Abstract
Previous studies have identified a central role for HLA-B alleles in influencing control of HIV infection. An alternative possibility is that a small number of HLA-B alleles may have a very strong impact on HIV disease outcome, dominating the contribution of other HLA alleles. Here, we find that even following the exclusion of subjects expressing any of the HLA-B class I alleles (B*57, B*58, and B*18) identified to have the strongest influence on control, the dominant impact of HLA-B alleles on virus set point and absolute CD4 count variation remains significant. However, we also find that the influence of HLA on HIV control in this C-clade-infected cohort from South Africa extends beyond HLA-B as HLA-Cw type remains a significant predictor of virus and CD4 count following exclusion of the strongest HLA-B associations. Furthermore, there is evidence of interdependent protective effects of the HLA-Cw*0401-B*8101, HLA-Cw*1203-B*3910, and HLA-A*7401-B*5703 haplotypes that cannot be explained solely by linkage to a protective HLA-B allele. Analysis of individuals expressing both protective and detrimental alleles shows that even the strongest HLA alleles appear to have an additive rather than dominant effect on HIV control at the individual level. Finally, weak but significant frequency-dependent effects in this cohort can be detected only by looking at an individual's combined HLA allele frequencies. Taken together, these data suggest that although individual HLA alleles, particularly HLA-B, can have a strong impact, HIV control overall is likely to be influenced by the additive effect of some or all of the other HLA alleles present.
HIV-specific CD8+ T cells play a central role in resolution of primary viremia and the long-term suppression of viral replication (13). Supporting this notion is the observed correlation between possession of particular human leukocyte antigen (HLA) class I alleles and control of HIV, measured both directly by time-to-AIDS (5, 6) and indirectly via clinical markers of disease progression (viral load [VL] and CD4 count) (15, 26, 28). Specific HLA class I alleles have been associated with relatively successful control of viral replication and slow disease progression, most notably, alleles HLA-B*57 and HLA-B*27 (1, 7, 12, 15, 21, 23), and also with relatively ineffective control of viral replication and rapid disease progression [B*35(Px), B*5802, and B*18] (5, 15, 17, 23). In addition, general trends suggesting an HLA class I heterozygote advantage (5) and rare allele advantage (28) and, most recently, a correlation between levels of surface expression linked to certain HLA-Cw alleles (11, 27) and HIV control has also been described.
Among the different HLA class I loci, the HIV-specific CD8+ T-cell responses restricted by HLA-B alleles are thought to play the central role in determining disease outcome: the majority of detectable HIV-specific CD8+ T-cell responses are restricted by HLA-B alleles (3, 15, 16), HLA-B-restricted responses typically express a more effective “polyfunctional” phenotype (14), the strongest HLA-associations with either slow or rapid progression are with HLA-B alleles (5, 10, 11, 15), and HLA-B-restricted CD8+ T cells exert the strongest selection pressure on the virus (15, 19, 24). However, whether this apparent association between HIV immune control and HLA-B is a general and causal trend or, rather, is biased by the coincidence that the strongest HLA associations with either extreme of disease control happen, by chance, to involve HLA-B alleles remains uncertain.
In order to further investigate the correlation between HLA type and HIV infection control, we here examine a cohort now comprising >1,200 chronically HIV C-clade-infected, treatment-naïve subjects from Durban, South Africa, in an extended analysis following from our previous studies of a smaller cohort (15). We first address the question of whether the dominant role of HLA-B in this population compared to the roles of HLA-A or HLA-C results from the influence of HLA-B alleles in general or is dependent on a few known strong associations, such as that between HLA-B*57 alleles and low viremia. Second, in light of recent data (11, 27), we assess the impact of HLA-C alleles on HIV disease outcome and examine the effect of HLA haplotypes on observed HLA associations with disease control. Third, we investigate the question of whether the impact of certain HLA-B alleles on HIV outcome dominates that of other HLA-B alleles to negate the contribution of the latter or whether the impact of individual HLA alleles can be additive. Finally, we compare the impact of individual HLA alleles on HIV on immune control to the impact of heterozygote and rare allele advantage in this cohort.
MATERIALS AND METHODS
Study subjects.
A total of 1,211 treatment-naïve adults with C-clade HIV infection were recruited from Durban, South Africa, following voluntary counseling and testing in either antenatal or outpatient clinics, as previously described (16). HIV-1 RNA VLs were available for all 1,211 of these subjects, and absolute CD4 counts were available for 1.158 subjects. VL was measured with the Roche Amplicor assay, version 1.5, and the CD4 count was measured by flow cytometry.
HLA typing.
High-resolution HLA typing was undertaken with genomic DNA by single-stranded conformation polymorphism PCR (4). In a small minority of instances, resolution to only two-digit types was possible. In those cases where there was more than one four-digit HLA subtype for a given two-digit type present in the study population, the two-digit types were excluded. HLA haplotypes were inferred by linkage disequilibrium, with significance measured by Fisher's exact test and strength measured by calculating D′ [D′ = D/Dmax, where D = (frequency of the AB haplotype × frequency of non-AB haplotype) − (frequency of the A haplotype × frequency of the B haplotype)]. This study was approved by the University of KwaZulu-Natal Biomedical Research Ethics Committee, the Oxford Research Ethics Committee, and the Massachusetts General Hospital Institutional Review Board.
Statistical analysis.
To test for the differential contribution that HLA-A, -B, and -C alleles make toward variation in VL of the cohort as a whole, we first tested for differences in VLs between individuals grouped according to their HLA-A type using a nonparametric Kruskal-Wallis test of variance, which, in addition to testing significance, returns a Kruskal-Wallis statistic (H) that corresponds to the strength of the effect tested, in this case, that of grouping individuals by their HLA-A alleles. This was repeated, grouping individuals in turn by their HLA-B and HLA-C types. Analysis of individual HLA associations with VL was undertaken by comparison of VLs in subjects expressing the relevant allele with VLs in subjects not expressing that allele (unpaired t test). This analysis was undertaken for the 65 alleles expressed in ≥1% of the 1,211 study subjects. Stringent Bonferroni correction for 65 multiple tests generates a significance cutoff P value of <0.0007. The same steps were repeated to examine associations between HLA class I type and absolute CD4 count, although with a slightly reduced cohort of 1,158 individuals due to unavailable CD4 data. In order to control for the effect of the strongest associations, the entire analysis was then repeated, excluding the four HLA alleles with the strongest associations, HLA-B*57, -B*5801, -B*18, and -B*5802, which were the only significant associations with high or low viremia in the smaller cohort (n = 706) previously reported (4).
For the analysis of the effect of HLA allele frequency, a combined phenotypic HLA frequency was generated for each individual which corresponded to the combined frequencies of their six HLA class I alleles. In addition, the frequencies of each individual's pair of HLA-A, -B, or -C alleles were considered separately in order negate the complication of linkage disequilibrium between the HLA-A, -B, and -C loci. All homozygotes were excluded from this analysis (n = 173) to reduce any bias introduced by possible homozygous disadvantage (5). The effect of this combined allele frequency on both VL and CD4 count was then tested by Spearman's rank correlation and linear regression.
For the analysis of homozygote disadvantage only individuals with full four-digit typing were considered homozygotes as different HLA subtypes can be associated with very different disease outcomes (15, 22). Subjects with two-digit HLA typing were included in the heterozygote group only if they were unambiguously heterozygote. Thus, the data set for this analysis was reduced to 1,180 individuals for VL (1,002 heterozygotes and 178 homozygotes [68 homozygous at HLA-A, 68 homozygous at HLA-B, and 105 homozygous at HLA-C; 122 homozygous at only one allele, 38 homozygous at two alleles, and 11 homozygous at all three alleles]) and 1,129 for CD4 count (956 heterozygotes and 173 homozygotes [67 homozygous at HLA-A, 67 homozygous at HLA-B, and 101 homozygous at HLA-C; 118 homozygous at only one allele, 37 homozygous at two alleles and 11 homozygous at all three]).
LRT.
In order to allow the use of certain HLA alleles as covariates, a likelihood ratio test (LRT) was used with linear regression models, comparing a null model to an alternative model with/without the variable(s) of interest. Permutation tests were also used to verify the validity of the LRT in this setting. Because results closely matched the LRT, only LRT results are reported. Tests were performed with the additional covariates of B*5702, B*5703, B*5801, B*5802, and B*18 as well as without these alleles. The CD4 target variable was used in original units, whereas the natural log of VL was used for that target. For univariate HLA associations, a linear regression model was used with/without (alternative/null) each possible HLA allele. For omnibus tests on all alleles at a locus (i.e., A, B, or C), a linear regression model with/without all alleles at the locus of interest was used. For testing association with homozygosity, a linear regression model was used in different ways, each with a different encoding of the information and using each of these in turn: (i) the number of homozygous loci as a feature for the alternative model; (ii) a variable indicating whether a person was homozygous at just one allele or not, again at two alleles or not, and finally at three alleles or not; (iii) a variable indicating whether a person was homozygous at any locus or not.
RESULTS
HLA-B type most strongly correlates with disease control in the extended data set.
We analyzed HIV VL and absolute CD4 counts from a total of 1,211 HIV C-clade-infected South Africans living in KwaZulu-Natal, extending the study population from an initial cohort of 706 (15), to seek correlation between HLA class I genotype and well-characterized markers of disease progression (2, 20). Our earlier analysis was restricted to alleles with a phenotypic frequency of greater than 5% (a total of 38 different HLA alleles comprised of 15, 13, and 10 HLA-A, -B, and -C alleles, respectively), but the increased cohort size enabled all HLA alleles occurring at a frequency of greater than 1% to be considered, thus encompassing a total of 65 different HLA alleles (23 HLA-A, 25 HLA-B, and 17 HLA-C).
Our earlier findings were confirmed in this larger data set, with HLA-B type being the strongest predictor of VL and CD4 count. For VL, the Kruskal-Wallis statistic, H, which is a nonparametric measure of variation between data groups, equals 146 (P < 0.00001) for grouping by HLA-B type, compared to an H of 48 (P = 0.0008) for HLA-A and an H of 60 (P < 0.00001) for HLA-C (Fig. 1 A). For CD4 count, H was 160 (P < 0.00001) for HLA-B, compared to an H of 42 (P = 0.006) for HLA-A and an H of 83 (P < 0.00001) for HLA-C (Fig. 1C). Thus, although the HLA-A or HLA-C type does appear to have an influence on variation in VL and CD4 count in the cohort, this effect is less marked than that of HLA-B. In addition, much of this apparent effect, as described below, is likely to be the result of linkage disequilibrium between HLA-A or HLA-C alleles with the more potent HLA-B alleles (see Table S1 in the supplemental material).
FIG. 1.
(A and B) Association of HLA allele expression with steady-state viral load (A) and absolute CD4 count (B) in a cohort of 1,211 HIV C-clade chronically infected individuals from KwaZulu-Natal, South Africa. Left-hand panels show association of all alleles occurring at a phenotypic frequency of greater than 1%. Associations with a P value of <0.05 (as measured by a Mann-Whitney U test) are highlighted in yellow, and those retaining significance after Bonferroni correction for multiple comparisons are highlighted in blue. Right-hand panels show the relative contribution of HLA-A, -B, and -C alleles to variation in viral load and CD4 count as measured by the Kruskal-Wallis test statistic (H). (C and D) Association of HLA allele expression with steady-state viral load (C) and absolute CD4 count (D), excluding the four alleles most strongly associated with differences in viral load and CD4 count (HLA-B*57, -B*5801, -B*18, and -B*5802). A total of 682 subjects were analyzed.
Without taking into account a correction for multiple comparisons or linkage disequilibrium, 52% (13/25) of HLA-B alleles are associated with a significantly higher or lower VL, compared to 13% (3/23) for HLA-A and 29% (5/17) for HLA-C alleles (Table 1). The same trend is observed for CD4 count, with 48% (12/25) of HLA-B alleles associated with significantly higher/lower CD4 counts compared to 17% (4/23) of HLA-A and 29% (5/17) of HLA-C alleles (Table 2). This dominance of HLA-B is maintained following the stringent Bonferroni correction for multiple comparisons. Four of the five alleles remaining significantly associated with higher/lower VLs are HLA-B alleles, i.e., B*5703 and B*5801 with low VLs and B*5802 and B*18 with high VLs; 7 of the 10 alleles remaining associated with higher/lower CD4 counts are HLA-B alleles, i.e., B*5703, B*5702, B*5801, B*8101, and B*4403 with high CD4 counts and B*5802 and B*18 with low CD4 counts (Tables 1 and 2 and Fig. 1A and B). In each case the non-HLA-B alleles associated with higher/lower VLs and/or CD4 counts are HLA-C (Cw*0602 for VL and Cw*0401, Cw*0602, and Cw*1203 for CD4 counts, as shown in Tables 1 and 2, respectively; the Cw*0602 effects, as previously observed [4], are likely to be due to linkage disequilibrium of Cw*0602 with B*5802).
TABLE 1.
Association between HLA alleles and VL
| Allele | Association with entire cohorta |
Association with exclusion of top four allelesb |
||
|---|---|---|---|---|
| Median VL | P value | Median VL | P value | |
| HLA-A | ||||
| A0101 | 29,700 | 0.424 | 41,800 | 0.918 |
| A0201 | 75,100 | 0.108 | 64,500 | 0.141 |
| A0202 | 26,900 | 0.426 | 26,128 | 0.657 |
| A0205 | 18,400 | 0.001 | 54,400 | 0.859 |
| A0301 | 49,300 | 0.149 | 44,300 | 0.439 |
| A2301 | 50,450 | 0.169 | 44,369 | 0.382 |
| A2402 | 23,200 | 0.23 | 26,600 | 0.994 |
| A2601 | 51,100 | 0.927 | 51,100 | 0.883 |
| A29 | 38,300 | 0.643 | 30,000 | 0.601 |
| A3001 | 42,035 | 0.297 | 42,850 | 0.529 |
| A3002 | 44,100 | 0.654 | 31,750 | 0.503 |
| A3004 | 53,500 | 0.195 | 28,300 | 0.702 |
| A3201 | 1E+05 | 0.32 | 162,000 | 0.154 |
| A33 | 26,050 | 0.137 | 17,868 | 0.093 |
| A3402 | 38,200 | 0.927 | 39,061 | 0.844 |
| A3601 | 38,550 | 0.631 | 15,100 | 0.582 |
| A4301 | 47,850 | 0.204 | 33,800 | 0.524 |
| A6601 | 60,400 | 0.152 | 14,700 | 0.018 |
| A6801 | 20,000 | 0.004 | 23,200 | 0.065 |
| A6802 | 73,900 | 0.456 | 88,150 | 0.25 |
| A7401 | 41,000 | 9E-04 | 40,250 | 0.327 |
| A6602 | 19,000 | 0.573 | 25,954 | 0.825 |
| A8001 | 92,250 | 0.108 | ||
| HLA-B | ||||
| B0702 | 28,300 | 0.21425 | 26,500 | 0.281 |
| B0705 | 100,000 | 0.1044 | 143,000 | 0.085 |
| B0801 | 50,950 | 0.10076 | 61,088 | 0.02 |
| B13 | 12,700 | 0.02711 | 22,500 | 0.222 |
| B1401 | 27,600 | 0.10719 | 14,360 | 0.14 |
| B1402 | 57,700 | 0.67348 | 26,128 | 0.643 |
| B1503 | 53,350 | 0.0167 | 55,300 | 0.004 |
| B1510 | 66,350 | 0.03362 | 61,900 | 0.037 |
| B1516 | 19,050 | 0.67152 | 12,010 | 0.307 |
| B18 | 92,500 | 0.00005 | ||
| B35 | 37,900 | 0.80593 | 30,000 | 0.527 |
| B3910 | 18,400 | 0.02571 | 12,000 | 0.008 |
| B4101 | 129,000 | 0.17934 | 129,000 | 0.227 |
| B4201 | 26,150 | 0.00884 | 26,000 | 0.028 |
| B4202 | 92,700 | 0.30147 | 92,700 | 0.173 |
| B4403 | 25,500 | 0.01917 | 24,579 | 0.091 |
| B4501 | 76,200 | 0.01804 | 63,900 | 0.136 |
| B4901 | 91,000 | 0.08138 | 52,700 | 0.38 |
| B51 | 70,900 | 0.38697 | 45,798 | 0.896 |
| B5301 | 33,550 | 0.89029 | 24,363 | 0.916 |
| B5702 | 14,145 | 0.00285 | ||
| B5703 | 5,528 | 1.30E-08 | ||
| B5801 | 18,400 | 0.0002 | ||
| B5802 | 75,900 | 4.60E-08 | ||
| B8101 | 19,400 | 0.00864 | 20,500 | 0.052 |
| HLA-C | ||||
| Cw02 | 44,795 | 0.08726 | 42,585 | 0.24 |
| Cw0302 | 15,000 | 0.01133 | ||
| Cw0304 | 55,900 | 0.63502 | 45,000 | 0.944 |
| Cw0401 | 29,850 | 0.03646 | 25,500 | 0.041 |
| Cw0501 | 38,200 | 0.31217 | ||
| Cw0602 | 64,200 | 5.90E-07 | 31,200 | 0.464 |
| Cw0701 | 31,400 | 0.06595 | 44,369 | 0.079 |
| Cw0702 | 31,600 | 0.57478 | 32,700 | 0.817 |
| Cw0704 | 84,900 | 0.0122 | ||
| Cw0801 | 43,369 | 0.94714 | 42,300 | 0.874 |
| Cw0802 | 34,750 | 0.42796 | 26,864 | 0.497 |
| Cw0804 | 55,800 | 0.26745 | 40,600 | 0.359 |
| Cw1203 | 14,750 | 0.001 | 6,055 | 1E-04 |
| Cw1601 | 75,000 | 0.12564 | 76,900 | 0.074 |
| Cw1701 | 1E+05 | 0.12763 | 33,200 | 0.245 |
| Cw1801 | 33,500 | 0.08234 | 35,750 | 0.637 |
| Cw1604 | 24,157 | 0.16371 | ||
Exact P values for association between HLA alleles and steady-state viral load are given. All P values represent Mann-Whitney U tests between all individuals possessing and all individuals lacking a given HLA allele. All associations significant at a P value of <0.05 are highlighted in boldface, and those that remain significant following Bonferroni correction are underlined.
Mann-Whitney U tests were carried out following exclusion of the four strongest HLA associations: HLA-B*57, HLA-B*5801, HLA-B*18, and HLA-B*5802.
TABLE 2.
Association between HLA alleles and absolute CD4 count
| Allele | Association with entire cohorta |
Association with exclusion of top four allelesb |
||
|---|---|---|---|---|
| Median CD4 count | P value | Median CD4 count | P value | |
| HLA-A | ||||
| A0101 | 414.5 | 0.23697 | 431 | 0.16141 |
| A0201 | 336 | 0.43918 | 376 | 0.86264 |
| A0202 | 447 | 0.26649 | 447 | 0.54073 |
| A0205 | 413.5 | 0.0025 | 382 | 0.34923 |
| A0301 | 328 | 0.03674 | 308 | 0.00792 |
| A2301 | 368 | 0.2267 | 370 | 0.53177 |
| A2402 | 360.5 | 0.74159 | 361.5 | 0.63343 |
| A2601 | 445 | 0.13058 | 423 | 0.28828 |
| A29 | 375 | 0.95601 | 396 | 0.45789 |
| A3001 | 362.5 | 0.3199 | 368.5 | 0.39151 |
| A3002 | 367 | 0.86507 | 377.5 | 0.82533 |
| A3004 | 341.5 | 0.29507 | 352 | 0.42233 |
| A3201 | 347 | 0.21075 | 347 | 0.25617 |
| A33 | 472.5 | 0.00859 | 480 | 0.00731 |
| A3402 | 415.5 | 0.04498 | 419 | 0.14516 |
| A3601 | 307 | 0.34355 | ||
| A4301 | 351 | 0.65896 | 363 | 0.86161 |
| A6601 | 307 | 0.05149 | 413 | 0.73452 |
| A6801 | 363 | 0.05781 | 316 | 0.08029 |
| A6802 | 310 | 0.81802 | 265.5 | 0.50581 |
| A7401 | 377 | 0.29202 | 371 | 0.6329 |
| A6602 | 399 | 0.69335 | 383 | 0.27032 |
| A8001 | 352 | 0.28678 | ||
| HLA-B | ||||
| B0702 | 366 | 0.3095 | 363.5 | 0.17576 |
| B0705 | 288 | 0.2504 | 272 | 0.06472 |
| B0801 | 322.5 | 0.03477 | 316 | 0.00798 |
| B13 | 431 | 0.24477 | 421.5 | 0.40309 |
| B1401 | 419 | 0.05833 | 420.5 | 0.07777 |
| B1402 | 408 | 0.28425 | 448 | 0.3488 |
| B1503 | 346 | 0.02963 | 355.5 | 0.01608 |
| B1510 | 333 | 0.01858 | 358.5 | 0.12639 |
| B1516 | 333 | 0.389 | 333 | 0.60262 |
| B18 | 285 | 0.00002 | ||
| B35 | 443 | 0.05023 | 465.5 | 0.03043 |
| B3910 | 436 | 0.02863 | 448.5 | 0.06529 |
| B4101 | 230.5 | 0.00291 | 230.5 | 0.0251 |
| B4201 | 401.5 | 0.20828 | 397 | 0.64474 |
| B4202 | 347 | 0.37401 | 366.5 | 0.58821 |
| B4403 | 427.5 | 0.00061 | 430.5 | 0.00206 |
| B4501 | 324.5 | 0.13118 | 337.5 | 0.64015 |
| B4901 | 365 | 0.7138 | 340 | 0.405 |
| B51 | 360 | 0.76569 | 420.5 | 0.71869 |
| B5301 | 408.5 | 0.68474 | 412 | 0.92665 |
| B5702 | 704 | 0.00001 | ||
| B5703 | 503 | 4.80E-07 | ||
| B5801 | 429 | 0.0001 | ||
| B5802 | 318 | 2.20E-07 | ||
| B8101 | 455.5 | 0.0002 | 465 | 0.00325 |
| HLA-C | ||||
| Cw02 | 331 | 0.00176 | 352 | 0.02147 |
| Cw0302 | 428 | 0.20997 | ||
| Cw0304 | 377 | 0.69318 | 373.5 | 0.85974 |
| Cw0401 | 426 | 0.00002 | 441 | 0.00004 |
| Cw0501 | 217 | 0.0062 | ||
| Cw0602 | 336 | 0.00004 | 381 | 0.74994 |
| Cw0701 | 398.5 | 0.0639 | 356 | 0.11071 |
| Cw0702 | 334.5 | 0.05211 | 338.5 | 0.04463 |
| Cw0704 | 293 | 0.19293 | ||
| Cw0801 | 365 | 0.37584 | 376.5 | 0.39944 |
| Cw0802 | 408 | 0.39674 | 427 | 0.36661 |
| Cw0804 | 360 | 0.61808 | 356 | 0.63007 |
| Cw1203 | 452 | 0.0014 | 508 | 0.00053 |
| Cw1601 | 336 | 0.10239 | 322 | 0.0795 |
| Cw1701 | 230 | 0.96497 | 387 | 0.78642 |
| Cw1801 | 386.5 | 0.00002 | 449.5 | 0.06443 |
| Cw1604 | 461.5 | 0.11381 | ||
Exact P values for association between HLA alleles and absolute CD4 count are given. All P values represent Mann-Whitney U tests between all individuals possessing and all individuals lacking a given HLA allele. All associations significant at a P value of <0.05 are highlighted in boldface, and those that remain significant following Bonferroni correction are underlined.
Mann-Whitney U tests were carried out following exclusion of the four strongest HLA associations: HLA-B*57, HLA-B*5801, HLA-B*18, and HLA-B*5802.
Dominant effect of HLA-B is not dependent on HLA-B*57/HLA-B*58/HLA-B*18 alleles.
We next examined the possibility that the observed dominant effect of HLA-B on VL and CD4 count might be a coincidence of the fact that the main protective (B*57 and B*5801) and susceptible (B*5802 and B*18) alleles happen to belong to the HLA-B locus. Initially, we simply excluded all subjects possessing B*57, B*5801, B*5802, and B*18 from the data set and repeated the analysis on the remaining 682 subjects. Despite exclusion of these four alleles, HLA-B remains the strongest predictor of both VL and CD4 count (Fig. 1B and D). For VL, H was 45 (P = 0.0007) for HLA-B, compared to an H of 20 (P = 0.5) for HLA-A and an H of 27 (P = 0.009) for HLA-C. For absolute CD4 count, H was 51 for HLA-B (P < 0.0001) compared to an H of 28 (P = 0.12) HLA-A and an H of 41 (P < 0.0001) for HLA-C (Fig. 1B and D).
To confirm this observation, we next adopted a more sophisticated likelihood ratio test (LRT) that allowed us to use the top four alleles described above as covariates rather than eliminating them from the analysis. This method also identifies HLA-B as the strongest predictor of both VL and CD4 count. When all alleles are considered, HLA-B gives the highest delta log likelihood (δ) values, which are approximate to the Kruskal-Wallis statistic (H); VL (δ = 119; P = 0) and CD4 (δ = 95; P = 0) are highest for HLA-B, followed by HLA-C (VL, δ = 59 and P = 2.53 × 10−11; CD4, δ = 41 and P = 4.43 × 10−6), and HLA-A (VL, δ = 25 and P = 0.2; CD4, δ = 31 and P = 0.02). This effect persists when HLA-B*57, -B*5801, -B*18, and -B*5802 are used as covariates, with the remaining HLA-B alleles giving δ values of 45 (P = 0.003) and 53 (P = 5.04 × 10−6) for VL and CD4, respectively, compared to those for HLA-C (VL, δ = 22 and P = 0.08; CD4, δ = 39 and P = 1.68 × 10−5) and HLA-A (VL, δ = 17 and P = 0.8; CD4, δ = 17 and P = 0.8).
These data suggest, first, that the dominant role of HLA-B in controlling HIV infection extends beyond the strongest four HLA-B associations and, second, that much of the apparent impact of HLA-A alleles on VL is related to linkage disequilibrium with HLA-B alleles, as grouping by HLA-A type no longer significantly explains any variation in VL or CD4 count when the top four HLA-B alleles are either excluded or used as covariates.
Impact of HLA-C genotype and HLA haplotypes on viral control.
Notably, HLA-C type remains significantly associated with variation in VL and CD4 count after the HLA-B*57/5801/5802/18 alleles are taken into account, albeit to a lesser extent than HLA-B. Furthermore, following the exclusion of these alleles, the only HLA individually associated with significantly higher/lower VL and/or CD4 count following Bonferroni correction are HLA-C alleles: HLA-Cw*0401 and HLA-Cw*1203. Both alleles are in linkage disequilibrium with HLA-B alleles, which is extremely tight in the case of the B*3910-Cw*1203 haplotype (D′ of 0.865, where perfect linkage disequilibrium gives a D′ of 1; P < 0.00001) and to a lesser extent with B*8101-Cw*0401 (D′ of 0.23; P < 0.00001) (see Table S1 in the supplemental material for cohort linkage disequilibrium calculations). However, in both cases, it is not possible to ascribe the protective effect to the HLA-B allele alone. In the case of the B*8101-Cw*0401 haplotype, the association with a lower VL and higher CD4 count holds only when the alleles are expressed together (Fig. 2), with neither allele being protective when it is expressed in isolation. For the B*3910-Cw*1203 haplotype, B*3910 appears to be protective only in association with Cw*1203. However, whether this represents a benefit of the B*3910-Cw*1203 haplotype or just of the Cw*1203 allele alone is hard to assess as only four subjects express this allele in the absence of B*3910 (Fig. 2).
FIG. 2.
Effect of HLA haplotype on steady-state viral load (A) and absolute CD4 count (B). The presence (+) or absence (−) of alleles is indicated. All P values were generated by a nonparametric Mann-Whitney U test.
Another example of the potentially additive effect of an HLA haplotype can be seen in the HLA-A*7401-B*5703 haplotype (D′ of 0.35; P < 0.0001). Although B*5703, unlike B*8101 and B*3910, remains protective in the absence of A*7401, there is a significantly lower VL in those individuals that express both alleles (for HLA-B*5703 and A*7401, a median VL of 1,650 [n = 23]; for B*5703 alone, a median VL of 10,400 [n = 22]; P = 0.005). A*7401 in the absence of B*5703 is not significantly associated with protection in this cohort (median VL, 29,500; P = 0.3) (Fig. 2).
Additive effect of protective and detrimental HLA alleles.
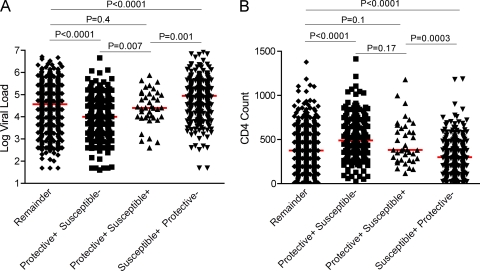
Although data presented here and elsewhere show that individual HLA alleles can have a highly significant effect on HIV immune control, the identification of potentially protective HLA haplotypes suggests that there might also be additive effects between HLA alleles. In order to investigate this further, we next examined the effect of coexpression of both protective and susceptible alleles to ascertain if they had a dominant or an additive effect. Study subjects were divided into the following categories: protective-positive /susceptible-negative group for individuals expressing at least one beneficial allele and no harmful allele, protective-positive/susceptible-positive group for individuals expressing one beneficial and one harmful allele, and susceptible-positive/protective-negative group for individuals expressing at least one harmful and no beneficial alleles. All other individuals were grouped together as the remainder (i.e., individuals expressing none of the protective or susceptible alleles identified). Only alleles that remained significantly associated with a higher or lower VL and/or CD4 count following Bonferroni correction were considered; HLA-B*5703, HLA-B*5801, and the HLA-B*3910-Cw*1203 and HLA-B*8101-Cw*0401 haplotypes were defined as protective alleles, and HLA-B*5802 and HLA-B*18 were defined as susceptible alleles.
This analysis shows that subjects with protective and no susceptible alleles have significantly lower VLs and higher CD4 counts than those individuals possessing both protective and susceptible alleles (10,105 versus 25,600 [P = 0.007] for VL and 491 versus 382 [P = 0.17] for CD4 count), who, in turn, have lower VLs and higher CD4 counts than those with only susceptible alleles (25,600 versus 89,700 [P = 0.001] and 382 versus 299 [P = 0.0003]) (Fig. 3). There are no significant differences between individuals possessing both protective and susceptible alleles and the remainder of individuals that express neither (25,600 versus 36,750 [P = 0.4] and 382 versus 376 [P = 0.1]). These data imply that even individual HLA alleles exerting a strong influence on disease progression, such as HLA-B*5703 and HLA-B*5802, do not dominate the immune response (dictating either a successful or unsuccessful immune response, respectively) but, rather, suggest that the contribution of the individual HLA alleles is additive.
FIG. 3.
The effect of coexpression of protective and harmful HLA alleles on steady-state viral load (A) and absolute CD4 count (B). Protective/susceptible alleles are taken as those that remain significantly associated with viral load and/or CD4 count following stringent Bonferroni corrections for multiple comparisons (protective, HLA-B*57, -B*5801 and the -B*8101-Cw*0401 and -B*3910-Cw*1203 haplotypes; susceptible, HLA-B*18 and -B*5802). Remainder includes all the individuals in the cohort expressing none of these alleles. All P values were generated by a Mann-Whitney U test. Median VL/CD4 counts for each group are indicated by the red line.
Weak frequency-dependent effects of HLA are additive.
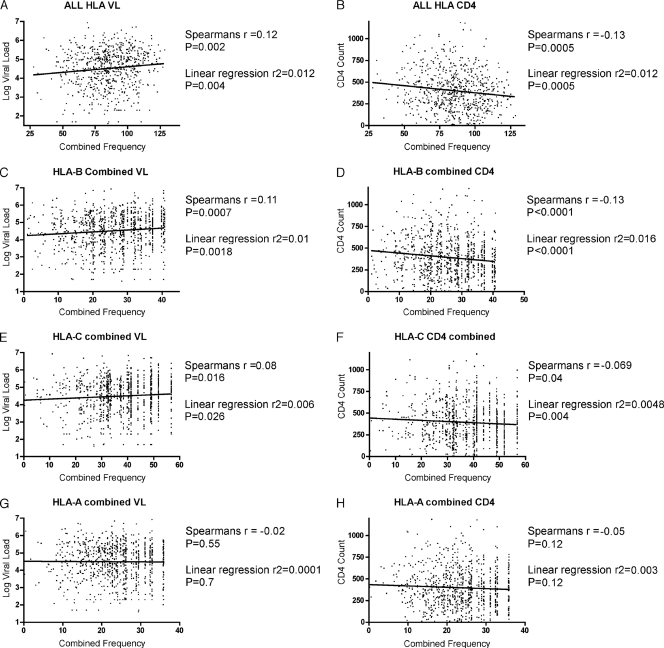
Next, we analyzed our data set for evidence of rare allele advantage, which has been described previously in relation to HIV (28). No significant correlation is observed between individual HLA allele frequency and median VL or CD4 count (r = −0.005 and P = 0.5 by Spearman's rank correlation; r2 = 0.00003 and P = 0.3 by linear regression) (data not shown). However, these analyses are complicated by the fact that at the individual level the beneficial effects of possessing one rare allele may be confounded by the presence of common alleles at any of the other five HLA class I loci, especially in light of the data presented here arguing for the additive affect of HLA type. To address this issue, we assigned each subject in our data set an overall HLA allele frequency, equivalent to the sum of the frequencies of all their individual HLA class I alleles. These overall frequency values give a weak, though significant, correlation with both VL and CD4 count (r = 0.12 and P = 0.002 by Spearman's rank; r2 = 0.012 and P = 0.004 by linear regression; r = −0.13 [P = 0.0005] and r2 = 0.012 [P = 0.0005] for VL and CD4 count, respectively) (Fig. 4 A and B). Although this analysis seeks to address the impact of overall allele frequency at the individual level, it is complicated by the fact that, as discussed, HLA alleles are often in very strong linkage disequilibrium. Consequently, the chance of encountering any given set of alleles (and, thus, from the point of view of the pathogen, adapting to them) is related to both the individual allele frequencies and the strength of linkage between them. In order to compensate for this, we examined the effect of combining each individual's HLA-A, -B, and -C alleles separately, as these allele pairs cannot be in linkage. In doing this we see weak, but significant, correlations between combined HLA-B and HLA-C allele frequency and VL and CD4 count, supporting the existence of an additive affect between these allele pairs. Perhaps unsurprisingly, the strongest and most significant effects are seen in correlation with the HLA-B frequency only (Fig. 4C to H), which supports the hypothesis that HLA-B has the strongest impact on viral control. However, it is again important to note that the trend is a weak one as the r and r2 values are very low.
FIG. 4.
Correlation between steady-state viral load and absolute CD4 count and HLA frequency. Panels A and B show the correlation of viral load (A) and CD4 count (B) with the total combined HLA frequency of each individual in the cohort, that is, the sum of the frequencies of each of the individual's HLA class I alleles. The Spearman's rank correlation coefficient, r, and the linear regression r2 values are given with their corresponding P values. Panels C to H show the correlation of the combined frequencies of the HLA-B (C and D), HLA-C (E and F), and HLA-A (G and H) alleles with viral load and CD4 count.
No evidence for HLA class I heterozygote advantage.
A linked hypothesis to frequency-dependent selection is that of heterozygote advantage, based on the principal that individuals heterozygous at HLA loci are able to present a greater variety of antigenic peptides than are homozygotes, resulting in a more effective immune response to a wide diversity of pathogens (8). The phenomena are linked as the more common an allele, the more frequent homozygotes will be. However, no significant differences in either VL or CD4 count were observed between the homozygote and heterozygote groups in our data set (Fig. 5 A and B) (P of 0.24 and 0.32, respectively). Indeed, although not significant, the median VL of the homozygotes is somewhat lower than that of the heterozygote group (31,292 compared to 41,200). Consistent with the hypothesized preeminence of HLA-B-restricted cytotoxic T lymphocytes (CTL), an earlier study found that homozygosity at the HLA-B locus was more detrimental than at either HLA-A or -C (5). However, in this cohort no significant difference was observed between individuals homozygous for HLA-A, -B, or -C (Fig. 5A and B). The effects of cumulative homozygosity were also examined by grouping individuals homozygous at one, two, or all three class I loci (denominated single, double, and triple homozygotes). No differences were observed between heterozygotes and individuals homozygous at one or two loci (median VLs and CD4 counts of 31,292 and 373, respectively, compared to 31,292 and 384, and 25,800 and 400) (Fig. 5C and D). Individuals homozygous for all three do tend to have higher median VLs (54,650) and lower CD4 counts (276); however, the difference is not significant, possibly due to sample size as only 11 triple homozygotes were identified. Finally, we repeated all of the above comparisons using an LRT to take into account the possibility that any homozygous effect was being masked by the presence of individual strong alleles. However, no significant differences were observed by this analysis, either with or without using HLA-B*57, HLA-B*5801, HLA-B*5802, and HLA-B*18 as covariates.
FIG. 5.
Effect of homozygosity on steady-state viral load (A and C) and absolute CD4 count (B and D). Panels A and B compare all heterozygotes to individuals homozygous for at least one HLA allele. Homozygotes are further grouped into those individuals homozygous for the HLA-A, -B, or -C locus. In panels C and D, homozygotes are grouped according to whether they are homozygous at only one, two, or all three HLA class I alleles. No significant differences are observed between any groups. Median VL/CD4 counts for each group are indicated by the red line.
DISCUSSION
The data presented here confirm and extend our earlier findings (15) that in this cohort of HIV C-clade-infected individuals from South Africa, of the three HLA class I loci, HLA-B plays the dominant role in controlling viral replication, as indicated by steady-state VL and absolute CD4 count. The majority of individual HLA alleles associated with significant differences in VL and/or CD4 count are HLA-B, and, examined as a whole, variation in these clinical markers of disease progression are best explained grouping by HLA-B. Furthermore, in this extended data set we are able to demonstrate that the effect is not due to a few protective and susceptible alleles that happen to be HLA-B since when the four most protective/susceptible alleles [HLA-B*57, HLA-B*5801 (protective), HLA-B*5802, and HLA-B*18 (susceptible)] are either excluded from the data set or used as covariates, HLA-B type remains the strongest predictor of VL and CD4 count. In addition, HLA-B alleles contribute most to the weak frequency- dependent effects we observe in our data set. Taken together with other recent publications (3, 14, 16), these data strengthen the hypothesis that HLA-B-restricted CD8+ T-cell responses play a dominant role in the immune control of HIV.
However, we also find several lines of evidence suggesting that, in this cohort at least, the influence of HLA on HIV extends beyond a few dominant HLA-B alleles and is likely to result from the additive effect of some or all of the HLA alleles an individual possesses. First, HLA-C type remains a significant contributor to variation in VL and CD4 count after the HLA-B57/58/18 alleles have been accounted for, either through exclusion or by using these alleles as covariates. Second, we identified three protective HLA haplotypes, B*3910-Cw*1203, B*8101-Cw*0401, and A*7401-B*5703, whereby protection is either dependent on or enhanced by coexpression of the HLA-B and non-B alleles. Similar effects of HLA haplotype in HIV infection have been observed previously (17, 26), and their existence is consistent with the hypothesis that HLA type has an additive effect on immune control. Third, we find that in individuals that coexpress both protective and harmful HLA alleles, neither appear to dominate and dictate the success of the immune response to HIV. And, finally, significant, albeit weak, frequency-dependent effects in this cohort can be detected only by considering an individual's combined HLA frequency rather than the frequency of each of the subject's alleles separately.
One caveat of this study is that cross-sectional analysis is much less sensitive than a longitudinal study of time-to-AIDS as not only do VL and CD4 measurements fluctuate, but also subjects with low VLs and high CD4 counts are known progress to AIDS somewhat unpredictably (5, 9). In addition, if, as hypothesized, HLA alleles have an additive effect, unless an HLA exerts a very strong effect, it is likely to be masked by the host's other class I alleles. Furthermore, these data are drawn from a chronically infected cohort, and it is possible that some HLA-driven effects maybe more readily detectable during the acute phase of disease. For these reasons it is unsurprising that even in this large cohort, only a fraction of the HLA alleles expressed have an effect on HIV control that can be detected. Finally, there are other potentially confounding host genetic factors, including KIR (killer-cell immunoglobulin-like receptor) type (18) and coreceptor polymorphisms (25). The implication of this is twofold. First, the fact that it is possible to identify highly significant associations with certain HLA alleles and haplotypes, despite all the confounding factors, would indicate that these have a strong influence on viral control. Second, due to the insensitivity of this approach, the fact that we detect no effect of homozygosity on immune control of HIV should not necessarily be taken as evidence against this hypothesis. The original study identifying an advantage for heterozygous alleles observed associations between heterozygosity at the HLA class I loci and slowed progression to AIDS and AIDS-related death (5). This type of longitudinal analysis is more sensitive than the cross-sectional study of surrogate markers of disease progression conducted here (9). However, these data do suggest that, in this chronically infected study cohort at least, the heterozygote advantage is less significant overall than the effect of individual beneficial or harmful alleles and/or HLA haplotypes.
In conclusion, the data here further confirm that in this cohort of C-clade-infected South Africans, HLA-B type has the strongest influence on control of HIV and that this effect extends beyond the few most protective and harmful alleles previously identified. However, although single HLA alleles do have a strong effect on disease control, we also find evidence that an individual's disease status is likely the result of the subject's combined HLA makeup rather than a dominant effect of a single protective or harmful HLA allele.
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health (2RO1AI46995), the South African AIDS Vaccine Initiative, and the Wellcome Trust (to A.L. and P.J.R.G.). P.J.R.G. is an Elizabeth Glaser Pediatric AIDS Foundation Scientist. T.N. holds the South African Research Chair in Systems Biology of HIV/AIDS.
Footnotes
The authors have paid a fee to allow immediate free access to this article.
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 21 July 2010.
REFERENCES
- 1.Altfeld, M., M. M. Addo, E. S. Rosenberg, F. M. Hecht, P. K. Lee, M. Vogel, X. G. Yu, R. Draenert, M. N. Johnston, D. Strick, T. M. Allen, M. E. Feeney, J. O. Kahn, R. P. Sekaly, J. A. Levy, J. K. Rockstroh, P. J. Goulder, and B. D. Walker. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581-2591. [DOI] [PubMed] [Google Scholar]
- 2.Badri, M., L. G. Bekker, C. Orrell, J. Pitt, F. Cilliers, and R. Wood. 2004. Initiating highly active antiretroviral therapy in sub-Saharan Africa: an assessment of the revised World Health Organization scaling-up guidelines. AIDS 18:1159-1168. [DOI] [PubMed] [Google Scholar]
- 3.Bihl, F., N. Frahm, L. Di Giammarino, J. Sidney, M. John, K. Yusim, T. Woodberry, K. Sango, H. S. Hewitt, L. Henry, C. H. Linde, J. V. Chisholm, 3rd, T. M. Zaman, E. Pae, S. Mallal, B. D. Walker, A. Sette, B. T. Korber, D. Heckerman, and C. Brander. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 176:4094-4101. [DOI] [PubMed] [Google Scholar]
- 4.Bunce, M. 2003. PCR-sequence-specific primer typing of HLA class I and class II alleles. Methods Mol. Biol. 210:143-171. [DOI] [PubMed] [Google Scholar]
- 5.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 6.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535-551. [DOI] [PubMed] [Google Scholar]
- 7.Costello, C., J. Tang, C. Rivers, E. Karita, J. Meizen-Derr, S. Allen, and R. A. Kaslow. 1999. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS 13:1990-1991. [DOI] [PubMed] [Google Scholar]
- 8.Dean, M., M. Carrington, and S. J. O'Brien. 2002. Balanced polymorphism selected by genetic versus infectious human disease. Annu. Rev. Genomics Hum. Genet. 3:263-292. [DOI] [PubMed] [Google Scholar]
- 9.Dolan, M. J., H. Kulkarni, J. F. Camargo, W. He, A. Smith, J. M. Anaya, T. Miura, F. M. Hecht, M. Mamtani, F. Pereyra, V. Marconi, A. Mangano, L. Sen, R. Bologna, R. A. Clark, S. A. Anderson, J. Delmar, R. J. O'Connell, A. Lloyd, J. Martin, S. S. Ahuja, B. K. Agan, B. D. Walker, S. G. Deeks, and S. K. Ahuja. 2007. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat. Immunol. 8:1324-1336. [DOI] [PubMed] [Google Scholar]
- 10.Emu, B., E. Sinclair, H. Hatano, A. Ferre, B. Shacklett, J. N. Martin, J. M. McCune, and S. G. Deeks. 2008. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J. Virol. 82:5398-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellay, J., K. V. Shianna, D. Ge, S. Colombo, B. Ledergerber, M. Weale, K. Zhang, C. Gumbs, A. Castagna, A. Cossarizza, A. Cozzi-Lepri, A. De Luca, P. Easterbrook, P. Francioli, S. Mallal, J. Martinez-Picado, J. M. Miro, N. Obel, J. P. Smith, J. Wyniger, P. Descombes, S. E. Antonarakis, N. L. Letvin, A. J. McMichael, B. F. Haynes, A. Telenti, and D. B. Goldstein. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 13.Goulder, P. J., and D. I. Watkins. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harari, A., C. Cellerai, F. B. Enders, J. Kostler, L. Codarri, G. Tapia, O. Boyman, E. Castro, S. Gaudieri, I. James, M. John, R. Wagner, S. Mallal, and G. Pantaleo. 2007. Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype. Proc. Natl. Acad. Sci. U. S. A. 104:16233-16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 16.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8(+) T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46-53. [DOI] [PubMed] [Google Scholar]
- 17.Lazaryan, A., E. Lobashevsky, J. Mulenga, E. Karita, S. Allen, J. Tang, and R. A. Kaslow. 2006. Human leukocyte antigen B58 supertype and human immunodeficiency virus type 1 infection in native Africans. J. Virol. 80:6056-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, M. P., X. Gao, J. H. Lee, G. W. Nelson, R. Detels, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, J. Trowsdale, M. Wilson, S. J. O'Brien, and M. Carrington. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429-434. [DOI] [PubMed] [Google Scholar]
- 19.Matthews, P. C., A. Prendergast, A. Leslie, H. Crawford, R. Payne, C. Rousseau, M. Rolland, I. Honeyborne, J. Carlson, C. Kadie, C. Brander, K. Bishop, N. Mlotshwa, J. D. Mullins, H. Coovadia, T. Ndung'u, B. D. Walker, D. Heckerman, and P. J. Goulder. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 82:8548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 21.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngumbela, K. C., C. L. Day, Z. Mncube, K. Nair, D. Ramduth, C. Thobakgale, E. Moodley, S. Reddy, C. de Pierres, N. Mkhwanazi, K. Bishop, M. van der Stok, N. Ismail, I. Honeyborne, H. Crawford, D. G. Kavanagh, C. Rousseau, D. Nickle, J. Mullins, D. Heckerman, B. Korber, H. Coovadia, P. Kiepiela, P. J. Goulder, and B. D. Walker. 2008. Targeting of a CD8 T cell env epitope presented by HLA-B*5802 is associated with markers of HIV disease progression and lack of selection pressure. AIDS Res. Hum. Retroviruses 24:72-82. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien, S. J., X. Gao, and M. Carrington. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7:379-381. [DOI] [PubMed] [Google Scholar]
- 24.Rousseau, C. M., M. G. Daniels, J. M. Carlson, C. Kadie, H. Crawford, A. Prendergast, P. Matthews, R. Payne, M. Rolland, D. N. Raugi, B. S. Maust, G. H. Learn, D. C. Nickle, H. Coovadia, T. Ndung'u, N. Frahm, C. Brander, B. D. Walker, P. J. Goulder, T. Bhattacharya, D. E. Heckerman, B. T. Korber, and J. I. Mullins. 2008. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype C proteome: immune escape and viral load. J. Virol. 82:6434-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, M. W., M. Dean, M. Carrington, C. Winkler, G. A. Huttley, D. A. Lomb, J. J. Goedert, T. R. O'Brien, L. P. Jacobson, R. Kaslow, S. Buchbinder, E. Vittinghoff, D. Vlahov, K. Hoots, M. W. Hilgartner, and S. J. O'Brien. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science 277:959-965. [DOI] [PubMed] [Google Scholar]
- 26.Tang, J., S. Tang, E. Lobashevsky, A. D. Myracle, U. Fideli, G. Aldrovandi, S. Allen, R. Musonda, and R. A. Kaslow. 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas, R., R. Apps, Y. Qi, X. Gao, V. Male, C. O'HUigin, G. O'Connor, D. Ge, J. Fellay, J. N. Martin, J. Margolick, J. J. Goedert, S. Buchbinder, G. D. Kirk, M. P. Martin, A. Telenti, S. G. Deeks, B. D. Walker, D. Goldstein, D. W. McVicar, A. Moffett, and M. Carrington. 2009. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 41:1290-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber, E. Hayes, R. Funkhouser, M. Fugate, J. Theiler, Y. S. Hsu, K. Kunstman, S. Wu, J. Phair, H. Erlich, and S. Wolinsky. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928-935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.