Abstract
Efforts to develop adenovirus vectors suitable for genetic interventions in humans have identified three major limitations of the most frequently used vector prototype, human adenovirus serotype 5 (Ad5). These limitations—widespread preexisting anti-Ad5 immunity in humans, the high rate of transduction of normal nontarget tissues, and the lack of target-specific gene delivery—justify the exploration of other Ad serotypes as vector prototypes. In this paper, we describe the development of an alternative vector platform using simian Ad serotype 24 (sAd24). We found that sAd24 virions formed unstable complexes with blood coagulation factor X and, because of that, transduced the liver and other organs at low levels when administered intravenously. The overall pattern of biodistribution of sAd24 particles was similar, however, to that of Ad5, and the intravenously injected sAd24 was cleared by Kupffer cells, leading to their depletion. We modified the virus's fiber protein to design a Her2-specific derivative of sAd24 capable of infecting target human tumor cells in vitro. In the presence of neutralizing anti-Ad5 antibodies, Her2-mediated infection with targeted sAd24 compared favorably to that with the Ad5-derived vector. When used to target Her2-expressing tumors in animals, this fiber-modified vector achieved a higher level of gene transfer to metastasis-containing murine lungs than to tumor-free lungs. In aggregate, these studies provide important insights into sAd24 biology, identify its advantages and limitations as a vector prototype, and are thus essential for further development of an sAd24-based gene delivery platform.
Ads are a highly diverse family of nonenveloped, DNA-containing viruses isolated from humans, various mammals, and other vertebrates. Despite this diversity, the structure of a typical Ad particle is highly conserved among various Ad serotypes. The double-stranded DNA genome of Ad associated with the core proteins is encapsidated in an icosahedral protein shell. The most abundant structural component of an Ad particle is the hexon protein that forms the facets of the capsid. The other two major capsid proteins, the penton base and the fiber, associate at a 5-to-3 ratio to form the penton capsomers located at each of the apexes of the virion.
Efforts to develop efficient Ad vectors for gene delivery in humans have centered primarily on human Ad serotype 5, owing to its well-elucidated biology. However, testing of Ad5 vectors in preclinical studies and gene therapy clinical trials identified several important drawbacks: widespread preexisting anti-Ad5 immunity in humans (12, 55), an unacceptably large amount of damage to normal nontarget tissues, which causes severe toxicity (28, 37), and low levels of target-specific gene delivery (15, 18, 19). Collectively, these findings warranted the search for Ad vector prototypes better suited for gene transfer in humans.
Recent improvements in vector technology have exploited the unique biological properties of Ad serotypes other than Ad5. For instance, the low seroprevalence of certain Ads in humans has justified the use of these viruses as alternative vector prototypes to overcome preexisting anti-Ad5 immunity. Indeed, it has been shown that the efficacy of in vivo gene delivery by vectors derived from simian Ads is minimally affected by Ad5-neutralizing antibodies (16, 45, 55), making these viruses promising vector prototypes.
The use of vectors based on Ads other than Ad5 may also offer the solution to the problem of the undesired in vivo transduction of nontarget tissues. Recent studies showed that the high-affinity association of FX with the hexon of systemically injected Ad5 directs the virus to FX receptors abundantly expressed in the liver, causing massive hepatic transduction (22, 56). Importantly, some Ad serotypes, such as Ad26 and Ad48, do not bind FX and cause no detectable transduction of the liver on vascular delivery (56). These findings provided the rationale for the recently reported improvements of Ad5 vectors through genetic engrafting into their hexons of either the hypervariable regions or the individual amino acids from the hexons of these FX-nonbinding serotypes (2, 42). They also provided an additional justification for developing such Ad serotypes as alternative gene vector platforms.
While the use of alternative Ad serotypes promises to solve two of the main drawbacks of Ad5 vectors, it does not address the problem of inefficient gene delivery to target tissues. Thus, development of an Ad vector targeting strategy applicable to these promising vector prototypes would constitute an important step toward the use of Ads in humans.
Most attempts to design gene delivery vectors with selectivity for specific markers on target cells have involved genetic alteration of the Ad5 fiber protein. The fiber protein is the mediator of the initial high-affinity interaction in vitro between the Ad virion and the primary receptor (14, 41) that is followed by secondary contact between the Ad penton base protein and cellular integrins (58, 59), which triggers internalization of Ad by the cell. The primary receptor binding is facilitated by the globular, carboxy-terminal knob domain of the fiber (20, 29). The knob also initiates the fiber trimerization (36) that is essential for its encapsidation (38) through anchoring of the amino-terminal tail of the protein within the penton base pentamer. The knob is extended away from the capsid surface by the fiber's central shaft domain, which is structured as a β-spiral (54). The primary Ad receptors identified to date, such as CAR (8, 51), CD46 (17), sialic acid (4), and CD80 and CD86 (50), are suboptimal targets for gene therapeutics because their expression is not correlated with disease.
The approach to Ad targeting initially developed for the Ad5 fiber protein includes identification of the receptor-binding site within the knob (9, 23, 43), its inactivation by mutagenesis (24, 43), and introduction of a target-specific ligand within the structure of the modified knob (6, 15, 31, 33, 60, 61). The unique structures of Ad knobs and their receptor-binding sites, together with the limited tolerance of the knob structure for ligand incorporation, made it difficult to apply this strategy to underexplored fibers, including those of the Ads that are being developed as alternative vector prototypes.
To overcome these challenges, alternative strategies based on fiber knob replacement rather than modification have been proposed. In these approaches, the knob of the Ad5 fiber is deleted and trimerization of the knobless protein restored by fusing it with a trimerization moiety, such as the coiled-coil domain of the retrovirus envelope glycoprotein (53), the neck region peptide of human lung surfactant D (32, 48), or reovirus sigma-1 protein (35, 47, 52).
We previously reported successful targeting of the Ad5 fiber through replacement of its knob with trimeric fragments of the fibritin protein of phage T4. Fusing the peptide and polypeptide ligands to the fiber-fibritin chimeras allowed successful targeting of Ad5 to a designed artificial receptor (26) and to natural receptors such as CD40 (5) and Her2 (7).
Given the promise of non-Ad5 serotypes as gene vector prototypes, we sought to test whether the fiber of one of these Ads could be targeted by knob replacement. We applied this strategy to the fiber of sAd24 (also known as Pan7), which is being developed as a vector for genetic immunization owing to its low seroprevalence in humans (45, 64) and its antigenic distinction from Ad5 (44-46, 64). The sAd24 fiber is an uncharacterized protein, and neither its native receptor nor its receptor-binding site have been identified. Nevertheless, as we report here, it has been possible to target this protein entirely on the basis of its predicted domain structure. The designed sAd24 fiber chimera efficiently incorporated in sAd24 particles and made viral infection dependent on the presence of the intended target receptor, Her2, which was chosen on the basis of the established correlation of its expression by human tumors, the aggressiveness of those tumors, and the poor prognosis for patients. We also demonstrated in this study the poor stability of FX-sAd24 complexes, which resulted in greatly diminished liver transduction by the sAd24-derived vector compared to that of the Ad5-derived vectors. In addition, we studied aspects of sAd24 biology directly relevant to the in vivo use of sAd24-derived vectors and tested an Her2-targeted version of sAd24 in two animal models.
Collectively, these studies provide important insights into sAd24 biology and identify its advantages and limitations as a vector prototype and are therefore essential for further development of an sAd24-based gene delivery platform.
MATERIALS AND METHODS
Abbreviations.
aa, amino acid; Ab, antibody; Ad, adenovirus; Ad5, adenovirus serotype 5; ANOVA, analysis of variance; CAR, coxsackievirus and Ad receptor; CT, computed tomography; EGFP, enhanced green fluorescent protein; FEAU, fluoro-5-ethyl-1-β-d-arabinofuranosyluracil; Fluc, firefly luciferase; FX, blood coagulation factor X; Her2, human epidermal growth factor receptor type 2; hRluc, codon-optimized Renilla luciferase; HVR, hypervariable region; IL, interleukin; IFN, interferon; kd, dissociation rate constant; KD, equilibrium dissociation constant; KCs, Kupffer cells; MAb, monoclonal antibody; MCP, monocyte chemotactic protein; MOI, multiplicity of infection; ORF, open reading frame; PBS, phosphate-buffered saline solution; PET, positron emission tomography; qPCR, quantitative PCR; Rluc, Renilla luciferase; ROI, region of interest; sAd24, simian adenovirus serotype 24; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TNF, tumor necrosis factor; TL, genetic fusion of HSV tk and Fluc; HSV tk, herpes simplex virus thymidine kinase; VP, viral particles; wt, wild type.
Cells and antibodies.
Cell lines 293, 293T, HCC1954, SKOV3, BT474, MDA-MB-361, and SK-BR-3 (all from American Type Culture Collection, Manassas, VA), human breast carcinoma cell line MDA-MB-231 (provided by Janet Price, the University of Texas M. D. Anderson Cancer Center), 293/Her2 cells (7), and 293/F28 cells (5) were maintained as described previously.
Cell line MDA-MB-231/hRluc-EGFP was made to express a hRluc-EGFP fusion protein by transduction of MDA-MB-231 cells with a retroviral vector encoding this protein. To generate this cell line, MDA-MB-231 cells were incubated overnight with medium containing the hRluc/EGFP-expressing retroviral vector LhRluc/EGFP (described below) and polybrene at a final concentration of 8 μg/ml. The transduced cells were selected by adding G418 at a final concentration of 1 mg/ml, and the GFP-positive MDA-MB-231/hRluc-EGFP cells were selected from the G418-resistant pool by four sequential rounds of cell sorting.
Cell line MDA-MB-231/Her2, a Her2-expressing derivative of MDA-MB-231/hRluc-EGFP cells, was made by transduction of MDA-MB-231/hRluc-EGFP cells with a Her2-expressing retrovirus. To generate this cell line, MDA-MB-231/hRluc-EGFP cells were incubated overnight with medium containing the Her2-expressing retroviral vector QCXIP.Her2 and polybrene. The transduced cells were selected in medium containing puromycin at a final concentration of 0.7 μg/ml. Her2 expression in individual puromycin-resistant clones was confirmed by fluorescence-activated cell sorting.
293/FsAd24 cells constitutively expressing wt sAd24 fiber were derived from 293 cells through transfection with the pFXsAd24 plasmid (described below) containing the wt sAd24 fiber ORF. Hybridoma line 20C1, expressing anti-sAd24 fiber MAb, was generated using recombinant N100sAd24 protein (described below) in a standard protocol established at the Monoclonal Antibody Facility of the M. D. Anderson Cancer Center.
Anti-fiber tail mouse MAb 4D2 (21) was a kind gift from Jeff Engler (University of Alabama, Birmingham). Anti-Her2 mouse MAb 3B5 was purchased from Merck KGaA (Darmstadt, Germany). Anti-fibritin mouse MAb 5E1 was described in our previous report (7).
Genetic engineering. (i) Plasmid for N100sAd24 protein expression.
To express the amino-terminal segment of the sAd24 fiber protein for the subsequent generation of MAbs, a fragment of this fiber ORF containing codons 2 through 102 was PCR amplified and cloned in pET20b (EMD Biosciences, Gibbstown, NJ). In the resultant plasmid, pET20b.6H.N100sAd24, this ORF was fused at its 5′ end with the six-histidine-tag-encoding sequence.
(ii) Plasmids for expression of wt and modified sAd24 fiber proteins.
These plasmids were designed for the subsequent generation of cell line 293/FsAd24 and for transient expression of the fiber chimera. To this end, the ORF of the wt Ad5 fiber was deleted from the previously described pVSII (25) by using inverse PCR and was replaced with either the ORF of the wt sAd24 or the ORF of the sAd24 fiber chimera, FsAd2411FHer2:7, yielding pFXsAd24 and pFusion2724, respectively. FsAd2411FHer2:7 was designed to contain the tail and the shaft domains of the sAd24 fiber (aa 1 to 264 [GenBank accession number AY530878]) followed by the carboxy-terminal fragment of the phage T4 fibritin protein comprising the last two a-helical repeats of the stalk and the foldon domain (aa 265 to 359 of the chimera, corresponding to aa 393 to 487 of the fibritin [GenBank accession number L43611]), the (Gly4Ser)3 linker (aa 360 to 374), and the Her2-specific affibody Zher2:7 (aa 375 to 432 [62]).
(iii) sAd24 shuttle plasmids.
The shuttle plasmid for replacement of the E1 region, pE1sAd24002, is a derivative of pZeRo2 (Invitrogen, Carlsbad, CA), which contains the previously described TL-expressing gene cassette (7) driven by the immediate-early cytomegalovirus promoter, which is flanked by the segments of the wt sAd24 genome that are immediately adjacent to the E1 region. The left and right flanks correspond to positions 1 to 474 and 3420 to 5893 in the wt sAd24 genome, respectively. The fiber gene shuttle plasmid, pFs24027, also a derivative of pZeRo2, contains the FsAd2411FHer2:7-encoding sequence flanked by the sequences of the wt sAd24 genome that surround the wt fiber gene (positions 29999 to 32111 and 33423 to 34306 in the wt sAd24 genome, respectively).
(iv) sAd24 rescue plasmids.
The master rescue plasmid containing the unmodified sAd24 genome was designed by cloning full-sized sAd24 genomic DNA in place of the Ad5 genome in the previously made pVK50 (27), yielding pRs24000. To design pRs24002, the TL-expressing cassette was transferred into the sAd24 genome by recombination between pRs24000 linearized with SnaBI and the pE1sAd24002 shuttle. Similarly, the wt fiber gene in pRs24002 was replaced with the FsAd2411FHer2:7 gene, using the pFs24027 shuttle and SwaI-cut pRs24002, thus yielding pRs24004. Ad genomes in all rescue vectors are flanked with PacI sites that are used to release these genomes for subsequent virus rescue by transfection.
(v) Retrovirus plasmid vectors.
The genome of retroviral vector LhRluc/EGFP, which expresses the fusion of hRluc and EGFP, was obtained by transferring the hRluc coding sequence from phRL null (Promega, Madison, WI) into the pLEGFP-N1 plasmid (BD Biosciences, San Jose, CA). The genome of retroviral vector QCXIP.Her2, which expresses Her2, was obtained by transferring the ORF of Her2 from the pIRES.neo3-c-erbB2 plasmid vector (7) into pQCXIP (Clontech, Mountain View, CA).
Additional experimental details, maps, and sequences of the designed DNA molecules are available upon request.
Viruses.
Stock of sAd24 was obtained from the American Type Culture Collection (VR-593) and expanded in 293 cells. The genomes of recombinant sAd24 vectors were assembled in plasmid vectors using homologous recombination in Escherichia coli strain BJ5183 between the corresponding shuttle and rescue plasmids as previously described (11). The genes of the E1 region (positions 475 to 3419 in the sAd24 genome) within these genomes were replaced with a cytomegalovirus promoter-driven expression cassette containing the TL transgene. The viruses were rescued by transfecting either 293 or 293/FsAd24 cells with the Ad genomes released from the corresponding rescue plasmids by restriction endonuclease digestion. All Ads were purified by double banding in CsCl gradients, subjected to dialysis against a buffer containing 10 mM Tris (pH 8.0), 50 mM NaCl, 2 mM MgCl, and 10% glycerol, and stored at −80°C. The titers of the viral preparations were determined by using the total protein concentration as previously described (7).
hRluc/EGFP- and Her2-expressing retroviruses were rescued by transfecting the packaging 293GPG cells (40) with pLhRluc/EGFP and pQCXIP.Her2 retroviral plasmids, respectively, using Lipofectamine reagent (Invitrogen). The media containing retroviruses was collected at 72 to 96 h after transfection, filtered through a 0.45-μm-pore-size filter, and stored at −80°C.
Surface plasmon resonance experiments.
Biosensor studies were done essentially as described by Kalyuzhniy et al. (22). In brief, purified Ad virions were immobilized on a CM5 sensor chip (GE Healthcare, Piscataway, NJ) by using an amine-coupling reaction as described by the manufacturer. Affinity measurements were done using a Biacore 3000 instrument (GE Healthcare). The chip was probed with murine factor X (Haematologic Technologies, Essex Junction, VT) in an HBSP running buffer (0.01 M HEPES [pH 7.4], 0.15 M NaCl, and 0.005% [vol/vol] Surfactant P20) supplemented with 1 mM CaCl2, 0.5 mM MgCl2, and 0.1 mg/ml of bovine serum albumin. The 5-min injection of FX was followed by 40 min of dissociation. The chips were regenerated after each binding cycle, using 30-s pulses of buffer containing 3 mM EDTA. The data were evaluated with BIAevaluation software version 3.1 (GE Healthcare) with the application of a simple 1:1 binding mass transfer model. The obtained sensorgrams were fitted globally over the whole range of injected concentrations and simultaneously over the association and dissociation phases. Equilibrium dissociation constants were calculated from the measured rate constants.
Expression and purification of recombinant proteins.
The N100sAd24 protein was expressed in E. coli Rosetta2(DE3)pLysS cells (EMD Biosciences) transformed with pET20b.6H.N100sAd24. Protein expression was induced by growing the culture in Overnight Express medium (EMD Biosciences) per the vendor's recommendations. The bacteria were collected by centrifugation and subjected to lysis by sonication, and the six-histamine-tagged N100sAd24 was purified from the lysate by immobilized metal ion affinity chromatography on a HisTrap column (GE Healthcare).
Methods used for bacterial expression and purification of Her2-specific affibodies were described elsewhere (7).
In vitro gene transfer.
Transduction of cells was done as previously described (7). Briefly, cells grown in 24-well plates were rinsed with fresh medium and incubated for 10 min with either medium alone or medium containing the Her2-binding Zher2:4 affibody. Next, the cells were infected for 30 min with one of the Ad vectors. The medium was replaced with fresh medium, and the incubation was continued for 24 h, after which the cells were subjected to lysis, using reporter lysis buffer (Promega). The Fluc activity in the lysate was measured in a tube luminometer (Sirius; Berthold, Pforzheim, Germany) using a luciferase assay system (Promega).
In the virus neutralization assay, the viruses were mixed with the diluted anti-Ad5 sera prior to being added to cells.
Flow cytometry.
Cell attachment by the targeted fiber was detected according to a previously described protocol (7). In brief, the cells, suspended in PBS with 0.1% bovine serum albumin and 0.01% NaN3 were incubated with aliquots of cleared lysates of 293T cells either mock transfected or transfected with the chimera-expressing pFXsAd24 plasmid. The cell-bound chimera was detected by anti-fibritin stalk MAb 5E1 (7) and secondary goat anti-mouse IgG Ab labeled with Alexa Fluor 488 (Invitrogen).
Western blotting.
Fibers, either transiently expressed in 293T cells or contained in purified viruses, were detected by Western blotting essentially as previously reported (7). However, because the anti-sAd24 fiber MAb 20C1 recognizes its epitope only in a fully denatured protein, detection of the bands corresponding to nondenatured wt sAd24 fiber and the FsAd2411FHer2:7 fiber chimera required a modified, two-step procedure. First, these proteins were separated by electrophoresis without prior denaturation; second, after electrotransfer of the proteins on a membrane, they were denatured by boiling the membrane in the transfer buffer for 10 min and were then probed with antibodies.
Studies with mice and experiments using murine tissues.
All experiments involving animals were done according to protocols approved by the Institutional Animal Care and Use Committee of the M. D. Anderson Cancer Center. Only replication-deficient Ad vectors with E1 deleted were used in these studies.
In vivo gene expression.
To study the in vivo patterns of transgene expression by Ad vectors, 6- to 8-week-old female NCr nu/nu mice (Taconic, Hudson, NY) were injected intravenously (via the tail vein) with either PBS or the tested Ad vector diluted in 100 μl of PBS. Forty-eight hours later, the mice were anesthetized by isoflurane inhalation and injected in the tail vein with a solution of the Fluc substrate d-luciferin (Caliper, Hopkinton, MA) in PBS (30 mg/ml, 100 μl per injection). The animals were transferred into the chamber of an IVIS 200 imaging system (Caliper) and imaged while under continuous anesthesia. The region of interest was drawn over the body area corresponding to the liver. The bioluminescence signals measured within the region of interest were then analyzed using Living Image software version 3.0 (Caliper). Twenty-four hours later, the mice were euthanized by CO2 inhalation, and individual organs were collected.
Pieces of the tissue samples were weighed and homogenized in PBS, using TissueLyser (Qiagen, Valencia, CA) at 30 Hz for 1 min. Aliquots of homogenized tissues were subjected to lysis in reporter lysis buffer (Promega); the samples were cleared of debris by centrifugation, and the Fluc activity in the supernatants was measured as already described under “In vitro gene transfer.”
The Ad-treated mice were also subjected to PET scans 48 h after being injected with the virus. The animals were anesthetized and injected via the tail vein with 18F-labeled FEAU (100 μCi in 100 μl of PBS) prepared as previously described (1). Two hours later, anesthetized mice were secured on custom holders to minimize motion across modalities and underwent a 10-min session of static PET imaging done in two-dimensional mode using an R4 microPET scanner (Concorde Microsystems, Knoxville, TN). The images were reconstructed using an ordered subset expectation maximization algorithm. Immediately after the PET scan, each animal underwent CT scanning in an RS-9 Micro-CT instrument (General Electric, London, Ontario) using the following parameters: 80 kVp, 450 A, 100 ms per frame, and calibration images (bright and dark fields). Next, PET data were registered to the reference anatomical CT data through a combination of interactive translations and rotations to match major structures such as body outlines that are visible on both modalities. This initial registration was then further refined through the multiresolution iterative maximization of a normalized mutual-information cost function (30). Once registered, the PET and CT data sets were fused together, processed, and visualized as described elsewhere (10) to localize tracer signals in relation to the anatomy.
Ad biodistribution experiments.
To establish patterns of biodistribution of Ad particles, the mice were injected intravenously (tail vein) with 1011 VP of one of the tested Ads and were killed at either 1 h or 24 h after injection. The organs were collected, weighed, and homogenized in PBS as described above. Proteinase K, SDS, and EDTA were added to aliquots of these homogenates to final concentrations of 200 μg/ml, 5%, and 100 mM, respectively, and the samples were incubated overnight at 56°C. Proteinase K was inactivated by incubating the lysates at 95°C for 10 min; 100-fold dilutions of these lysates in water were used as templates for qPCR (3 μl per 25 μl of reaction mixture). Ad DNA was detected using primers and probes complementary to the E4 regions of the respective genomes. Ad5 genomes were detected with primers Ad5.F (TGG CTT CGG GTT CTA TGT AAA CTC) and Ad5.R (TTC TGC GGT GGT GGA TGT TA) and probe Ad5.Pr (TTC ATG CGC CGC TGC CCT G), which was labeled with 6-carboxyfluorescein (6FAM); sAd24 genomes were detected with primers sAd24.F (TGG CCG GCG TGA ATG) and sAd24.R (TCG CGG ACG CCA ATG) and a 6FAM-labeled probe, sAd24.Pr (TTC GAC AAG ATG AAT ACA CCC CCG GA). Reactions were run in an ABI 7500 instrument, and the data were processed using ABI 7500 software. Statistical comparison of Ad DNA copy numbers per organ for the different vectors was performed separately for each organ at each time point. No multiple-comparison adjustment was implemented for these analyses. DNA copy numbers were analyzed on the logarithmic scale by two-sample t tests. The tests were two sided, and P values of 0.05 or less were considered statistically significant. Statistical analysis was carried out using SAS version 9 software (SAS Institute, Cary, NC).
Sequestration of Ad by KCs.
For immunofluorescent detection of Ad virions and KCs, livers were collected from mice injected with 4 × 1010 VP of Ads (3 mice per group) at either 10 min or 16 h after intravenous administration of the vectors. The livers were fixed and embedded in paraffin. The 5-μm sections were prepared using a rotary microtome Leica RM2255 instrument (Leica Microsystems, Bannockburn, IL). To detect KCs, the sections were incubated with rat anti-mouse IgG Ab (BD Biosciences) to F4/80, a recognized biomarker of murine macrophages, followed by the secondary goat anti-rat IgG Ab labeled with Alexa Fluor 488 (Invitrogen). The cells containing Ad virions of both serotypes were detected with rabbit polyclonal anti-Ad5 Ab (Abcam, Cambridge, MA) and stained with Alexa Fluor 546-labeled goat anti-rabbit IgG Ab (Invitrogen). Nuclei were stained with Hoechst 33342. Images of liver sections were acquired using an Olympus BX51 microscope (Olympus America, Center Valley, PA) and were processed with Olympus Micro software. To quantify KCs, they were counted by two readers in five randomly selected field images (per animal) acquired at ×40. A linear mixed model was used to estimate the number of KCs for each vector and to compare between vectors. The linear mixed model took into account that KCs in each field were measured twice. A Tukey-Kramer adjustment was used for all pairwise comparisons between vectors to control the overall type I error rate at 5%. The tests were two sided, and P values of 0.05 or less were considered statistically significant. Statistical analysis was carried out using SAS version 9 software.
Cytokine release study.
For the cytokine response study, C57BL/6 mice were injected intravenously with Ads (4 × 1010 VP per injection), and blood samples were collected in heparinized tubes (Multivette 600; Sarstedt, Newton, NC). Blood cells were pelleted by centrifugation (2,000 × g; 10 min), and plasma samples were frozen. Plasma samples diluted 1:4 in assay diluent were analyzed in a FACSCalibur flow cytometer using a mouse inflammation cytometric bead array kit (all from BD Biosciences) according to the protocol provided with the kit. Concentrations of cytokines and chemokines in samples were determined by processing the raw data with FCAP Array software. The duplicate concentration measurements obtained from the same mouse were averaged to obtain a single measurement for each animal. A two-sample t test with unequal variance was used to compare the average final concentrations of Ad5 and sAd24 by cytokine and time point. All tests were two sided, and P values of 0.05 or less were considered statistically significant.
Generation of neutralizing anti-Ad5 Ab.
To raise anti-Ad5 antibodies, a group of six 6-week-old C57BL/6 mice were injected in the tail veins with 1010 VP of Ad5TL (7). On postinjection day 26, blood samples were collected by cardiac puncture and pooled. The sera obtained from these pooled samples were used as a source of polyclonal anti-Ad5 antibodies.
In vivo transduction studies.
To target tumor cells in circulation, nude mice were injected intravenously with MDA-MB-231/Her2 cells (106 cells/injection) and, 5 min later, with either nontargeted sAd24 vector, sAd24TL, or its Her2-targeted counterpart, sAd24TL.11FHer2:7, (4 × 1010 VP/injection). The patterns of the Rluc reporter activities in these animals were established by using whole-body bioluminescence imaging as described above. Mice were killed and their organs were collected at 26 h after the administration of the virus.
To establish the target metastases in the lungs, the animals were injected with MDA-MB-231/Her2 cells as outlined in the previous paragraph. The development of metastases was monitored by the whole-body imaging of Rluc activity and was also corroborated by visual examination of lungs isolated from randomly selected mice showing bioluminescence in the thoracic area. Viral injections in tumor-positive mice started on day 66 after administration of the cells, at which point each animal was given 1011 VP intravenously. A second dosing of 1011 VP was given 24 h later. The same two-injection regimen was used in control, tumor-free mice. Animals in both groups were imaged and killed 48 h after the second administration of the virus. Homogenates of isolated lungs were prepared, and measurements of Fluc activity were done as described under “In vitro gene transfer.”
For statistical analysis, the measurements of Fluc activity were transformed to the logarithmic scale and a one-way ANOVA was carried out to compare measurements between treatment groups. All tests were two sided, and P values of 0.05 or less were considered statistically significant. Pairwise comparisons were performed only if the overall likelihood ratio test for groups was significant. Statistical analysis was carried out using SAS software.
RESULTS
Complexes formed through association of FX with virions of sAd24 are unstable.
Two recent studies have demonstrated a direct correlation between the efficacy of binding of blood coagulation factor FX to particles of a particular Ad serotype and that serotype's ability to transduce the liver (22, 56). Given these findings, we wished to assess the stability of FX in a complex with the virions of sAd24. We conducted a surface plasmon resonance study on immobilized sAd24 particles using purified FX as an analyte. Similarly prepared, biosensor-bound virions of Ad5 were used as controls.
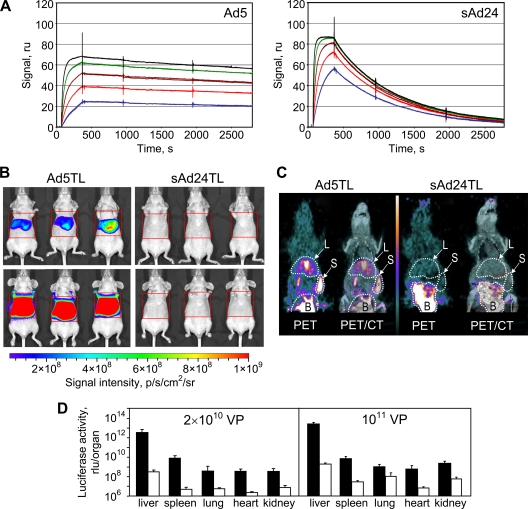
This experiment showed that compared to the expected stable complexes of FX with Ad5 particles, the FX-sAd24 association is significantly weak (as evidenced by rapid dissociation of the FX-sAd24 complex [Fig. 1A]). In particular, while the equilibrium dissociation constants, KDs, measured in this experiment were not dramatically different (6.95 × 10−10 M for FX-Ad5 and 3.08 × 10−9 M for FX-sAd24), the dissociation rate constant, kd, whose value inversely correlates with the stability of the complex, was much higher for the FX-sAd24 complex (1.17 × 10−3 s−1) than for the FX-Ad5 complex (7.16 × 10−5 s−1).
FIG. 1.
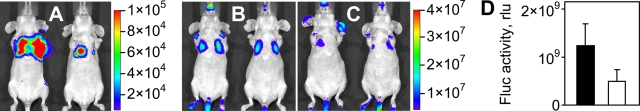
Instability of FX-sAd24 complexes results in reduced liver transduction by sAd24-derived vector. (A) The interactions of Ad5 and sAd24 virions with FX were analyzed using surface plasmon resonance. Viral particles bound to the biosensor chip (Ad5 at a coating level of 1,387 resonance units [ru] and sAd24 at 1,699 ru) were probed with soluble FX. Colored lines correspond to the signals obtained in duplicate runs at each tested concentration of FX (blue, 12.5 nM; red, 25 nM; brown, 50 nM; green, 100 nM; and black, 200 nM). As evidenced by the dissociation part of the sensorgrams, the FX-sAd24 complexes were much less stable than the FX-Ad5 complexes. (B) Whole-body images of female nude mice that were injected intravenously (tail vein) with an Ad5- or an sAd24-derived vector, each expressing a dual-modality TL reporter (HSV tk fused with Fluc). Mice injected with 2 × 1010 VP (upper panels) and 1011 VP (lower panels) of either Ad5TL (left panels) or sAd24TL (right panels) are shown. The intensities of the light signals in the selected regions of interest (red squares) are shown as pseudocolor overlays. Here and in Fig. 8 and 9, the pseudocolor scale shows the signal intensity range. Notably, the relative infectivities of Ad5TL and sAd24TL, which had been determined by the spot assay in 293/Her2 cells in vitro, were 10 VP/infectious units and 53 VP/infectious units, respectively. p/s/cm2/sr, photons/s/cm2/steradian. (C) On the left on each panel, PET images (coronal slices) of Ad-injected animals enabled by 18F-labeled FEAU are shown. On the right, these PET images are superimposed on the CT images that provide anatomic landmarks. The white-dotted contours show the tracer uptake by the livers (L) and the spleens (S) of the animals. The signals in the bladders (B) are due to physiologic accumulation of the small-molecule tracer in the urine rather than vector-expressed HSV tk activity. The pseudocolor scale between the panels shows the PET signal intensity range, from 0 (black) to 6 times the mean muscle uptake value (white). (D) Activities of Fluc reporter in homogenates of tissue samples collected from mice injected with 2 × 1010 VP or 1011 VP of either Ad5TL (black bars) or sAd24TL (white bars) is shown. Each bar represents an average signal intensity (shown in relative light units [rlu] per entire organ) measured in organ samples obtained from five animals. Error bars indicate the standard deviations calculated for duplicate data points.
The relative instability of the FX-sAd24 complex suggested that systemically delivered sAd24 may transduce the liver to a lesser extent than does Ad5.
Systemically administered sAd24-based gene vectors greatly reduced transduction of liver.
Liver transduction by Ad5 and sAd24 was compared in vivo by monitoring reporter transgene expression in mice that were intravenously injected with either the Ad5-derived vector (Ad5TL) or the sAd24-derived vector (sAd24TL). These vectors, designed to express Fluc, were administered to nude mice through tail vein injection, and the pattern of in vivo reporter expression was established 48 h later by noninvasive bioluminescence imaging. Comparison of the reporter expression patterns for Ad5TL and sAd24TL revealed a dramatic serotype-related difference in the strength of bioluminescence signals in the livers of the vector-injected mice. On average, those signals were 1.9 × 103-fold weaker in mice that were given 2 × 1010 VP of sAd24TL (7.7 × 106 photon/s/ROI) than in animals injected with the same dose of Ad5TL (1.4 × 1010 photon/s/ROI) (Fig. 1B, upper row of images). When the viral dose was elevated to 1011 VP, the average liver-localized signals increased to 2.5 × 107 photon/s/ROI and 6.5 × 1011 photon/s/ROI in sAd24TL- and Ad5TL-injected mice, respectively (Fig. 1B, lower row of images). These bioluminescence data were further corroborated by [18F]-FEAU-enabled PET imaging, which was facilitated by the HSV tk activity of the vector-encoded TL reporter (Fig. 1C).
The results of these imaging studies were then confirmed by measurements of luciferase activity in homogenates of organs isolated from the vector-treated animals after image acquisition. These measurements demonstrated that the levels of bioluminescence were significantly higher in all tested organs from the Ad5TL-injected mice than in organs from sAd24TL-injected animals (Fig. 1D and Table 1).
TABLE 1.
Reporter activities in homogenates of tissues isolated from Ad-injected micea
| Vector (dose) | Reporter activities in indicated organs per organ or gram of tissue (rlu)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Liver |
Spleen |
Lung |
Heart |
Kidney |
||||||
| Per organ | Per gram of tissue | Per organ | Per gram of tissue | Per organ | Per gram of tissue | Per organ | Per gram of tissue | Per organ | Per gram of tissue | |
| Ad5TL (2 × 1010 VP) | 3.6 × 1012 | 2.8 × 1012 | 8.1 × 109 | 6.0 × 1010 | 3.8 × 108 | 1.2 × 109 | 3.4 × 108 | 2.5 × 109 | 3.5 × 108 | 1.0 × 109 |
| sAd24TL (2 × 1010 VP) | 3.1 × 108 | 2.2 × 108 | 5.0 × 106 | 5.3 × 107 | 5.5 × 106 | 1.6 × 107 | 2.3 × 106 | 1.8 × 107 | 7.4 × 106 | 2.4 × 107 |
| Signal ratiob | 1.2 × 104 | 1.3 × 104 | 1.6 × 103 | 1.1 × 103 | 6.9 × 10 | 7.4 × 10 | 1.5 × 102 | 1.3 × 102 | 4.7 × 10 | 4.2 × 10 |
| Ad5TL (1011 VP) | 2.8 × 1013 | 1.6 × 1013 | 7.0 × 109 | 6.3 × 1010 | 1.1 × 109 | 2.9 × 109 | 6.2 × 108 | 3.3 × 109 | 2.4 × 109 | 6.8 × 109 |
| sAd24TL (1011 VP) | 2.0 × 109 | 1.2 × 109 | 3.0 × 107 | 3.4 × 108 | 1.1 × 108 | 4.1 × 108 | 7.0 × 106 | 4.7 × 107 | 5.9 × 107 | 1.7 × 108 |
| Signal ratiob | 1.4 × 104 | 1.3 × 104 | 2.4 × 102 | 1.8 × 102 | 9.9 | 7.0 | 8.8 × 10 | 7.1 × 10 | 4.1 × 10 | 3.1 × 10 |
| PBS | 1.1 × 107 | 8.5 × 106 | 7.3 × 105 | 8.4 × 106 | 3.1 × 106 | 9.3 × 106 | 1.4 × 106 | 9.6 × 106 | 3.3 × 106 | 1.0 × 107 |
The activities of the reporter averaged for groups of five animals is shown in relative light units (rlu) per entire organ and per gram of tissue.
Ratios of bioluminescence signals measured in Ad5TL-injected mice to those measured in sAd24TL-injected mice.
The pattern of in vivo transgene expression by an sAd24 vector does not correlate with the virus biodistribution profile.
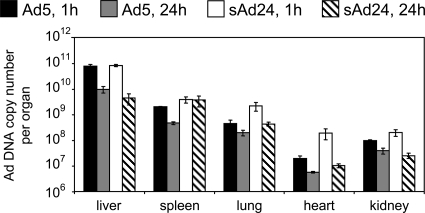
Prior study of the in vivo distribution of systemically administered Ads representing groups A through F identified significant interserotype differences (3). Because the biodistribution of sAd24 particles has never been explored, we wished to establish it by injecting sAd24TL intravenously in mice and measuring the amount of viral DNA in organs by qPCR at 1 h and 24 h after injection. In parallel, a control group of animals was injected with Ad5TL, and this vector DNA was quantified using Ad5-specific primers and an Ad5-specific probe. This experiment showed significantly greater sAd24 DNA content at both time points in the heart, the lungs, and the spleen (Fig. 2). These organs contained 1.7 (heart, 24 h after injection) to 10 times (heart, 1 h after injection) more of the sAd24 genomes than of the Ad5 genomes (all P values are below 0.02). At the 1-h time point, the kidneys of sAd24-injected mice contained 2 times the amount of viral DNA seen in the kidneys of mice that received the Ad5 vector (P = 0.014). No statistically significant difference in DNA content was seen in kidneys 23 h later. The liver was the main site of viral DNA accumulation. At 1 h after injection, the livers contained 79% of injected Ad5 virions and 85% of injected sAd24 virions; at the 24-h time point, these levels had decreased to 9.9% and 4.5%, respectively.
FIG. 2.
Ad5 and sAd24 virions show similar patterns of in vivo distribution. Shown are the copy numbers of Ad5 and sAd24 viral genomes per organ detected by qPCR in isolated murine organs at 1 h and 24 h after systemic administration of the vectors. Error bars indicate the standard deviations calculated for duplicate data points.
This noted discrepancy between the low level of reporter expression by the sAd24 vector in the liver and the high concentration of its virions in the liver strongly suggested an involvement of virus retention mechanisms that do not yield gene transfer.
Sequestration of intravenously injected sAd24 virions leads to rapid depletion of KCs in the liver.
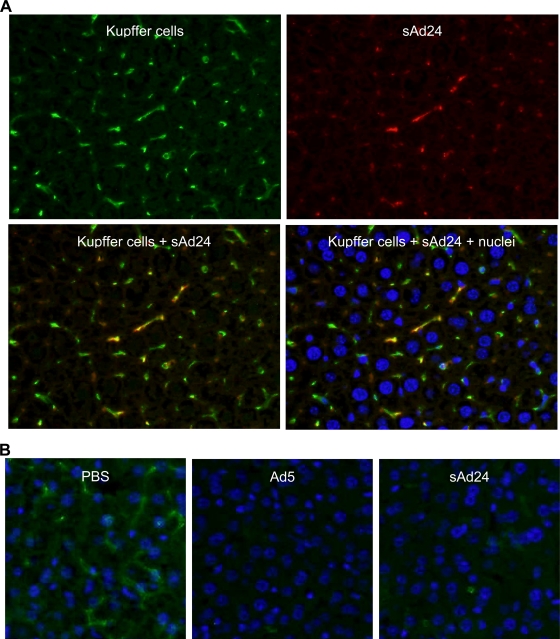
Sequestration of systemically delivered Ad particles by KCs, macrophages residing in the liver sinusoids, has been reported for several Ads, including human serotypes 3, 5, 31, 37, and 41 and simian Ad23 (3). The uptake of Ad5 virions by KCs is mediated primarily by the scavenger receptors, with the natural Abs and complement playing secondary roles (63). Accumulation of Ad5 virions in KCs very shortly after virus administration rapidly kills KCs (34) through as-yet-unknown molecular mechanisms. Given these findings, we wished to examine whether KCs play any significant role in the clearance of intravenously delivered sAd24 and, in doing so, contribute to the elimination of sAd24 vector particles from the gene delivery process.
To this end, we injected mice with sAd24TL and analyzed the livers collected from these animals 10 min and 16 h later by fluorescence microscopy, using an Ab to an established macrophage biomarker, F4/80, and anti-Ad Ab, each labeled with a different fluorophore. As shown in Fig. 3A, 10 min after vector administration all Ad-associated fluorescence signals colocalized with the KCs, thus confirming efficient clearance of the virus by these liver macrophages. In concordance with the data previously reported for other Ad serotypes, we also saw massive depletion of KCs by sAd24 (Fig. 3B). Notably, 16 h after virus injections, the rate of KC depletion caused by sAd24 (89%) was somewhat lower than that caused by the control Ad5 vectors (94%) (P = 0.018).
FIG. 3.
Uptake of sAd24 virions by KCs in the liver leads to KC depletion. (A) Immunofluorescence staining of the liver collected from a mouse injected with sAd24 vector 10 min after injection. The upper panels show the liver section stained with either anti-F4/80 (left, green fluorescence) or anti-Ad Ab (right, red fluorescence). The lower panels show the overlay of the upper images, either alone (left) or merged with the staining of nuclei (right, blue staining). KCs containing sAd24 particles are orange. (B) Immunofluorescence staining of the livers collected from mice injected with PBS (left), Ad5 (center), or sAd24 (right) 16 h after injection. All images show Hoechst-stained nuclei (blue).
The profile of the acute inflammatory response to sAd24 is different from that caused by Ad5.
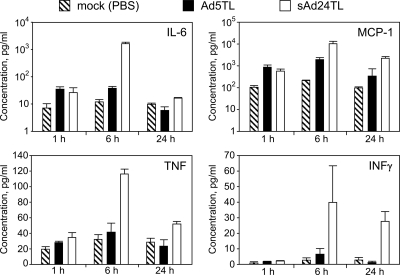
To further characterize sAd24 interaction with the host, we established both the temporal profile and the magnitude of the acute inflammatory response to sAd24 virions, using cytometric bead array technology. In particular, concentrations of six cytokines (IL-6, IL-10, IL-12p70, IFN-γ, TNF, and MCP-1) were measured in plasma samples collected from sAd24-injected mice at 1 h, 6 h, or 24 h after injection. Comparison of these data with the similarly measured cytokine concentrations in plasma of Ad5-injected mice showed significant differences for IL-6, MCP-1, TNF, IFN-γ (Fig. 4), and IL-12p70 (data not shown). The differences in concentrations of IL-6, MCP-1, and TNF induced by sAd24 and Ad5 vectors were most significant at the 6-h time point and, to a lesser extent, at the 24-h time point; IFN-γ and IL-12p70 concentrations were different only at the 24-h time point (P values of 0.02 and 0.047, respectively). Importantly, cytokine release by the sAd24-injected mice was invariably more robust than that by the animals that received Ad5. Compared to those for mock-injected animals, no statistically significant changes in the concentrations of IL-10 were seen in mice injected with either serotype of Ad (data not shown).
FIG. 4.
Comparison of cytokine release in response to intravenous injections of Ad5 and sAd24. Concentrations of IL-6, MCP-1, TNF, and IFN-γ in plasma samples collected from mice at 1 h, 6 h, or 24 h after injection with either Ad5- or sAd24-derived vectors are shown in pg/ml. Three mice were injected with a given Ad to generate data for each time point shown. Error bars indicate the standard deviation calculated for duplicate data points corresponding to each animal in the group.
The modification of sAd24 fiber by knob replacement strategy yields protein chimera suitable for Ad vector targeting.
We previously succeeded in altering the receptor specificity of Ad5 through genetic replacement of the CAR-binding knob domain of its fiber with a trimerization domain and a targeting ligand (5, 7, 26). We then reasoned that because of evolutionary conservation of the overall Ad fiber structure, which is very similar in most known fiber proteins, the same approach should also be applicable to the sAd24 fiber. Importantly, application of this strategy to sAd24 did not require identification of its natural fiber receptor, which is not yet known, and, if successful, would further support the applicability of this strategy to fibers of other Ads.
To test this assumption, we designed a recombinant gene encoding the tail and shaft domains of the sAd24 fiber protein fused to the 12th α-helical coiled-coil of the phage T4 fibritin protein, a peptide linker, and the Her2-specific affibody Zher2:7 and transiently expressed it in 293T cells. The product of this expression (designated FsAd2411FHer2:7) was analyzed by Western blotting to test whether the designed chimera assumes the trimeric configuration that is both characteristic of Ad fibers and essential for their encapsidation.
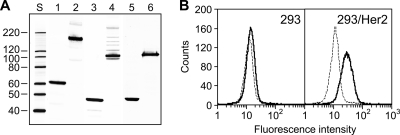
The results of this analysis, however, were unconvincing: in contrast to the well-defined bands corresponding to the trimeric wt Ad5 fiber (Fig. 5A, lane 2), the sAd24 fiber chimera failed to produce such a pattern. Instead, the major band seen in the nondenatured sample of FsAd2411FHer2:7 migrated considerably faster than had been expected (Fig. 5A, lane 4) and corresponded to a protein with an apparent molecular mass of approximately 105 kDa, not the 136 kDa predicted for the trimeric FsAd2411FHer2:7. The fact that the same sample, when fully denatured, clearly showed the monomeric chimera of the expected size (Fig. 5A, lane 3) ruled out the possibility that the smaller-than-expected size of the nondenatured protein was due to degradation or truncation of its subunits. Also, comparison of the electrophoretic mobilities of the FsAd2411FHer2:7 chimera and the wt sAd24 fiber showed that the nonboiled samples of both proteins produced very similar patterns of gel migration (Fig. 5A, lanes 4 and 6). On the basis of this comparison, we concluded that the trimer of FsAd2411FHer2:7 was sufficiently stable to be incorporated into sAd24 virions.
FIG. 5.
sAd24 fiber-derived targeting chimera forms stable trimers and binds to cell-associated Her2. (A) Western blotting of lysates of 293T cells transiently expressing the fiber constructs. Lanes 1 and 2, wt Ad5 fiber; lanes 3 and 4, FsAd2411FHer2:7, lanes 5 and 6, wt sAd24 fiber; lane S, protein standards with the molecular masses shown in kDa (here and in Fig. 6). Samples in lanes 1, 3, and 5 were boiled prior to being loaded onto the gel and thus show proteins in their fast-migrating monomeric form; samples in lanes 2, 4, and 6 were not boiled. Fibers were detected with either the anti-Ad5 fiber tail MAb 4D2 (wt Ad5 fiber), the anti-fibritin MAb 5E1 (FsAd2411FHer2:7), or the anti-sAd24 fiber MAb 20C1 (wt sAd24 fiber) followed by a fluorescence-labeled secondary antibody. Predicted molecular masses for trimeric wt Ad5, FsAd2411FHer2:7, and wt sAd24 fiber are 185 kDa, 136 kDa, and 142 kDa, respectively. (B) Flow cytometry detected binding of the transiently expressed FsAd2411FHer2:7 chimera (shown by solid black line) to Her2-expressing 293/Her2 cells but no binding above the background to Her2-negative 293 cells. The background signals generated in both cell lines by the lysate of mock-transfected 293T cells are shown by the dotted lines.
The FsAd2411FHer2:7 protein was designed to mediate efficient attachment of sAd24 viral particles to Her2 and, as a prerequisite to this, was expected to bind the cell-associated Her2. To test this function of FsAd2411FHer2:7, the transiently expressed protein was used in a flow cytometry experiment to probe the surfaces of cells either expressing or not expressing Her2. As shown in Fig. 5F, the strength of the fluorescence signal measured in Her2-positive cell targets was significantly higher than that measured in cells lacking the target receptor.
Collectively, our data confirm both the expected trimeric configuration of the FsAd2411FHer2:7 protein and the ability of this molecule to recognize its intended target receptor on the cell surface, thereby providing a rationale for development of an sAd24 vector containing this chimera.
sAd24 virions incorporating designed fiber chimeras infect cells in a Her2-dependent manner.
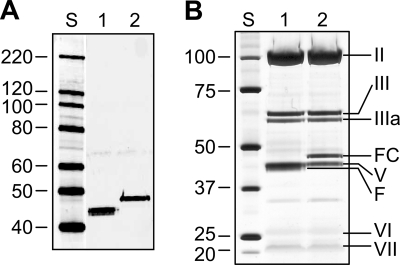
The final proof of the FsAd2411FHer2:7 chimera's functionality was obtained by using this protein to replace the wt fiber in sAd24 virions. To this end, we constructed a replication-deficient genome of sAd24 with the E1 region deleted and containing the ORF of FsAd2411FHer2:7 in place of the wt fiber-coding sequence; the virus was rescued and propagated using the two-step approach described in Materials and Methods. The efficacy of incorporation of FsAd2411FHer2:7 into sAd24 virions was gauged and compared with that of the wt sAd24 fiber by using Western blotting of purified viral particles. As Fig. 6A illustrates, the outcome of the trimerization assays of the transiently expressed FsAd2411FHer2:7 correctly predicted efficient encapsidation of the designed protein. This efficient encapsidation was in agreement with the high yield of the modified sAd24 vector (5,500 VP/infected cell), which was comparable to the yield for sAd24 containing the wt fibers (5,000 VP/infected cell). Comparison of the protein compositions of the virions of both types showed no differences other than the presence of the expected fiber proteins, the wild type or the fiber chimera, in the respective vector particles (Fig. 6B), thus confirming the proper assembly and maturation of the modified sAd24 particles.
FIG. 6.
Efficient encapsidation of FsAd2411FHer2:7 protein yields fully matured sAd24 virions. (A) Western blot of sAd24TL (1010 VP, lane 1) and sAd24TL.11FHer2:7 (1010 VP, lane 2) virions. The fully denatured fibers were detected with the anti-sAd24 fiber tail MAb 20C1. (B) SDS-PAGE-resolved proteins of sAd24TL (2 × 1010 VP, lane 1) and sAd24TL.11FHer2:7 (2 × 1010 VP, lane 2) virions stained with silver using a PageSilver kit (Fermentas, Glen Burnie, MD). Indicated are the migration positions of the following sAd24 proteins: hexon (II), penton base (III), peripentonal protein (IIIa), FsAd2411FHer2:7 fiber chimera (FC), major core protein (V), fiber (F), hexon-associated protein (VI), and minor core protein (VII).
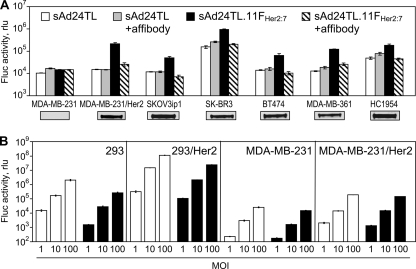
Further, in good agreement with the cell-binding data shown in Fig. 5B, our subsequent gene transfer experiments demonstrated that the FsAd2411FHer2:7 chimera contained in sAd24TL.11FHer2:7 virions enables Her2-dependent transduction of target cells (Fig. 7A). The Her2 specificity of sAd24TL.11FHer2:7 infection was confirmed by transducing two isogenic cell lines, MDA-MB-231 and MDA-MB-231/Her2, that differ from each other by their Her2 phenotypes. In addition, augmentation of gene transfer to the target receptor-expressing cells by the tropism-modified sAd24 was demonstrated through transduction of a panel of Her2-positive human tumor cell lines. In this experiment, gene delivery by the targeted sAd24 vector was 3.7 to 14 times more efficient than delivery by the vector bearing unmodified wt fibers. Importantly, by using the free Her2-binding affibody competitor we showed selective inhibition of transduction by sAd24TL.11FHer2:7 but not by the nontargeted control Ad (Fig. 7A).
FIG. 7.
Gene transfer by the targeted sAd24TL.11FHer2:7 vector is Her2 dependent. (A) In vitro transduction of human tumor cell lines by sAd24TL and sAd24TL.11FHer2:7 vectors. Viral infections were done in either standard medium (white and black bars) or in medium containing free affibody Zher2:4 used at a concentration of 100 μg/ml (gray and striped bars). Shown are the activities of Fluc in lysates of infected cells. Inserts below the graph show the signals detected in each of the tested cell lines (7 × 103 cells/lane) by Western blotting with anti-Her2 antibody. (B) Comparison of the efficacies of gene transfer by Her2-targeted vectors derived from Ad5 (Ad5TL.11FHer2:7, white bars) and sAd24 (sAd24TL.11FHer2:7, black bars). Levels of transgene expression in the lysates of cells infected with Ads at the MOIs indicated below the graph (in VP/cell) are shown. Error bars indicate the standard deviations calculated for triplicate data points.
The efficacy of this gene transfer was then compared with that by the previously designed Her2-targeted derivative of Ad5 (7) (Fig. 7B). In this experiment, two pairs of cell lines in which the parental line was Her2 negative (293 and MDA-MB-231) and the derivative line was Her2 expressing (293/Her2 and MDA-MB-231/Her2) were transduced with either Ad5TL.11FHer2:7 or sAd24TL.11FHer2:7 at various MOIs. With the exception of the transduction of MDA-MB-231/Her2 at an MOI of 10, at which both Ads were equally efficient, the Ad5-derived vector was more efficient in transducing both Her2-expressing cell lines. The differences between the levels of gene expression directed by the vectors depended on the vectors' replication. Specifically, in 293/Her2 cells, which complement deletion of the E1 genes in both the Ad5- and the sAd24-derived vectors, reporter expression by Ad5TL.11FHer2:7 was 3 to 6 times more efficient. In comparison, in MDA-MB-231/Her2 cells, which do not complement deletion of the E1 genes, reporter expression by Ad5TL.11FHer2:7 was only 1.3 to 1.6 times that by sAd24TL.11FHer2:7. Notably, the relative infectivities of the tested vectors, which had been determined by the spot assay in 293/Her2 cells, were 51 VP/infectious unit for sAd24TL.11FHer2:7 and 33 VP/infectious unit for Ad5TL.11FHer2:7.
In light of the potential use of sAd24TL.11FHer2:7-derived vectors for gene delivery in humans with preexisting anti-Ad5 antibodies, moreover, we used the established virus neutralization assay to test whether gene delivery by this vector would be affected by such antibodies. This test showed that sAd24TL.11FHer2:7 largely evaded inactivation by Ad5-specific antibodies and retained most of its infectivity in their presence (data not shown).
In vivo testing of the fiber-modified sAd24 vector.
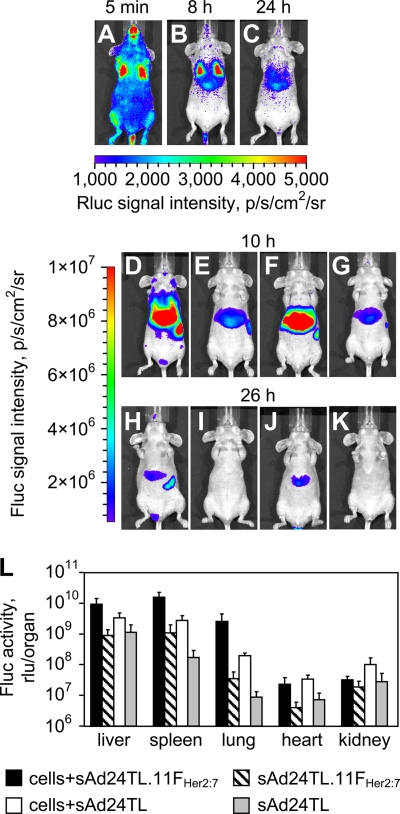
Having established the Her2 specificity of sAd24TL.11FHer2:7, we next wished to test this vector's ability to accomplish transduction in vivo. To this end, we injected mice intravenously with MDA-MB-231/Her2 cells, allowed them to distribute in the bloodstream (Fig. 8A), and targeted these cells with the vector, which was also administered intravenously. Another group of cell-injected animals was given the nontargeted control vector, sAd24TL. Two groups of mice that had received one of the vectors, but no tumor cells, were used as additional controls. To monitor the spread of cells and the patterns of Ad-mediated transduction, we took advantage of luciferase reporters expressed by the MDA-MB-231/Her2 cells (Rluc) and the virus (Fluc).
FIG. 8.
Transduction of circulating tumor cells by a Her2-targeted sAd24 vector. (A, B, and C) Patterns of Rluc expression at 5 min (A), 8 h (B), and 24 h (C) after intravenous injection of MDA-MB-231/Her2 cells into mice. (D to K) Fluc-enabled luminescence in Ad-injected mice at 10 h (D to G) and 26 h (H to K) after vector administration. The 2-h intervals between imaging of Rluc and Fluc activities were allowed for the Rluc signals to decay to background level. Mice shown in panels D and H were injected with MDA-MB-231/Her2 and with sAd24TL.11FHer2:7 5 min after administration of cells. Control mice shown in panels E and I were injected with sAd24TL.11FHer2:7 but did not receive tumor cells. Panels F and J show mice that were injected with the cells and with sAd24TL; the corresponding control mice, which were injected with sAd24TL alone, are shown in panels G and K. Each experimental group contained four animals. (L) Activities of the vector-encoded Fluc reporter in individual organs isolated from experimental mice that were injected with both the cells and the virus and from control mice injected with the virus alone. Error bars indicate the standard deviations calculated for duplicate data points.
As expected, the whole-body imaging of Rluc activity in the control animals, which did not receive tumor cells, did not reveal any bioluminescence above background throughout the duration of the study (data not shown). Imaging of Rluc activity in mice that had received both the cells and the viruses, done at 8 h after injections, showed that by that time point most tumor cells had cleared the circulation and were localized to the liver and the lungs (Fig. 8B). Imaging of the sAd24TL.11FHer2:7-expressed Fluc activity shortly thereafter revealed a 10.7-fold difference in signal intensity between the MDA-MB-231/Her2-injected mice (Fig. 8D) and the control mice (Fig. 8E) (P < 0.0001). Importantly, the Fluc-enabled signals largely localized within the regions of Rluc-caused luminescence, thereby linking the observed increase in vector-expressed reporter activity with the presence of tumor cells in a particular locale. However, a similar 9.6-fold difference was seen between the groups of mice that received the nontargeted control vector sAd24TL in combination with the tumor cells (Fig. 8F) or alone (Fig. 8G) (P = 0.0001). Comparison of Fluc signals measured in cell-injected animals showed a 1.7-fold-higher level of activity in mice that received sAd24TL.11FHer2:7 than in mice injected with sAd24TL. This difference was, however, statistically insignificant (P = 0.22).
Sixteen hours later, the strength of the Rluc signals in mice injected with MDA-MB-231/Her2 cells decreased substantially, suggesting their continuous clearance (Fig. 8C). As expected, this depletion of tumor cells resulted in a decrease of the vector-expressed Fluc activity. At this time point, the ratios of signal intensities measured during the whole-body imaging in the experimental and control groups of mice were 23.1 for groups of mice injected with sAd24TL.11FHer2:7 (Fig. 8H and I) (P = 0.0007) and 14.1 for groups of mice injected with sAd24TL (Fig. 8J and K) (P = 0.0008). While the ratio of Fluc activity in the group of mice injected with the cells and targeted Ad to that in the group that received the cells and sAd24TL increased to 2.9, it remained statistically insignificant (P = 0.28).
To establish a more detailed pattern of Ad vector reporter activity at this time point, we measured Fluc luminescence in the homogenates of organs collected from mice in all groups (Fig. 8L). In agreement with the whole-body imaging of Fluc activity, these measurements confirmed invariably higher levels of transduction in animals injected with the cells and the viruses than in animals treated with the Ads only. They also showed that the livers, spleens, and lungs were the primary sites of viral transduction. In these organs, sAd24TL.11FHer2:7- caused transduction was 2.8-fold (livers; P = 0.07), 5.7-fold (spleens; P = 0.003), and 13.5-fold (lungs; P = 0.003) higher than transduction caused by sAd24TL. This difference in the liver was, however, statistically insignificant, as was the difference in reporter expression in the hearts (P = 0.25). The kidneys of cell-injected animals that received sAd24TL contained a 3.3-fold-higher level of Fluc activity than kidneys in mice injected with the cells and sAd24TL.11FHer2:7 (P = 0.04).
We also tested the performance of sAd24TL.11FHer2:7 in an in vivo model of metastatic Her2-expressing cancer. For this study, we established target metastases in the lungs of mice by systemically injecting the animals with MDA-MB-231/Her2 cells (Fig. 9A). Next, the tumor-bearing animals and the tumor-free control animals were injected intravenously with sAd24TL.11FHer2:7 at a dose of 1011 VP/injection, using the two-injection regimen described in Materials and Methods. Bioluminescence imaging of Fluc activity done 48 h after the second vector administration detected weak signals in the thoracic areas of mice in both groups (Fig. 9B and C). Measurements in homogenates of the lungs isolated from animals in both groups showed that, on average, Fluc activity was 2.5 times higher in mice with metastases than in tumor-free mice (Fig. 9D). This difference was statistically significant (P = 0.005).
FIG. 9.
Gene delivery by sAd24TL.11FHer2:7 in mice with metastatic tumors. (A) Whole-body images of mice bearing Her2- and Rluc-expressing breast cancer metastases in the lungs. (B and C) Fluc bioluminescence in the mice shown in panel A (B) and in control tumor-free mice (C) 48 h after animals in both groups were injected intravenously with sAd24TL.11FHer2:7. (D) Activity of Fluc reporter in the lungs isolated from Ad-injected mice after image acquisition. The black and white bars show the averaged values of signals in the lungs of tumor-bearing and control mice, respectively. Each experimental group contained six animals. Error bars indicate the standard deviations calculated for duplicate data points.
DISCUSSION
Prior work on modifying the tropism of Ad vectors was focused mostly on Ad5-derived vectors and was based primarily on modification of the fiber knob domain with ligands. The natural diversity of Ad serotypes and the advantages offered by some of those Ads as targeted vector prototypes remain underexplored. In this study, we sought to test whether the Ad tropism alteration approach based on fiber knob domain replacement, which we had previously developed for Ad5, could be applied to another Ad serotype. The rationale for this work was that its success would support the feasibility of developing a versatile targeting approach that would be applicable to many Ads, thus making it possible to fully exploit the potential benefits offered by underexplored Ad serotypes.
Toward these goals, we sought to make a tropism-modified vector based on sAd24, which was chosen for this study for the following reasons. First, owing to the widespread preexisting immunity to Ad5 in humans (12, 55), the serological distinction of sAd24 from Ad5 makes this virus a preferred alternative to Ad5 as a vector platform. Second, the fiber of sAd24 is an unexplored molecule and is thus a reasonable starting point to test an approach that is expected to be applicable to many fibers, including the uncharacterized ones. This protein has never been studied in detail; at the level of the primary structure it shares little homology with the Ad5 fiber and other well-characterized Ad fibers. Neither its primary receptor nor the receptor-binding site within its knob has been identified. Against this background, successful targeting of the sAd24 fiber through the knob replacement strategy would yield a vector prototype of potential clinical utility and would also suggest the feasibility of targeting of other Ads.
sAd24 would be an even stronger candidate as a vector for gene delivery in humans should it avoid liver tissue on systemic delivery. The massive transduction of the liver by the Ad5 vectors causes severe systemic toxicity and raises serious safety concerns. Recent studies have identified the high-affinity binding of FX to the Ad5 hexon protein as the molecular basis for hepatic transduction (22, 56). These studies also showed that the particles of several Ad serotypes bind FX poorly or do not bind it at all and that the stability of the resultant complexes correlates with the degree of liver transduction by the Ads. Given these findings, we studied the association of FX with the virions of sAd24 and showed that FX-sAd24 complexes are rather unstable. In particular, while the equilibrium dissociation constant, KD, for the FX-sAd24 complexes was only slightly higher than the KD for the control FX-Ad5 complex, the FX-sAd24 dissociation rate constant, kd, was 16 times the FX-Ad5 kd. To put these findings in perspective, recent successes in drug development have more often correlated with improvements of the kd values of the most efficacious drugs rather than with improvements of their KD values. In particular, the kd values correlated much better with the agent-target complex residence time, its dissociative half-life, and target selectivity (13). On this basis, the dissociation rate constant is now seen as a more meaningful parameter in assessing important biological interactions.
The observed instability of the FX-sAd24 complexes could be explained by variations in the primary sequences of the HVRs of the Ad5 and sAd24 hexons. Notably, recent studies have collectively suggested that FX binding to the Ad5 hexon involves HVRs 3, 5, and 7 (2, 22). At the level of the primary amino acid sequence, these HVRs within the sAd24 hexon share little homology with their Ad5 counterparts (the identity levels are 0%, 29%, and 33% for HVRs 3, 5, and 7, respectively). In addition, of the five amino acid residues within the Ad5 hexon HVR7 that have been confirmed to contribute to FX binding (22, 56), only two, T404 and E431, are conserved within the sAd24 hexon structure. It is thus reasonable to speculate that these differences in the structures of the Ad5 and sAd24 HVRs contribute to the much weaker association of FX with the sAd24 hexon.
The instability of FX-sAd24 complexes results in much lower hepatic transduction by sAd24 than by Ad5 upon systemic delivery to mice. Imaging of the vector-injected mice detected reporter expression in the livers of the sAd24-injected animals only at very low levels, while injections with the Ad5 vector yielded massive liver transduction. Notably, these findings correlate well with the earlier report by Roy et al., who used histochemical staining of murine livers to show reduced transgene expression by an intravenously administered sAd24 vector (45).
Our optical imaging data were corroborated by PET-CT results and by measurements of luciferase activity in homogenates of tissues isolated from vector-treated mice. With its greater sensitivity, the latter assay has confirmed very significant differences in reporter expression by the tested Ads in those tissues, where signals were undetectable by whole-body imaging. Depending on the particular tissue assayed, the reporter activities expressed by the sAd24 vector (per organ) were 10 to 14,000 times less than those induced by the Ad5 vector.
Measurements of viral DNA content in organs isolated from Ad-injected mice showed that, like Ad5 virions, sAd24 particles accumulated primarily in the liver shortly after vector administration. This accumulation of sAd24 did not yield any significant reporter expression, suggesting that most of the injected viral dose was sequestered by the sinusoidal liver macrophages, KCs, which had been shown to clear other serotype Ads after systemic delivery (3). Examination of the livers of sAd24-injected mice by fluorescence microscopy confirmed the rapid uptake of the vector by KCs and the nearly total depletion of these cells 24 h after vector administration.
Next, by measuring the levels of six inflammatory cytokines in the blood of animals treated with Ads, we have shown that expression of five of these cytokines was elevated in sAd24-injected animals to significantly higher levels than in mice that had received the Ad5 vector. These data are in line with the previously reported differences in inflammatory responses to Ads, which were found to be Ad serotype related (3). More-detailed studies of immune responses to sAd24 are now warranted to identify the challenges related to the use of this serotype for gene therapy and to develop strategies to overcome those challenges.
Together the greatly reduced transduction of nontarget tissues by sAd24 revealed by our studies and the low seroprevalence of this Ad in humans reported by others (45, 64) provided a rationale for developing a tropism-modified derivative of sAd24 as a prototype of targeted gene vectors for clinical use. This was accomplished by applying the fiber knob replacement strategy that we had previously developed for the Ad5 fiber to the sAd24 fiber protein. We reasoned that together the evolutionary conservation of the domain-based structure of the Ad fiber proteins and the fact that the same functions are carried out by the same domains in various fibers suggest the promise of this approach.
Despite the fact that the sAd24 fiber protein has never been studied before and its structure has not been elucidated, the established general principles of the Ad fiber structure could be applied to this protein's primary structure to map its domains. Thus, the amino-terminal aa 1 to 44 and the carboxy-terminal aa 265 to 443 could be predicted to correspond to the protein's tail and knob, respectively. On the basis of the established folding pattern for an Ad fiber shaft (54), the central 220-aa-long shaft of the sAd24 fiber could be modeled to consist of 13 pseudorepeats. The sAd24 fiber shaft appears to contain an extended third repeat, the feature known to make the fiber flexible, and in this respect is similar to the Ad5 fiber. Furthermore, both fibers share a substantial similarity near their carboxy termini: not only do they both contain the KLGxGLxFD/N consensus motif seen in the last full repeat of most Ad fibers, but this homology region also covers most of the downstream shaft-to-knob transition region, with 62% of the amino acid residues here being identical in both Ads. These structural similarities were supportive of the rationale for modifying the sAd24 fiber by the knob replacement that had been successfully applied before to the Ad5 fiber. However, a potential confounding factor was the difference between the two fibers' shaft lengths. According to the model proposed by van Raaij et al., nine intersubunit hydrogen bonds are formed within each repeat of the Ad fiber shaft, and therefore each repeat contributes to the overall stability of the trimeric fiber (54). With the sAd24 shaft being much shorter than the Ad5 shaft (354 aa, 22 repeats), the former could thus form less-stable trimers. Whether fusion of this relatively short shaft with the fibritin would yield a trimer stable enough for encapsidation was unclear. In spite of this concern, our testing of the designed FsAd2411FHer2:7 chimera confirmed that its trimer was stable and thus predicted its successful encapsidation.
In our flow cytometry experiments, this protein showed binding to Her2-expressing cells but not to Her2-negative cells. The magnitude of this binding was lower, however, than that we saw previously for the similarly designed Ad5 fiber-derived chimera (7). Additional experiments showed that this difference was due not to the weaker binding of the FsAd2411FHer2:7 to Her2 but likely to suboptimal detection of cell-bound fibers with the anti-fibritin MAb 5E1, which was the only available MAb suitable for this comparative-binding assay. When used in parallel with anti-Ad5 fiber tail MAb to detect the cell-attached Ad5 fiber-derived chimera, the 5E1 MAb produced a much weaker signal (unpublished data), perhaps because of steric hindrance caused by the proximity of its epitope to the affibody-Her2 interface.
Successful rescue and propagation of the sAd24 vector in which its wt fiber gene was replaced with the gene for the targeting chimera and the efficient encapsidation of the chimera have further confirmed the structural equivalency of the designed protein to the wt fiber. The functional adequacy of this protein was then demonstrated through successful Her2-dependent gene delivery by this vector to Her2-expressing tumor cells in vitro. The efficacy of this gene delivery was invariably and substantially greater than that of the sAd24 vector carrying unmodified fibers and was comparable to that of the previously designed Ad5-based vector targeted to Her2. Compared with unmodified Ad5 vector in the presence of neutralizing anti-Ad5 antibodies, moreover, the Her2-targeted sAd24 revealed a much lower degree of inactivation, thus suggesting that a tropism-modified sAd24 would be better suited for gene interventions in patients with Her2-expressing tumors than are the Ad5-based vectors.
Testing of the fiber-modified sAd24 in vivo by intravenous injection into immunocompromised mice suggested that the virus could transduce Her2-expressing human tumor cells present in murine vasculature. In particular, the whole-body imaging of the reporter activities expressed by the target cells and the vector showed colocalization of these signals. This imaging also showed a significant increase in Ad-produced luminescence in animals injected with both the cells and the vector compared to that in mice treated with the vector alone. However, despite our encouraging data on the Her2 dependence of sAd24TL.11FHer2:7 gene transfer in vitro, the in vivo comparison of this vector with the control nontargeted sAd24 showed a marginal increase in transduction that was not statistically significant. By measuring reporter activity in organs isolated from the experimental animals, we have been able to show statistically significant enhancement of gene transfer by sAd24TL.11FHer2:7 to two of the three organs containing the highest levels of Ad-expressed reporter, the lungs (13.5-fold) and the spleens (5.7-fold), while the 2.8-fold enhancement in the livers was not statistically significant.
The same approach was also used to assess the levels of gene transfer by the tropism-modified sAd24 vector to lung-localized Her2-expressing tumor metastases. In this scenario, we saw low levels of lung transduction in both the tumor-free and the tumor-bearing animals. Notably, however, the statistical significance of the higher level of reporter activity measured in the metastasized lungs was established.
Taken together, the results of these in vivo experiments suggest that the selectivity of the fiber-modified sAd24 vector for Her2, although clearly shown in vitro, was insufficient to support robust targeted transduction of tumor cells under the challenging conditions of systemic administration. While the demonstrated substantial and statistically significant enhancement of transduction in the lungs and spleens of animals systemically injected with the tumor cells and sAd24TL.11FHer2:7 is encouraging, future improvements of the vector are needed to achieve the desired levels of target-specific transduction on vascular delivery.
Given the diversity of soluble molecules encountered by the blood-borne Ad vectors, some level of nonspecific transduction resulting from interactions between the virus and these molecules, both known (22, 49) and yet unknown, appears practically unavoidable. The goal of vector targeting is, therefore, to achieve a level of target-specific gene transfer that is substantially higher than that of off-target transduction. In a recent study, the efficacy of on-target gene transfer by a systemically injected Ad vector was significantly improved by increasing the affinity of the virus's fiber protein for its receptor (57). This success suggests that the in vivo target selectivity of an sAd24-derived vector can also be augmented by using ligands with higher affinity for the target receptor. To this end, the next version of the Her2-targeted sAd24 can be designed to contain a fiber-fibritin chimera fused with a higher-affinity affibody, which is now available. While the Zher2:7 affibody ligand used in our study has an affinity for Her2 in the midnanomolar range (KD = 1.4 × 10−7 M) (62), Her2-specific affibodies with much higher affinities (up to 2.2 × 10−11 M) have recently been developed (39).
It could be speculated that once the desired level of target specificity is achieved for sAd24, similarly designed targeted vectors could be derived from other Ad serotypes that have been shown to evade neutralization by anti-Ad5 antibodies. Availability of multiserotype panels of vectors targeted to the same disease biomarker would provide a means of overcoming vector-induced immunity in patients and would thus make repeated gene delivery in humans more efficient. However, additional studies are needed to see whether the knob replacement approach would produce functional derivatives of those serotypes' fibers.
In conclusion, as the first developmental step toward an sAd24 vector suitable for targeted gene delivery in humans, we demonstrated that the systemically administered virions of sAd24 transduce normal tissues at decreased levels compared with those of Ad5 particles; studied the in vivo biodistribution of intravenously injected sAd24 virions and showed active sequestration of the virus by Kupffer cells; compared the inflammatory responses to sAd24 and Ad5 vectors; designed a proof-of-concept, fiber-modified sAd24 vector and confirmed its altered tropism in vitro; studied in vivo transduction by the systemically administered, modified sAd24; and identified the limitations of this novel vector.
The results of this work add significantly to the rather limited knowledge of sAd24 biology and also suggest a potential use of sAd24-derived vectors beyond the previously proposed genetic vaccination.
Acknowledgments
We thank the personnel of the Monoclonal Antibody Facility, the DNA Analysis Facility, the Vector Core Laboratory, the Radiochemistry Section of the Department of Experimental Diagnostic Imaging, and the Small Animal Imaging Facility of the M. D. Anderson Cancer Center for their help with producing hybridoma cell lines and antibodies to the sAd24 fiber, with DNA sequencing, with production and purification of wt sAd24, with production of 18F-labeled tracer, and with animal imaging, respectively. We are grateful to Andrey Volgin and Amer Najjar (both with the M. D. Anderson Cancer Center) and the personnel of the Flow Cytometry and Cellular Imaging Core Facility for their help with the cell-sorting experiments and to Oleksandr Kalyuzhniy (University of Washington, Seattle) for his advice on the setup of surface plasmon resonance studies. Michael Hulsey (BD Biosciences) is thanked for processing of the flow cytometry data. Kathryn Hale and Sunita Patterson are thanked for editing the manuscript.
This work was supported by Public Health Service grants R01 CA119387 (to C.L.) and R01 CA116621 and R01 CA128807 (to V.K.). Additional support was provided by the University Cancer Foundation of the University of Texas M. D. Anderson Cancer Center (to V.K.), Cancer Center Support Grant P30CA16672, and Experimental Cancer Imaging Research Program CA126577.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Alauddin, M. M., P. S. Conti, and J. D. Fissekis. 2003. A general synthesis of 2′-deoxy-2′-[18F]fluoro-5-methyl-1-β-d-arabinofuranosyluracil and its 5-substituted nucleosides. J. Label. Comp. Radiopharm. 46:285-289. [Google Scholar]
- 2.Alba, R., A. C. Bradshaw, A. L. Parker, D. Bhella, S. N. Waddington, S. A. Nicklin, N. van Rooijen, J. Custers, J. Goudsmit, D. H. Barouch, J. H. McVey, and A. H. Baker. 2009. Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: effect of mutagenesis on FX interactions and gene transfer. Blood 114:965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appledorn, D. M., A. Kiang, A. McBride, H. Jiang, S. Seregin, J. M. Scott, R. Stringer, Y. Kousa, M. Hoban, M. M. Frank, and A. Amalfitano. 2008. Wild-type adenoviruses from groups A-F evoke unique innate immune responses, of which HAd3 and SAd23 are partially complement dependent. Gene Ther. 15:885-901. [DOI] [PubMed] [Google Scholar]
- 4.Arnberg, N., K. Edlund, A. H. Kidd, and G. Wadell. 2000. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 74:42-48. [PMC free article] [PubMed] [Google Scholar]
- 5.Belousova, N., N. Korokhov, V. Krendelshchikova, V. Simonenko, G. Mikheeva, P. L. Triozzi, W. A. Aldrich, P. T. Banerjee, S. D. Gillies, D. T. Curiel, and V. Krasnykh. 2003. Genetically targeted adenovirus vector directed to CD40-expressing cells. J. Virol. 77:11367-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belousova, N., V. Krendelchtchikova, D. T. Curiel, and V. Krasnykh. 2002. Modulation of adenovirus vector tropism via incorporation of polypeptide ligands into the fiber protein. J. Virol. 76:8621-8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belousova, N., G. Mikheeva, J. Gelovani, and V. Krasnykh. 2008. Modification of adenovirus capsid with a designed protein ligand yields a gene vector targeted to a major molecular marker of cancer. J. Virol. 82:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 9.Bewley, M. C., K. Springer, Y. B. Zhang, P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 10.Bidaut, L. M., R. Pascual-Marqui, J. Delavelle, A. Naimi, M. Seeck, C. Michel, D. Slosman, O. Ratib, D. Ruefenacht, T. Landis, N. de Tribolet, J. R. Scherrer, and F. Terrier. 1996. Three- to five-dimensional biomedical multisensor imaging for the assessment of neurological (dys) function. J. Digit. Imaging 9:185-198. [DOI] [PubMed] [Google Scholar]
- 11.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chirmule, N., K. Propert, S. Magosin, Y. Qian, R. Qian, and J. Wilson. 1999. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 6:1574-1583. [DOI] [PubMed] [Google Scholar]
- 13.Copeland, R. A., D. L. Pompliano, and T. D. Meek. 2006. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 5:730-739. [DOI] [PubMed] [Google Scholar]
- 14.Defer, C., M. T. Belin, M. L. Caillet-Boudin, and P. Boulanger. 1990. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J. Virol. 64:3661-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 72:9706-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farina, S. F., G. P. Gao, Z. Q. Xiang, J. J. Rux, R. M. Burnett, M. R. Alvira, J. Marsh, H. C. Ertl, and J. M. Wilson. 2001. Replication-defective vector based on a chimpanzee adenovirus. J. Virol. 75:11603-11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 18.Hasenburg, A., D. C. Fischer, X. W. Tong, A. Rojas-Martinez, R. H. Kaufman, I. Ramzy, P. Kohlberger, M. Orlowska-Volk, E. Aguilar-Cordova, and D. G. Kieback. 2002. Adenovirus-mediated thymidine kinase gene therapy for recurrent ovarian cancer: expression of coxsackie-adenovirus receptor and integrins alphavbeta3 and alphavbeta5. J. Soc. Gynecol. Invest. 9:174-180. [PubMed] [Google Scholar]
- 19.Hemminki, A., N. Belousova, K. R. Zinn, B. Liu, M. Wang, T. R. Chaudhuri, B. E. Rogers, D. J. Buchsbaum, G. P. Siegal, M. N. Barnes, J. Gomez-Navarro, D. T. Curiel, and R. D. Alvarez. 2001. An adenovirus with enhanced infectivity mediates molecular chemotherapy of ovarian cancer cells and allows imaging of gene expression. Mol. Ther. 4:223-231. [DOI] [PubMed] [Google Scholar]
- 20.Henry, L. J., D. Xia, M. E. Wilke, J. Deisenhofer, and R. D. Gerard. 1994. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J. Virol. 68:5239-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong, J. S., and J. A. Engler. 1996. Domains required for assembly of adenovirus type 2 fiber trimers. J. Virol. 70:7071-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalyuzhniy, O., N. C. Di Paolo, M. Silvestry, S. E. Hofherr, M. A. Barry, P. L. Stewart, and D. M. Shayakhmetov. 2008. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:5483-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirby, I., E. Davison, A. J. Beavil, C. P. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 2000. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J. Virol. 74:2804-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirby, I., E. Davison, A. J. Beavil, C. P. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 1999. Mutations in the DG loop of adenovirus type 5 fiber knob protein abolish high-affinity binding to its cellular receptor CAR. J. Virol. 73:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korokhov, N., G. Mikheeva, A. Krendelshchikov, N. Belousova, V. Simonenko, V. Krendelshchikova, A. Pereboev, A. Kotov, O. Kotova, P. L. Triozzi, W. A. Aldrich, J. T. Douglas, K. M. Lo, P. T. Banerjee, S. D. Gillies, D. T. Curiel, and V. Krasnykh. 2003. Targeting of adenovirus via genetic modification of the viral capsid combined with a protein bridge. J. Virol. 77:12931-12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krasnykh, V., N. Belousova, N. Korokhov, G. Mikheeva, and D. T. Curiel. 2001. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J. Virol. 75:4176-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krasnykh, V., I. Dmitriev, G. Mikheeva, C. R. Miller, N. Belousova, and D. T. Curiel. 1998. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J. Virol. 72:1844-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieber, A., C. Y. He, L. Meuse, D. Schowalter, I. Kirillova, B. Winther, and M. A. Kay. 1997. The role of Kupffer cell activation and viral gene expression in early liver toxicity after infusion of recombinant adenovirus vectors. J. Virol. 71:8798-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louis, N., P. Fender, A. Barge, P. Kitts, and J. Chroboczek. 1994. Cell-binding domain of adenovirus serotype 2 fiber. J. Virol. 68:4104-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maes, F., A. Collignon, D. Vandermeulen, G. Marchal, and P. Suetens. 1997. Multimodality image registration by maximization of mutual information. IEEE Trans. Med. Imaging 16:187-198. [DOI] [PubMed] [Google Scholar]
- 31.Magnusson, M. K., P. Henning, S. Myhre, M. Wikman, T. G. Uil, M. Friedman, K. M. Andersson, S. S. Hong, R. C. Hoeben, N. A. Habib, S. Stahl, P. Boulanger, and L. Lindholm. 2007. Adenovirus 5 vector genetically re-targeted by an Affibody molecule with specificity for tumor antigen HER2/neu. Cancer Gene Ther. 14:468-479. [DOI] [PubMed] [Google Scholar]
- 32.Magnusson, M. K., S. S. Hong, P. Boulanger, and L. Lindholm. 2001. Genetic retargeting of adenovirus: novel strategy employing “deknobbing” of the fiber. J. Virol. 75:7280-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magnusson, M. K., S. See Hong, P. Henning, P. Boulanger, and L. Lindholm. 2002. Genetic retargeting of adenovirus vectors: functionality of targeting ligands and their influence on virus viability. J. Gene Med. 4:356-370. [DOI] [PubMed] [Google Scholar]
- 34.Manickan, E., J. S. Smith, J. Tian, T. L. Eggerman, J. N. Lozier, J. Muller, and A. P. Byrnes. 2006. Rapid Kupffer cell death after intravenous injection of adenovirus vectors. Mol. Ther. 13:108-117. [DOI] [PubMed] [Google Scholar]
- 35.Mercier, G. T., J. A. Campbell, J. D. Chappell, T. Stehle, T. S. Dermody, and M. A. Barry. 2004. A chimeric adenovirus vector encoding reovirus attachment protein sigma1 targets cells expressing junctional adhesion molecule 1. Proc. Natl. Acad. Sci. U. S. A. 101:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitraki, A., A. Barge, J. Chroboczek, J. P. Andrieu, J. Gagnon, and R. W. Ruigrok. 1999. Unfolding studies of human adenovirus type 2 fibre trimers. Evidence for a stable domain. Eur. J. Biochem. 264:599-606. [DOI] [PubMed] [Google Scholar]
- 37.Muruve, D. A., M. J. Barnes, I. E. Stillman, and T. A. Libermann. 1999. Adenoviral gene therapy leads to rapid induction of multiple chemokines and acute neutrophil-dependent hepatic injury in vivo. Hum. Gene Ther. 10:965-976. [DOI] [PubMed] [Google Scholar]
- 38.Novelli, A., and P. A. Boulanger. 1991. Assembly of adenovirus type 2 fiber synthesized in cell-free translation system. J. Biol. Chem. 266:9299-9303. [PubMed] [Google Scholar]
- 39.Orlova, A., M. Magnusson, T. L. Eriksson, M. Nilsson, B. Larsson, I. Hoiden-Guthenberg, C. Widstrom, J. Carlsson, V. Tolmachev, S. Stahl, and F. Y. Nilsson. 2006. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 66:4339-4348. [DOI] [PubMed] [Google Scholar]
- 40.Ory, D. S., B. A. Neugeboren, and R. C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. U. S. A. 93:11400-11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philipson, L., K. Lonberg-Holm, and U. Pettersson. 1968. Virus-receptor interaction in an adenovirus system. J. Virol. 2:1064-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts, D. M., A. Nanda, M. J. Havenga, P. Abbink, D. M. Lynch, B. A. Ewald, J. Liu, A. R. Thorner, P. E. Swanson, D. A. Gorgone, M. A. Lifton, A. A. Lemckert, L. Holterman, B. Chen, A. Dilraj, A. Carville, K. G. Mansfield, J. Goudsmit, and D. H. Barouch. 2006. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441:239-243. [DOI] [PubMed] [Google Scholar]
- 43.Roelvink, P. W., G. Mi Lee, D. A. Einfeld, I. Kovesdi, and T. J. Wickham. 1999. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing Adenoviridae. Science 286:1568-1571. [DOI] [PubMed] [Google Scholar]
- 44.Roy, S., G. Gao, D. S. Clawson, L. H. Vandenberghe, S. F. Farina, and J. M. Wilson. 2004. Complete nucleotide sequences and genome organization of four chimpanzee adenoviruses. Virology 324:361-372. [DOI] [PubMed] [Google Scholar]
- 45.Roy, S., G. Gao, Y. Lu, X. Zhou, M. Lock, R. Calcedo, and J. M. Wilson. 2004. Characterization of a family of chimpanzee adenoviruses and development of molecular clones for gene transfer vectors. Hum. Gene Ther. 15:519-530. [DOI] [PubMed] [Google Scholar]
- 46.Roy, S., G. P. Kobinger, J. Lin, J. Figueredo, R. Calcedo, D. Kobasa, and J. M. Wilson. 2007. Partial protection against H5N1 influenza in mice with a single dose of a chimpanzee adenovirus vector expressing nucleoprotein. Vaccine 25:6845-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schagen, F. H., F. M. Wensveen, J. E. Carette, T. S. Dermody, W. R. Gerritsen, and V. W. van Beusechem. 2006. Genetic targeting of adenovirus vectors using a reovirus sigma1-based attachment protein. Mol. Ther. 13:997-1005. [DOI] [PubMed] [Google Scholar]
- 48.Sebestyen, Z., J. de Vrij, M. Magnusson, R. Debets, and R. Willemsen. 2007. An oncolytic adenovirus redirected with a tumor-specific T-cell receptor. Cancer Res. 67:11309-11316. [DOI] [PubMed] [Google Scholar]
- 49.Shayakhmetov, D. M., A. Gaggar, S. Ni, Z. Y. Li, and A. Lieber. 2005. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J. Virol. 79:7478-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Short, J. J., A. V. Pereboev, Y. Kawakami, C. Vasu, M. J. Holterman, and D. T. Curiel. 2004. Adenovirus serotype 3 utilizes CD80 (B7.1) and CD86 (B7.2) as cellular attachment receptors. Virology 322:349-359. [DOI] [PubMed] [Google Scholar]
- 51.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. U. S. A. 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuruta, Y., L. Pereboeva, J. N. Glasgow, C. L. Luongo, S. Komarova, Y. Kawakami, and D. T. Curiel. 2005. Reovirus sigma1 fiber incorporated into adenovirus serotype 5 enhances infectivity via a CAR-independent pathway. Biochem. Biophys. Res. Commun. 335:205-214. [DOI] [PubMed] [Google Scholar]
- 53.van Beusechem, V. W., A. L. van Rijswijk, H. H. van Es, H. J. Haisma, H. M. Pinedo, and W. R. Gerritsen. 2000. Recombinant adenovirus vectors with knobless fibers for targeted gene transfer. Gene Ther. 7:1940-1946. [DOI] [PubMed] [Google Scholar]
- 54.van Raaij, M. J., A. Mitraki, G. Lavigne, and S. Cusack. 1999. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401:935-938. [DOI] [PubMed] [Google Scholar]
- 55.Vogels, R., D. Zuijdgeest, R. van Rijnsoever, E. Hartkoorn, I. Damen, M. P. de Bethune, S. Kostense, G. Penders, N. Helmus, W. Koudstaal, M. Cecchini, A. Wetterwald, M. Sprangers, A. Lemckert, O. Ophorst, B. Koel, M. van Meerendonk, P. Quax, L. Panitti, J. Grimbergen, A. Bout, J. Goudsmit, and M. Havenga. 2003. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 77:8263-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waddington, S. N., J. H. McVey, D. Bhella, A. L. Parker, K. Barker, H. Atoda, R. Pink, S. M. Buckley, J. A. Greig, L. Denby, J. Custers, T. Morita, I. M. Francischetti, R. Q. Monteiro, D. H. Barouch, N. van Rooijen, C. Napoli, M. J. Havenga, S. A. Nicklin, and A. H. Baker. 2008. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell 132:397-409. [DOI] [PubMed] [Google Scholar]
- 57.Wang, H., Y. Liu, Z. Li, S. Tuve, D. Stone, O. Kalyushniy, D. Shayakhmetov, C. L. Verlinde, T. Stehle, J. McVey, A. Baker, K. W. Peng, S. Roffler, and A. Lieber. 2008. In vitro and in vivo properties of adenovirus vectors with increased affinity to CD46. J. Virol. 82:10567-10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wickham, T. J., E. J. Filardo, D. A. Cheresh, and G. R. Nemerow. 1994. Integrin αvβ5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 60.Wickham, T. J., P. W. Roelvink, D. E. Brough, and I. Kovesdi. 1996. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 14:1570-1573. [DOI] [PubMed] [Google Scholar]
- 61.Wickham, T. J., E. Tzeng, L. L. Shears II, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wikman, M., A. C. Steffen, E. Gunneriusson, V. Tolmachev, G. P. Adams, J. Carlsson, and S. Stahl. 2004. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng. Des Sel. 17:455-462. [DOI] [PubMed] [Google Scholar]
- 63.Xu, Z., J. Tian, J. S. Smith, and A. P. Byrnes. 2008. Clearance of adenovirus by Kupffer cells is mediated by scavenger receptors, natural antibodies, and complement. J. Virol. 82:11705-11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhi, Y., J. Figueredo, G. P. Kobinger, H. Hagan, R. Calcedo, J. R. Miller, G. Gao, and J. M. Wilson. 2006. Efficacy of severe acute respiratory syndrome vaccine based on a nonhuman primate adenovirus in the presence of immunity against human adenovirus. Hum. Gene Ther. 17:500-506. [DOI] [PubMed] [Google Scholar]