peptide that we found that could elicit CD8+ T cell responses at 1 × 10−7 M was included in the pools of peptide tested (21). In any case, our data emphasize (i) the importance of careful characterization of putative epitopes, requiring particular attention to the concentrations required for activity in assays, and (ii) the technical difficulty of detecting every possible hit when screening large pools of peptides. Having noted this, we would not suggest that finding a single missed peptide invalidates large-scale approaches but rather that such a finding would help researchers to interpret them by placing a limit on their sensitivity. With respect to the value of current predictive algorithms, we used the ANN method (24) to predict MHC binding peptides from A47, and TMMINPFMI and YAHINALEYI rank first and second in the list of H-2Db binders (YAHINLEY ranks eighth), whereas AAFEFINSL ranks second among predicted H-2Kb binders. In this respect, our findings here support the use of such tools.
The finding that the total CD8+ T cell response to VACV is larger than previously thought is timely, given a recent upward revision of the estimated number of CD8+ precursors that can recognize this virus in naïve mice (32). In addition, a published experiment that used GzmB alone suggested that the total size of the CD8+ T cell response to VACV may be higher than that predicted by other estimates (13). Likewise, the total response to lymphocytic choriomeningitis virus (LCMV) has also recently been found to be higher than was previously appreciated (19). Our new higher estimate of the total response to VACV is of importance for at least three reasons. (i) It aids our understanding of the natural history of VACV infection and how effectively this virus evades, or does not evade, the generation of CD8+ T cell immunity. The finding that such a large percentage of CD8+ T cells responds to the virus at the peak of infection suggests that, if the virus is able to damp the priming of this response, then either the immune system has overcome this mechanism or it is relatively ineffective. This finding certainly makes it very difficult to sustain the argument that VACV substantially evades the priming of CD8+ T cells by inhibiting dendritic cell function (11). (ii) It is required for placing the VACV epitopes mapped thus far into the proper context. While it is possible that responses to currently mapped peptides are simply underestimated by ICS and tetramer assays, it seems unlikely that these methods are revealing only half of the T cells with the appropriate specificity. Indeed, in the case of acute LCMV infection, ICS for IFN-γ after stimulation with 10 known peptides can account for up to 80% of all CD8+ T cells, and this approach limits underestimation by this method to far less than 2-fold (8, 18, 19, 22). This, and the apparent ease with which we found a peptide missed by two genome-wide mapping efforts, suggests to us that more VACV CD8+ T cell epitopes remain to be mapped. If this is the case, the most dominant peptide, namely, B820, is not as dominant as previously thought. Instead of accounting for around 20 to 25% of the total response, it perhaps represents closer to 10% of the response. The finding of a higher total response also leaves room for the existence of another, equally dominant epitope. It also suggests that the prediction and screening method used to comprehensively map epitopes identified a smaller fraction of immunogenic epitopes than was previously reported (21). (iii) It also provides a context for the responses elicited by the recombinant antigens expressed by engineered VACV vaccines. The responses to recombinant antigens seen in mouse and human studies with VACV vaccines were always dwarfed by responses primed by endogenous VACV epitopes (15, 34). It would now seem that the disparity is even greater. However, in this context, it clearly shows the potential of VACV to stimulate very robust CD8+ T cell responses and, as noted above, suggests that VACV has a limited capacity for evading the priming of CD8+ T cells. Further, in previous work we found little evidence that responses to some epitopes are reduced by immunodomination in primary infections by VACV (14, 43). Overall, we would suggest that an inherently high immunogenicity of VACV for CD8+ T cells is encouraging for recombinant vaccines, especially if these strong responses can be further directed toward recombinant antigens. However, it remains to be shown whether all the anti-viral CD8+ T cells we detected with our method are of equal quality or functionality. This would be important to determine in future studies.
Finding higher-than-expected responses to VACV and LCMV suggests that it may be fruitful to revisit estimations of the total size of CD8+ T cell responses to other viruses. This is especially the case where the best estimate of total response was determined using virus-infected cells as stimulators in in vitro assays, the method that was the gold standard for VACV investigations. It might also be most appropriate for studies of larger viruses, where the mapping of all epitopes is a far more difficult task.
In summary, while no role has been found for A47L in the replication or pathogenesis of the virus, we confirm that it is indeed an immunoprevalent antigen for CD8+ T cells. Further, our studies have led to a reevaluation of the total size of the anti-VACV CD8+ T cell response that has important ramifications for our understanding of the immunology of VACV and VACV-vectored vaccines.
Acknowledgments
We thank Stewart Smith for technical assistance, Bernard Moss for VACV WR and MVA, and Jon Yewdell and Jack Bennink for VACV NP-S-GFP.
This work was funded by grants to D.C.T. from the Australian NHMRC (Biomedical CDA 418108 and projects 389819 and 509199).
Footnotes
Published ahead of print on 28 July 2010.
REFERENCES
- 1.Abrahão, J. S., M. I. Guedes, G. S. Trindade, F. G. Fonseca, R. K. Campos, B. F. Mota, Z. I. Lobato, A. T. Silva-Fernandes, G. O. Rodrigues, L. S. Lima, P. C. Ferreira, C. A. Bonjardim, and E. G. Kroon. 2009. One more piece in the VACV ecological puzzle: could peridomestic rodents be the link between wildlife and bovine vaccinia outbreaks in Brazil? PLoS One 4:e7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcamí, A., and G. L. Smith. 1992. A soluble receptor for interleukin-1 beta encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell 71:153-167. [DOI] [PubMed] [Google Scholar]
- 3.Almazán, F., D. C. Tscharke, and G. L. Smith. 2001. The vaccinia virus superoxide dismutase-like protein (A45R) is a virion component that is nonessential for virus replication. J. Virol. 75:7018-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antón, L. C., U. Schubert, I. Bacik, M. F. Princiotta, P. A. Wearsch, J. Gibbs, P. M. Day, C. Realini, M. C. Rechsteiner, J. R. Bennink, and J. W. Yewdell. 1999. Intracellular localization of proteasomal degradation of a viral antigen. J. Cell Biol. 146:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assarsson, E., J. A. Greenbaum, M. Sundstrom, L. Schaffer, J. A. Hammond, V. Pasquetto, C. Oseroff, R. C. Hendrickson, E. J. Lefkowitz, D. C. Tscharke, J. Sidney, H. M. Grey, S. R. Head, B. Peters, and A. Sette. 2008. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc. Natl. Acad. Sci. U. S. A. 105:2140-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banham, A. H., and G. L. Smith. 1993. Characterization of vaccinia virus gene B12R. J. Gen. Virol. 74:2807-2812. [DOI] [PubMed] [Google Scholar]
- 7.Belz, G. T., W. Xie, and P. C. Doherty. 2001. Diversity of epitope and cytokine profiles for primary and secondary influenza A virus-specific CD8+ T cell responses. J. Immunol. 166:4627-4633. [DOI] [PubMed] [Google Scholar]
- 8.Butz, E. A., and M. J. Bevan. 1998. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity 8:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chantrey, J., H. Meyer, D. Baxby, M. Begon, K. J. Bown, S. M. Hazel, T. Jones, W. I. Montgomery, and M. Bennett. 1999. Cowpox: reservoir hosts and geographic range. Epidemiol. Infect. 122:455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dienes, L., and H. L. Naterman. 1937. The immunological response to vaccinia in guinea pigs. J. Infect. Dis. 61:279-290. [Google Scholar]
- 11.Engelmayer, J., M. Larsson, M. Subklewe, A. Chahroudi, W. I. Cox, R. M. Steinman, and N. Bhardwaj. 1999. Vaccinia virus inhibits the maturation of human dendritic cells: a novel mechanism of immune evasion. J. Immunol. 163:6762-6768. [PubMed] [Google Scholar]
- 12.Falkner, F. G., and B. Moss. 1990. Transient dominant selection of recombinant vaccinia viruses. J. Virol. 64:3108-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, M., and L. J. Sigal. 2006. Direct CD28 costimulation is required for CD8+ T cell-mediated resistance to an acute viral disease in a natural host. J. Immunol. 177:8027-8036. [DOI] [PubMed] [Google Scholar]
- 14.Flesch, I. E., W. P. Woo, Y. Wang, V. Panchanathan, Y. C. Wong, N. L. La Gruta, T. Cukalac, and D. C. Tscharke. 2010. Altered CD8+ T cell immunodominance after vaccinia virus infection and the naïve repertoire in inbred and F1 mice. J. Immunol. 184:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrington, L. E., R. van der Most, J. L. Whitton, and R. Ahmed. 2002. Recombinant vaccinia virus-induced T-cell immunity: quantitation of the response to the virus vector and the foreign epitope. J. Virol. 76:3329-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman, H. D., K. Takeda, C. N. Skon, F. R. Murray, S. E. Hensley, J. Loomis, G. N. Barber, J. R. Bennink, and J. W. Yewdell. 2008. Direct priming of antiviral CD8+ T cells in the peripheral interfollicular region of lymph nodes. Nat. Immunol. 9:155-165. [DOI] [PubMed] [Google Scholar]
- 17.Hogquist, K. A., S. C. Jameson, W. R. Heath, J. L. Howard, M. J. Bevan, and F. R. Carbone. 1994. T cell receptor antagonist peptides induce positive selection. Cell 76:17-27. [DOI] [PubMed] [Google Scholar]
- 18.Kotturi, M. F., I. Scott, T. Wolfe, B. Peters, J. Sidney, H. Cheroutre, M. G. von Herrath, M. J. Buchmeier, H. Grey, and A. Sette. 2008. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J. Immunol. 181:2124-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masopust, D., K. Murali-Krishna, and R. Ahmed. 2007. Quantitating the magnitude of the lymphocytic choriomeningitis virus-specific CD8 T-cell response: it is even bigger than we thought. J. Virol. 81:2002-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathew, A., M. Terajima, K. West, S. Green, A. L. Rothman, F. A. Ennis, and J. S. Kennedy. 2005. Identification of murine poxvirus-specific CD8+ CTL epitopes with distinct functional profiles. J. Immunol. 174:2212-2219. [DOI] [PubMed] [Google Scholar]
- 21.Moutaftsi, M., B. Peters, V. Pasquetto, D. C. Tscharke, J. Sidney, H. H. Bui, H. Grey, and A. Sette. 2006. A consensus epitope prediction approach identifies the breadth of murine TCD8+-cell responses to vaccinia virus. Nat. Biotechnol. 24:817-819. [DOI] [PubMed] [Google Scholar]
- 22.Murali-Krishna, K., J. D. Altman, M. Suresh, D. J. Sourdive, A. J. Zajac, J. D. Miller, J. Slansky, and R. Ahmed. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8:177-187. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, J. B. 1938. The behavior of poxviruses in the respiratory tract. I. The response of mice to the nasal instillation of vaccinia virus. J. Exp. Med. 68:401-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, M., C. Lundegaard, P. Worning, S. L. Lauemøller, K. Lamberth, S. Buus, S. Brunak, and O. Lund. 2003. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 12:1007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norbury, C. C., S. Basta, K. B. Donohue, D. C. Tscharke, M. F. Princiotta, P. Berglund, J. Gibbs, J. R. Bennink, and J. W. Yewdell. 2004. CD8+ T cell cross-priming via transfer of proteasome substrates. Science 304:1318-1321. [DOI] [PubMed] [Google Scholar]
- 26.Norbury, C. C., D. Malide, J. S. Gibbs, J. R. Bennink, and J. W. Yewdell. 2002. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat. Immunol. 3:265-271. [DOI] [PubMed] [Google Scholar]
- 27.Oseroff, C., B. Peters, V. Pasquetto, M. Moutaftsi, J. Sidney, V. Panchanathan, D. C. Tscharke, B. Maillere, H. Grey, and A. Sette. 2008. Dissociation between epitope hierarchy and immunoprevalence in CD8 responses to vaccinia virus Western Reserve. J. Immunol. 180:7193-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasquetto, V., H.-H. Bui, R. Giannino, C. Banh, F. Mirza, J. Sidney, C. Oseroff, D. C. Tscharke, K. Irvine, J. R. Bennink, B. Peters, S. Southwood, V. Cerundolo, H. M. Grey, J. W. Yewdell, and A. Sette. 2005. HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J. Immunol. 175:5504-5515. [DOI] [PubMed] [Google Scholar]
- 29.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, K. Limbach, E. K. Norton, and E. Paoletti. 1990. Vaccinia virus host range genes. Virology 179:276-286. [DOI] [PubMed] [Google Scholar]
- 30.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, E. K. Norton, and E. Paoletti. 1991. Deletion of 55 open reading frames from the termini of vaccinia virus. Virology 180:406-410. [DOI] [PubMed] [Google Scholar]
- 31.Price, N., D. C. Tscharke, and G. L. Smith. 2002. The vaccinia virus B9R protein is a 6 kDa intracellular protein that is non-essential for virus replication and virulence. J. Gen. Virol. 83:873-878. [DOI] [PubMed] [Google Scholar]
- 32.Seedhom, M. O., E. R. Jellison, K. A. Daniels, and R. M. Welsh. 2009. High frequencies of virus-specific CD8+ T-cell precursors. J. Virol. 83:12907-12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen, Z., G. Reznikoff, G. Dranoff, and K. L. Rock. 1997. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J. Immunol. 158:2723-2730. [PubMed] [Google Scholar]
- 34.Smith, C. L., F. Mirza, V. Pasquetto, D. C. Tscharke, M. J. Palmowski, P. R. Dunbar, A. Sette, A. L. Harris, and V. Cerundolo. 2005. Immunodominance of poxviral-specific CTL in a human trial of recombinant-modified vaccinia Ankara. J. Immunol. 175:8431-8437. [DOI] [PubMed] [Google Scholar]
- 35.Staib, C., I. Drexler, and G. Sutter. 2004. Construction and isolation of recombinant MVA, p. 77-99. In S. N. Isaacs (ed.), Vaccinia virus and poxvirology: methods and protocols. Humana Press, Clifton, NJ. [DOI] [PubMed]
- 36.Symons, J. A., A. Alcamí, and G. L. Smith. 1995. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 81:551-560. [DOI] [PubMed] [Google Scholar]
- 37.Terajima, M., J. Cruz, A. M. Leporati, W. E. Demkowicz, Jr., J. S. Kennedy, and F. A. Ennis. 2006. Identification of vaccinia CD8+ T-cell epitopes conserved among vaccinia and variola viruses restricted by common MHC class I molecules, HLA-A2 or HLA-B7. Hum. Immunol. 67:512-520. [DOI] [PubMed] [Google Scholar]
- 38.Tscharke, D. C., G. Karupiah, J. Zhou, T. Palmore, K. R. Irvine, S. M. M. Haeryfar, S. Williams, J. Sidney, A. Sette, J. R. Bennink, and J. W. Yewdell. 2005. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 201:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tscharke, D. C., P. C. Reading, and G. L. Smith. 2002. Dermal infection with vaccinia virus reveals roles for virus proteins not seen using other inoculation routes. J. Gen. Virol. 83:1977-1986. [DOI] [PubMed] [Google Scholar]
- 40.Tscharke, D. C., and G. L. Smith. 1999. A model for vaccinia virus pathogenesis and immunity based on intradermal injection of mouse ear pinnae. J. Gen. Virol. 80:2751-2755. [DOI] [PubMed] [Google Scholar]
- 41.Tscharke, D. C., W.-P. Woo, I. G. Sakala, J. Sidney, A. Sette, D. J. Moss, J. R. Bennink, G. Karupiah, and J. W. Yewdell. 2006. Poxvirus CD8+ T-cell determinants and cross-reactivity in BALB/c mice. J. Virol. 80:6318-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Upton, C., S. Slack, A. L. Hunter, A. Ehlers, and R. L. Roper. 2003. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J. Virol. 77:7590-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Y., I. E. A. Flesch, and D. C. Tscharke. 2009. Vaccinia virus CD8+ T cell dominance hierarchies cannot be altered by prior immunization with individual peptides. J. Virol. 83:9008-9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, R., A. J. Johnson, D. Liggitt, and M. J. Bevan. 2004. Cellular and humoral immunity against vaccinia virus infection of mice. J. Immunol. 172:6265-6271. [DOI] [PubMed] [Google Scholar]
- 45.Xu, R. H., S. Remakus, X. Ma, F. Roscoe, and L. J. Sigal. 2010. Direct presentation is sufficient for an efficient anti-viral CD8 T cell response. PLoS Pathog. 6:e1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yewdell, J. W. 2005. The seven dirty little secrets of major histocompatibility complex class I antigen processing. Immunol. Rev. 207:8-18. [DOI] [PubMed] [Google Scholar]