Abstract
Proteolysis of influenza virus hemagglutinin by host cell proteases is essential for viral infectivity, but the proteases responsible are not well defined. Recently, we showed that engineered expression of the type II transmembrane serine proteases (TTSPs) TMPRSS2 and TMPRSS4 allows hemagglutinin (HA) cleavage. Here we analyzed whether TMPRSS2 and TMPRSS4 are expressed in influenza virus target cells and support viral spread in the absence of exogenously added protease (trypsin). We found that transient expression of TMPRSS2 and TMPRSS4 resulted in HA cleavage and trypsin-independent viral spread. Endogenous expression of TMPRSS2 and TMPRSS4 in cell lines correlated with the ability to support the spread of influenza virus in the absence of trypsin, indicating that these proteases might activate influenza virus in naturally permissive cells. Indeed, RNA interference (RNAi)-mediated knockdown of both TMPRSS2 and TMPRSS4 in Caco-2 cells, which released fully infectious virus without trypsin treatment, markedly reduced the spread of influenza virus, demonstrating that these proteases were responsible for efficient proteolytic activation of HA in this cell line. Finally, TMPRSS2 was found to be coexpressed with the major receptor determinant of human influenza viruses, 2,6-linked sialic acids, in human alveolar epithelium, indicating that viral target cells in the human respiratory tract express TMPRSS2. Collectively, our results point toward an important role for TMPRSS2 and possibly TMPRSS4 in influenza virus replication and highlight the former protease as a potential therapeutic target.
Infection with influenza viruses—negative-stranded, segmented RNA viruses of the Orthomyxovirus family—is responsible for substantial morbidity and mortality, particularly among the young and the elderly (10). A hallmark of influenza A viruses is their ability to adapt to immune pressure, which allows constant circulation of these viruses in the human population (seasonal influenza) (8, 31). In addition, antigenically novel viruses, arising from the large pool of animal influenza viruses, are occasionally transmitted to humans and may spread in a pandemic manner (pandemic influenza) (8, 31). The high mutation rate of influenza viruses has major consequences for antiviral prevention and therapy. First, the vaccine against seasonal influenza needs to be reformulated almost annually and will not be effective against a new pandemic virus. Second, antiviral therapy, which targets viral proteins essential for uncoating and release, is plagued by the rapid development of viral resistance (32). Consequently, new targets for antiviral intervention are under investigation, and therapeutic inhibition of invariant host cell factors essential for influenza virus spread is an attractive strategy, since it may allow the suppression of resistance development.
The viral envelope protein hemagglutinin (HA) mediates the first essential steps in the viral life cycle: attachment to target cells and virus-cell fusion (14, 38). Attachment of virions to target cells is mediated by the surface unit of HA, HA1, while the fusion of the viral envelope with a target cell membrane is driven by the transmembrane unit, HA2 (14, 38). In infected cells, HA is initially synthesized as an inactive precursor protein, HA0, in which HA1 und HA2 are connected by a protease-sensitive linker sequence. Cleavage of the linker by host cell proteases generates mature HA1 and HA2 and is crucial for viral infectivity (23, 24, 26, 27), making the responsible proteases attractive targets for therapeutic intervention (2, 11). However, their exact identity has not been determined previously.
Highly pathogenic avian influenza viruses (HPAIV) harbor a cleavage site composed of several arginines and lysines (multibasic cleavage site), which is recognized by furin or related subtilisin proteases (40). Since these proteases are ubiquitously expressed, HPAIV can spread systemically and cause severe disease (1, 17, 21, 22, 28). In contrast, the cleavage site of low-pathogenic avian influenza viruses (LPAIV) consists of a single arginine or lysine residue (monobasic cleavage site) and is believed to be exclusively recognized by as yet uncharacterized proteases expressed in the respiratory and enteric tracts (1, 17, 21, 22, 28, 39). As with LPAIV, human influenza viruses contain a monobasic cleavage site, and the identity of the protease(s) activating these viruses is unclear.
It has been suggested that soluble proteases might activate human influenza viruses in the infected host (18, 19, 30, 43). However, analysis of cultured human respiratory epithelial cells revealed efficient HA cleavage during HA biogenesis and upon uptake of virions into target cells, suggesting that HA cleavage in the human host is a cell-associated process (47). Böttcher and colleagues amplified the type II transmembrane serine proteases (TTSPs) TMPRSS2 (transmembrane protease, serine 2) and HAT (human airway trypsin-like protease) from the human lung and showed that these proteases, upon engineered expression, support the spread of human influenza viruses (2-4). Employing lentiviral vectors to analyze HA activation, we have demonstrated previously that TMPRSS2 and another TTSP, TMPRSS4, activate the HA of the highly pathogenic 1918 influenza virus (5). These results suggest that TTSPs might contribute to the activation of influenza virus in the human host. However, it remained to be determined whether HA-activating TTSPs are endogenously expressed in viral target cells and whether they support viral spread in the absence of an exogenously added protease (trypsin).
We addressed the relevance of TMPRSS2 and TMPRSS4 for the activation of influenza virus in permissive cells. A combination of expression and knockdown analyses revealed that endogenous TMPRSS2 and TMPRSS4 activate influenza virus in cell lines and, in the case of TMPRSS2, most likely also in human alveolar epithelium, suggesting a major role for TMPRSS2 in viral replication in human hosts.
MATERIALS AND METHODS
Plasmids.
Expression plasmids for the 1918 influenza virus HA, the 1918 influenza virus neuraminidase (NA), TMPRSS2, TMPRSS3, and TMPRSS4 have been described previously (5, 15, 34, 36). cDNAs encoding full-length human hepsin (hMU004062), TMPRSS3 (KU004606), and TMPRSS6 (hMU004435) were provided by the Korean UniGene Information Center (http://kugi.kribb.re.kr/), Daejeon, South Korea. The cDNA encoding each gene was amplified by PCR using primer pairs 5′-GAGGCTAGCCACCATGGCGCAGAAGGAGGGTGG-3′ (forward) and 5′-GAGGCGGCCGCTCAGAGCTGGGTCACCATGC-3′ (reverse) for hepsin, 5′-GAGGCTAGCCACCATGGGGGAAAATGATCCGCCTGCT-3′ (forward) and 5′-GAGGCGGCCGCTCAGGTTTTTAGGTCTCTCT-3′ (reverse) for TMPRSS3, and 5′-GAGGCTAGCCACCATGTTGTTACTCTTCCACTCCAA-3′ (forward) and 5′-GAGGCGGCCGCTCAGGTCACCACTTGCTGGA-3′ (reverse) for TMPRSS6. The PCR products were then cloned into plasmid pcDNA3.1 (Invitrogen) by NheI and NotI digestion, and the integrity of the cloned cDNAs was verified by sequencing.
Cell culture.
The following media were used for cell culture: Dulbecco's modified Eagle's medium (DMEM; Invitrogen) (for culture of Vero E6, Huh-7, and 293T cells), Glutamax DMEM (DMEM enriched with glutamate; Invitrogen) (for culture of Caco-2 cells), and minimum essential medium (MEM; Invitrogen) (for culture of MDCK II cells). All media were supplemented with 10% fetal calf serum (FCS), penicillin, and streptomycin, and all cell cultures were maintained at 37°C under a 5% CO2 atmosphere.
VLPs.
For the production of virus-like particles (VLPs), 293T cells were transiently cotransfected with the HIV-1 Gag (p55)-encoding plasmid p96ZM651gag-opt, 1918 HA and NA expression plasmids, and expression plasmids for TTSPs or empty vector. At 48 h posttransfection, the supernatants were harvested and concentrated by ultrafiltration employing Vivaspin columns (Sartorius). As a control for HA cleavage, VLPs were treated either with phosphate-buffered saline (PBS) or with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Sigma), followed by incubation with soybean trypsin inhibitor (Sigma).
Generation of replication-competent influenza virus.
Ten-day-old pathogen-free embryonated hens' eggs were inoculated via the allantoic sac with 0.2 ml of virus dilutions. Eggs were incubated for 48 h in an incubator at 37°C with passive humidity. Thereafter, the allantoic fluids were collected, tested for hemagglutinating activity, pooled, aliquoted, and stored at −70°C. The titer of the virus was determined by the focus formation assay.
Infection experiments with replication-competent influenza viruses.
For infection experiments with replication-competent viruses, 293T cells, transiently transfected with plasmids encoding proteases or empty vector, were seeded in 6-well plates at a density of 1.2 ×105 cells/well, washed, and inoculated with PR8 (H1N1) or SC35M (H7N7) virus (contained in Dulbecco's PBS [DPBS] supplemented with 0.2% bovine serum albumin [BSA]) at a multiplicity of infection (MOI) of 0.01. Alternatively, Caco-2 cells were either mock treated; transfected with a nonsense small interfering RNA (siRNA), a TMPRSS2 siRNA, or a TMPRSS4 siRNA (Santa Cruz); or cotransfected with both TMPRSS2 and TMPRSS4 siRNAs. Then the cells were washed and infected as described above for 293T cells. Viruses were allowed to adsorb to target cells for 1 h. Subsequently, the infection medium was removed; MEM supplemented with 0.2% BSA and either 1 μg/ml TPCK-trypsin or PBS was added; and the cells were incubated for 24 h. Thereafter, the cell culture supernatants were collected and stored at −80°C for subsequent quantification of infectious virus particles.
Generation of HA-bearing pseudotypes.
Pseudotypes were generated essentially as described previously (5, 37). Briefly, 293T cells were cotransfected, by use of CaPO4, with the HIV-1-derived vector pNL4-3 E-R-Luc (7) and expression plasmids for 1918 HA, 1918 NA, or vesicular stomatitis virus glycoprotein (VSV-G). For analysis of HA activation by TTSPs, TMPRSS2, TMPRSS3, TMPRSS4, TMPRSS6, or hepsin was coexpressed with viral components during the production of pseudoparticles. At 16 h posttransfection, the culture medium was replaced by fresh medium, and the cultures were maintained at 37°C under 5% CO2. At 48 h posttransfection, the supernatants were harvested, passed through 0.45-μm-pore-size filters, and stored at −80°C. The concentration of HIV-1 capsid protein (p24) in virus stocks was determined by a p24 enzyme-linked immunosorbent assay (ELISA) (AIDS Research and Reference Reagent Program). For the production of pseudotypes in Caco-2 cells, cells were cotransfected with pNL4-3 E-R-Luc and an expression plasmid for 1918 HA, 1918 NA, or VSV-G by use of Lipofectamine 2000 (Invitrogen) and were processed as described above for 293T cells.
Infection experiments with HA-bearing pseudotypes.
For infection experiments with pseudotyped viruses, Huh-7, Caco-2, and 293T cells were seeded in 96-well plates at a density of 0.8 × 105 cells/well, washed, and incubated overnight with p24- or luciferase-normalized virus stocks pretreated with either PBS or TPCK-trypsin. After 72 h, the cells were lysed, and the luciferase activities in cell lysates were determined by employing a commercially available kit (Promega). Alternatively, 293T target cells were first transfected with protease-encoding plasmids and then infected as described above.
Detection of HA in cell lysates and virions by Western blotting.
For the detection of HA in cell lysates, 293T cells were cotransfected with 1918 HA and TTSP expression plasmids or empty vector. The medium was replaced with fresh culture medium at 16 h posttransfection. At 48 h posttransfection, the cells were harvested, treated with PBS or TPCK-trypsin, and lysed in 2× sodium dodecyl sulfate (SDS) loading buffer. For analysis of HA cleavage by endogenous proteases, Caco-2 cells were Lipofectamine transfected with 1918 HA and lysed at 48 h posttransfection in SDS loading buffer. For analysis of the incorporation of HA into VLPs and pseudotypes, the particle preparations were pelleted by centrifugation through a 20% sucrose cushion at 42,000 × g for 2 h at 4°C, and the pellets were lysed in SDS loading buffer. For immunoblotting, the lysates were separated by SDS-gel electrophoresis, and HA was detected by staining with a mouse anti-HA antibody (12) at a dilution of 1:500, followed by incubation with a horseradish peroxidase (HRP)-coupled anti-mouse secondary antibody. As a loading control, an anti-β-actin antibody (Sigma) or an anti-p24 hybridoma supernatant (183-H12-5C) was used.
Analysis of HA glycosylation.
For enzymatic deglycosylation of 1918 HA/NA-bearing VLPs produced in the presence of TMPRSS2 and hepsin, a commercially available kit (New England Biolabs) was used. For this purpose, the VLPs were concentrated via Vivaspin columns and were additionally ultracentrifuged through a 20% sucrose cushion at 42,000 × g for 2 h at 4°C. The resulting pellets were harvested with TNE buffer (Tris-HCl [pH 7.4], 0.15 M NaCl, and 10 mM EDTA) and digested by peptide:N-glycosidase F (PNGase F). The samples were analyzed by immunoblotting as described above.
Focus formation assay.
MDCK II cells (6 × 104/well) were seeded in 96-well culture plates and were incubated at 37°C under 5% CO2 for 24 h. On the next day, serial 10-fold dilutions of supernatants (collected from the infected cells) in DMEM containing 0.1% BSA and 2.5 μg/ml N-acetylated trypsin (NAT; Sigma) were prepared in a separate 96-well plate, and then 50 μl of each dilution was transferred to the confluent monolayers of MDCK II cells in 96-well culture plates. The plates were incubated at 37°C under 5% CO2 for 1 h with shaking at 20-min intervals. The inoculates were aspirated and replaced with 100 μl of a 1% Avicel overlay containing 0.1% BSA and 2.5 μg/ml NAT (Sigma). The infected cells were incubated at 37°C under 5% CO2 for 24 h. Subsequently, cells were washed twice with PBS and were then fixed with 4% formalin in PBS (100 μl/well) for 10 min at room temperature. The formalin was removed; the cells were first washed as described above and then incubated for 10 min with 100 μl/well Quencher (0.5% Triton X-100, 20 mM glycine in PBS). After 10 min, the cells were washed with wash buffer (WB) (0.5% Tween 20 in PBS) and were then blocked with 50 μl of blocking buffer (BB) (0.5% Tween, 20% BSA in PBS) at 37°C under 5% CO2 for 30 min. The primary antibody (a polyclonal goat antibody against the influenza virus nucleocapsid [NP]; Virostat) and the secondary antibody (an HRP-conjugated anti-goat antibody; Kirkegaard & Perry Laboratories, Gaithersburg, MD) were diluted 1:1,000 in BB. A 50-μl volume of the primary antibody was added to each well and was incubated at room temperature for 1 h. After 1 h of incubation, the cells were washed three times with WB, incubated with 50 μl of the secondary antibody for 45 min, washed again, and incubated with 50 μl of the substrate (True Blue; Kirkegaard & Perry Laboratories, Gaithersburg, MD) until blue spots appeared. Foci were counted, and viral titers were calculated as focus-forming units (FFU) per milliliter.
Immunohistochemistry.
Tissue samples, obtained with full ethical approval from the National Research and Ethics Service (Oxfordshire Research and Ethics Committee A; reference 04/Q1604/21), were stained with hematoxylin and eosin using standard techniques and were either immunostained for TMPRSS2 (mouse monoclonal antibody P5H9 [29]) or stained with Sambucus nigra lectin (Vector Laboratories, Peterborough, United Kingdom). Immunostaining was performed using a Bond Max immunostaining machine (Leica Microsystems Newcastle Ltd., Newcastle, United Kingdom) with the manufacturer's standard protocols and reagents for mouse primary antibodies. The biotinylated Sambucus nigra lectin was detected directly by means of the Bond Intense R kit (Leica Microsystems Newcastle Ltd., Newcastle, United Kingdom). Negative controls were as follows: standard immunostaining with a mouse monoclonal antibody against Melan-A (clone A103; Leica Microsystems Newcastle Ltd., Newcastle, United Kingdom) for the anti-TMPRSS2 antibody and the Bond Intense R kit alone for the biotinylated Sambucus nigra lectin. Stained sections were photographed with a Nikon DS-FI1 camera with a Nikon DS-L2 control unit (Nikon UK Limited, Kingston-upon-Thames, United Kingdom) and an Olympus BX40 microscope (Olympus UK Limited, Watford, United Kingdom).
Reverse transcription-PCR (RT-PCR) analysis of TTSP mRNA expression in cell lines.
mRNAs were extracted from different cell lines (293T, Caco-2, Huh-7, Vero E6), treated with DNase, and reverse transcribed employing commercially available kits (Qiagen and Invitrogen, Germany). Subsequently, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), TMPRSS2, TMPRSS4, and hepsin sequences were amplified by nested PCR. The following set of primers was used for amplification of TTSPs: TMPRSS2 outer primers 5′-TACCTGCATCAACCCCTCTAACTG-3′ (p5 TMPRSS2 out) and 5′-CTTCTGAGGTCTTCCCTTTCTCCT-3′ (p3 TMPRSS2 out), TMPRSS2 inner primers 5′-GCCTTTACGGACCAAACTTCATCC-3′ (p5 TMPRSS2 in) and 5′-CGCAAATGCCGTCCAATGCCATGG-3′ (p3 TMPRSS2 in), TMPRSS4 outer primers 5′-GAGAGCTGGACTGTCCCTTG-3′ (p5 TMPRSS4 out) and 5′-TCGTACTGGATGCTGACCTG-3′ (p3 TMPRSS4 out), TMPRSS4 inner primers 5′-GACGAGGAGCACTGTGTCAA-3′ (p5 TMPRSS4 in) and 5′-CTTCCCACAGGCAAGACAGT-3′ (p3 TMPRSS4 in), hepsin outer primers 5′-CCCTGCTACTTCTGACAGCCATC-3′ (p5 hepsin out) and 5′-TCGTTGCTGTTCTCCTCGCTGTT-3′ (p3 hepsin out), and hepsin inner primers 5′-GCACGTCGGGCTTCTTCTGTGTGG-3′ (p5 hepsin in) and 5′-CCACGGCACCGGCAAACACTCGC-3′ (p3 hepsin in).
Quantitative RT-PCR analysis of TMPRSS2 and TMPRSS4 mRNA expression in cell lines.
Total RNA (500 ng) was reverse transcribed using 50 U of BioScript RNase H Low reverse transcriptase (BIO-27036; Bioline) in 20-μl reaction mixtures. The enzyme was then inactivated for 10 min at 70°C. Aliquots (1 μl) of the cDNA samples generated (25 ng total RNA equivalents) were used for real-time PCR in 10-μl reaction mixtures with the ABI 7500 Fast real-time PCR system (Applied Biosystems). Specific amplification was ensured with TaqMan gene expression assays (catalog no. 4331182; Applied Biosystems) according to the manufacturer's recommendations. The following specific assays were used: Hs00237175_m1 (TMPRSS2), Hs00212669_m1 (TMPRSS4), and Hs99999908_m1 (β-glucuronidase [GUSB]). The average cycle threshold (CT) for each individual assay was calculated from triplicate measurements by means of the instrument's software in “auto Ct” mode (ABI 7500 Fast system software, version 1.3.0). Average CT values calculated for TMPRSS2 and TMPRSS4 were normalized by subtraction from the CT values obtained for GUSB (housekeeping reference). Template-free cDNA reaction mixtures were analyzed in parallel, using both TaqMan assays; no specific signal was detected in any of these experiments.
RESULTS
TMPRSS2, TMPRSS4, and hepsin cleave influenza virus hemagglutinin.
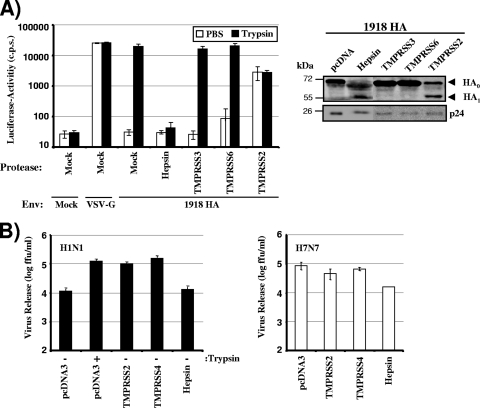
We have previously demonstrated that influenza virus HA is proteolytically processed by TMPRSS2 and TMPRSS4 (5). We now asked whether other TTSPs known to be expressed in lung tissue (41) also cleave HA. For this purpose, we coexpressed HA (from the 1918 influenza virus) with TMPRSS3, TMPRSS6, or hepsin in 293T cells and analyzed HA cleavage by Western blotting (Fig. 1A). Trypsin treatment and coexpression of TMPRSS2 and TMPRSS4 served as positive controls for HA cleavage; cells transfected with empty vector (pcDNA) or nontransfected cells (Mock) were employed as negative controls. As expected, trypsin treatment or coexpression of TMPRSS2 or TMPRSS4 facilitated HA cleavage, while HA was not cleaved when empty vector was cotransfected (Fig. 1A). No evidence for cleavage was observed upon coexpression of TMPRSS3 and TMPRSS6. In contrast, a band corresponding to HA1 appeared upon expression of hepsin, indicating that hepsin cleaves HA between HA1 and HA2.
FIG. 1.
Cleavage of influenza virus hemagglutinin by TMPRSS2, TMPRSS4, or hepsin. (A) Cleavage of hemagglutinin (HA) in 293T cells expressing TMPRSS2, TMPRSS4, or hepsin. The HA of the 1918 influenza virus and the protease indicated were transiently coexpressed in 293T cells. The cells were then treated with trypsin or PBS, and HA cleavage was detected by Western blot analysis of cell lysates. Detection of β-actin served as a loading control. Mock, cells transfected with empty vector alone; pcDNA, cells cotransfected with an HA expression plasmid and empty vector. (B) Incorporation of cleaved HA into virions. HA was expressed as described for panel A; however, cells were additionally cotransfected with plasmids encoding the HIV-1 Gag protein and the 1918 influenza virus NA. VLPs were concentrated from culture supernatants by centrifugation, and HA cleavage and Gag contents were analyzed by Western blotting. Mock, VLPs produced in cells transfected with a Gag expression plasmid and empty vector; pcDNA, VLPs produced in cells cotransfected with plasmids encoding Gag, HA, and NA and empty vector. (C) Expression of TMPRSS2 or hepsin, but not TMPRSS4, alters the glycosylation of HA1. 293T cells, transfected either with expression vectors for the indicated proteases or with empty vector, were used for the production of HA-bearing VLPs as described for panel B, and concentrated particles were subsequently treated with trypsin or PBS. Thereafter, the VLPs were either digested with PNGase F or mock treated, and HA cleavage was analyzed by Western blotting.
We then asked whether cleaved HA was incorporated into viral particles. For this purpose, we produced virus-like particles by using an HIV-1 Gag expression plasmid, and we analyzed the incorporation of HA and Gag into particles by Western blotting (Fig. 1B). Cleaved HA was present in particles produced in the presence of TMPRSS2 or TMPRSS4, in agreement with our previous results (5). Similarly, cleaved HA was found in particles produced in hepsin-expressing cells (Fig. 1B), indicating that cleavage by hepsin is compatible with the incorporation of HA into virions.
Closer inspection of the HA1 bands obtained upon HA processing by TMPRSS2, TMPRSS4, hepsin, or trypsin reproducibly revealed size differences: HA digestion by trypsin or TMPRSS4 yielded HA1 bands of identical sizes, as did HA digestion by TMPRSS2 or hepsin, and the latter bands migrated slightly faster than the former (Fig. 1A and B). We hypothesized that the size differences might be due to altered glycosylation of HA in hepsin- or TMPRSS2-transfected cells. To address this hypothesis, we digested virus-like particles (VLPs) from TMPRSS2-, TMPRSS4-, or hepsin-transfected cells, as well as PBS- or trypsin-treated control VLPs, with PNGase F, which removes all N-linked glycans. Indeed, the differences in HA1 size observed in mock-treated cells were not detected after treatment with PNGase F (Fig. 1C), indicating that engineered expression of TMPRSS2 and hepsin modulates HA glycosylation. In summary, our results add hepsin to the list of TTSPs that cleave influenza virus HA and indicate that the expression of particular TTSPs modulates HA glycosylation by a currently unknown mechanism.
Cleavage by TMPRSS2 and TMPRSS4, but not cleavage by hepsin, activates hemagglutinin.
Since hepsin, like TMPRSS2 and TMPRSS4, was able to cleave HA, we next asked whether cleavage by hepsin allows HA to transit into an activated state. A previous study by others (45) and our own published results (5) demonstrate that trans-complementation (pseudotyping) of env-defective retroviral vectors with influenza virus HA is an adequate system for the analysis of HA activation. We employed this approach to analyze HA activation by hepsin. For this purpose, we produced HA-bearing lentiviral pseudotypes in cells cotransfected with TTSP expression plasmids or empty vector, and we then analyzed whether the pseudotypes were infectious or whether they required trypsin treatment in order to acquire infectivity. In agreement with our previous results (5), viruses produced in the presence of TMPRSS2 or TMPRSS4 were fully infectious in the absence of trypsin treatment, while viruses generated in cells transfected with empty vector (pcDNA) required trypsin treatment in order to transit into an infectious form (Fig. 2A, left). Unexpectedly, coexpression of hepsin did not render viruses infectious (Fig. 2A, left), despite the incorporation of cleaved HA into virions (Fig. 2A, right), and infectivity was not acquired upon exposure to trypsin.
FIG. 2.
TMPRSS2 and TMPRSS4, but not hepsin, activate influenza virus hemagglutinin by cleavage. (A) Cleavage activation of lentiviral pseudotypes bearing HA. (Left) Lentiviral reporter viruses bearing 1918 HA and NA were generated in 293T cells coexpressing the indicated protease, treated with PBS or trypsin, and used for the infection of Huh7 target cells. Luciferase activities in cell lysates were determined at 72 h postinfection. The results of a representative experiment performed in triplicate are shown; error bars indicate standard deviations. Comparable results were obtained in two independent experiments. (Right) Incorporation of HA into lentiviral pseudotypes. Lentiviral pseudotypes generated in 293T cells coexpressing the indicated protease were pelleted through a sucrose cushion, and the incorporation of influenza virus HA and HIV-1 capsid protein (p24) into virions was assessed by Western blotting. (B) Cleavage activation of PR8 (H1N1) and SCM35 (H7N7). The indicated proteases were transiently expressed in 293T cells, and the cells were infected at an MOI of 0.01 with PR8 (H1N1) or SCM35 (H7N7) produced in hens' eggs. Subsequently, the cells were treated with PBS or trypsin as indicated, and at 24 h postinfection, viral spread was quantified as the release of infectious particles into the culture supernatants, as measured by a focus formation assay. The results of a representative experiment performed in triplicate are shown; error bars indicate standard deviations. Comparable results were obtained in a separate experiment.
We then determined whether the activation of HA pseudotypes reflects the activation of replication-competent influenza viruses. To this end, we analyzed whether transient expression of TMPRSS2, TMPRSS4, or hepsin facilitated the spread of PR8 (H1N1), a virus that has a monobasic cleavage site and requires exogenous trypsin for its activation (in most cell lines). 293T cells were chosen for these experiments, because this cell line is readily transfectable, and it is well documented that 293T cells do not express an endogenous HA-activating protease (5, 9). As a control, infection of 293T cells with SC35M (H7N7) (33) was analyzed. This influenza virus harbors a multibasic cleavage site and does not depend on trypsin treatment to acquire infectivity. TMPRSS2 or TMPRSS4 expression or trypsin treatment increased the spread (measured as the release of infectious virus particles into the supernatant) of PR8 (H1N1) about 10-fold compared to cells transfected with empty vector (Fig. 2B, left), indicating that these proteases were able to support the spread of influenza virus in the absence of an exogenous protease. In contrast, expression of hepsin did not augment viral spread, in agreement with the results obtained with pseudotypes (Fig. 2A, left). It should be noted in this context that PR8 (H1N1) was prepared in hens' eggs and thus contained activated HA. Therefore, the virus was able to undergo a single round of replication without proteolytic activation, resulting in the release of a readily quantifiable number (upon viral activation by trypsin) of infectious units in the supernatant of control-transfected 293T cells. Finally, as expected, the spread of SC35M (H7N7) was not modulated by the expression of TTSPs (Fig. 2B, right), with the exception of hepsin expression, which reduced viral spread about 10-fold. Collectively, these results demonstrate that engineered expression of TMPRSS2 or TMPRSS4 facilitates HA activation and allows the trypsin-independent spread of influenza virus.
Expression of TMPRSS2 and TMPRSS4 correlates with the trypsin-independent spread of influenza virus in cell lines.
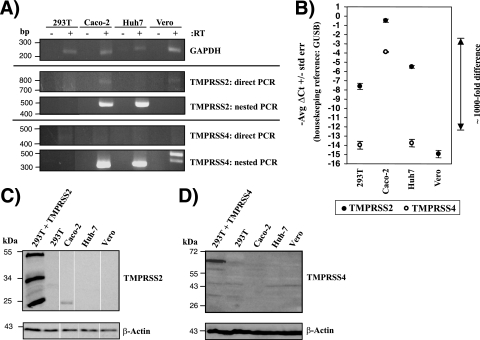
The activation of influenza virus HA by transfected TMPRSS2 and TMPRSS4 raised the question of whether these proteases are endogenously expressed by cell lines that allow trypsin-independent viral spread. To address this question, we investigated TMPRSS2 and TMPRSS4 expression in 293T, Huh-7, and Vero cells, which allow influenza virus to spread only in the presence of exogenous trypsin (5, 9, 16), and in Caco-2 cells, which support trypsin-independent viral spread (48). Direct RT-PCR analyses of TTSP mRNA expression revealed the presence of TMPRSS2 transcripts in Caco-2 cells, and a nested approach detected TMPRSS2 message in Huh-7 cells also, but not in the other cell lines analyzed (Fig. 3A). No TMPRSS4 mRNA could be detected by direct RT-PCR in any of the cell lines examined (Fig. 3A), and similar results were obtained for hepsin (data not shown). Nevertheless, nested PCR demonstrated the presence of TMPRSS4 message in Caco-2 and Huh-7 cells (Fig. 3A). Quantitative RT-PCR confirmed high levels of TMPRSS2 mRNA in Caco-2 cells but not in the other cell lines tested (Fig. 3B), and Western blot analysis demonstrated the expression of TMPRSS2 protein in this cell line (Fig. 3C). Notably, TMPRSS4 mRNA was also readily detectable in Caco-2 cells by quantitative RT-PCR, while the signal for TMPRSS4 mRNA was close to (293T and Huh-7 cells) or within (Vero cells) background levels in the other cell lines examined (Fig. 3B). Western blot analysis revealed TMPRSS4 expression only in transiently transfected 293T cells, not in Caco-2 cells or the other cell lines tested (Fig. 3D), and attempts to detect TMPRSS4 by immunofluorescence and fluorescence-activated cell sorting (FACS) yielded inconclusive results (not shown), precluding any conclusion on the expression of endogenous TMPRSS4 protein. In sum, these results revealed a correlation between TMPRSS2 mRNA and protein expression and the ability of cell lines to allow the spread of influenza virus in the absence of trypsin. A similar correlation was also observed for TMPRSS4 mRNA expression, but the results await confirmation on the protein level.
FIG. 3.
TMPRSS2 and TMPRSS4 mRNAs and TMPRSS2 protein are expressed in Caco-2 cells. (A) Analysis of TMPRSS2 and TMPRSS4 mRNA expression by RT-PCR. Total RNA was prepared from the indicated cell lines, treated with DNase, reverse transcribed, and used for the amplification of TMPRSS2, TMPRSS4, and GAPDH mRNAs by nested PCR. The results of single gels from which irrelevant lanes were removed are shown. (B) Analysis of TMPRSS2 and TMPRSS4 mRNA expression by quantitative RT-PCR. RNA was prepared as described for panel A, and TMPRSS2, TMPRSS4, and GUSB (housekeeping control) were amplified by TaqMan gene expression assays. Similar results were obtained in two independent experiments. No TMPRSS4 signal was detected in Vero E6 cells. (C and D) Expression of TMPRSS2 (C) or TMPRSS4 (D) in cell lines as determined by Western blotting. The indicated cell lines were lysed, and the expression of TMPRSS2 or TMPRSS4 was analyzed by Western blotting. Detection of β-actin served as a loading control. The results of a single gel from which irrelevant lanes were removed are shown in panel C. 293T + TMPRSS2, 293T cells transiently expressing TMPRSS2; 293T + TMPRSS4, 293T cells transiently expressing TMPRSS4.
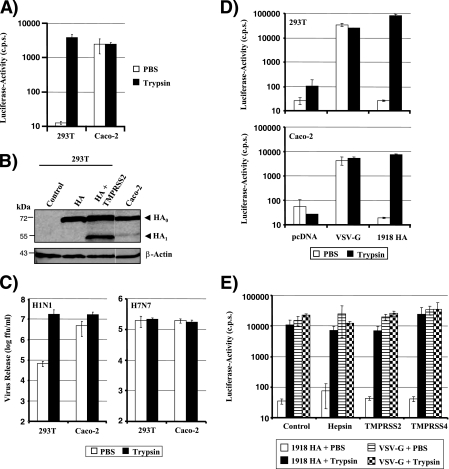
We next sought to confirm that Caco-2 cells indeed allow the trypsin-independent spread of influenza virus, as proposed by Zhirnov and coworkers (48). Examination of the infectivity of HA pseudotypes showed that viruses produced in 293T cells required trypsin treatment to acquire infectivity, while pseudotypes produced in Caco-2 cells were fully infectious in the absence of trypsin treatment (Fig. 4A). In addition, Western blot analysis of transfected cells revealed evidence for HA cleavage in Caco-2 cells but not in 293T cells (Fig. 4B), and replication of PR8 (H1N1) in the former but not the latter cell line was independent of trypsin treatment (Fig. 4C). These observations indicate that Caco-2 cells express a HA-activating activity, potentially TMPRSS2 and/or TMPRSS4. To further characterize HA activation in Caco-2 cells, we asked if the cellular location of HA proteolysis is limited to the secretory pathway or if activation can also occur during viral entry into target cells. Efficient infection of Caco-2 cells by HA-bearing pseudotypes was dependent on previous activation of these viruses by trypsin (Fig. 4D), indicating that the HA-activating enzyme either is not present or does not recognize HA in virion-containing endocytic vesicles in Caco-2 cells. Similar results were obtained upon overexpression of TMPRSS2 and TMPRSS4 in 293T cells (Fig. 4E), suggesting that HA activation in Caco-2 cells and in TMPRSS2-expressing 293T cells most likely occurs in the secretory pathway of productively infected cells.
FIG. 4.
Influenza virus HA is activated in Caco-2 cells in the absence of trypsin. (A) Activation of HA pseudotypes in Caco-2 cells. Caco-2 and 293T cells were transiently cotransfected with plasmids encoding a lentiviral vector and 1918 HA and NA. The supernatants were treated with trypsin or PBS and were then used to infect Huh7 target cells. Luciferase activity in cell lysates was determined at 72 h postinfection. The results ± standard deviations of a representative experiment performed in triplicate are shown and were confirmed in three separate experiments. (B) Analysis of HA cleavage in Caco-2 and 293T cells. 293T and Caco-2 cells were transfected with 1918 HA, or 293T cells were cotransfected with 1918 HA and TMPRSS2 expression plasmids, and HA cleavage in cell lysates was detected by Western blotting. The results of a single gel from which irrelevant lanes were removed are shown. (C) Activation of influenza viruses in Caco-2 cells. Caco-2 and 293T cells were first infected with PR8 (H1N1) or SC35M (H7N7) at an MOI of 0.01 and then treated with PBS or trypsin. The release of infectious particles in the supernatant was determined by a focus formation assay. Results of a representative experiment performed in triplicate are shown, and activation of PR8 (H1N1) in Caco-2 but not 293T cells was confirmed in two separate experiments. (D) Infection of Caco-2 and 293T cells with pseudotypes bearing nonactivated or trypsin-activated HA. Pseudotypes bearing the 1918 HA, the 1918 NA, or the G protein of VSV were first treated either with PBS or with trypsin and then used to infect 293T and Caco-2 cells. Luciferase activities in cell lysates were determined at 72 h postinfection. Similar results were obtained in two independent experiments. (E) Infection of TTSP-expressing 293T cells with pseudotypes bearing nonactivated or trypsin-activated HA. The indicated TTSPs were transiently expressed in 293T cells, and the cells were infected with HA-bearing pseudotypes as described for panel D.
TMPRSS2 and TMPRSS4 are required for the efficient trypsin-independent spread of influenza virus in Caco-2 cells.
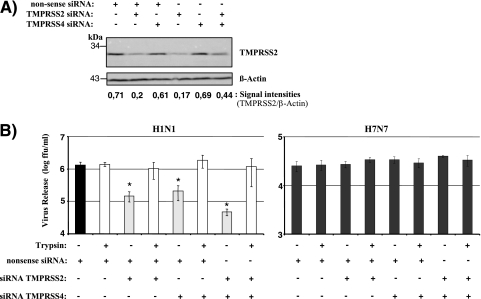
Our results obtained so far suggested that endogenous expression of TMPRSS2 and/or TMPRSS4 might contribute to the trypsin-independent spread of influenza virus in Caco-2 cells. To directly test this hypothesis, we examined the consequences of TMPRSS2 and TMPRSS4 knockdown for the spread of influenza virus in Caco-2 cells. For this purpose, Caco-2 cells either were transiently transfected with a TMPRSS2 or a TMPRSS4 siRNA or were cotransfected with both siRNAs, and expression of TMPRSS2 protein was analyzed by Western blotting (Fig. 5A). Transfection of a nonsense siRNA served as a negative control. Transfection of cells with a TMPRSS2-specific siRNA or with a 1:1 mixture of TMPRSS2- and TMPRSS4-specific siRNAs resulted in a decrease in TMPRSS2 expression, which was not observed with the nonsense siRNA or the TMPRSS4-specific siRNA alone (Fig. 5A), indicating that our protocol allowed efficient and specific reduction of TMPRSS2 expression. The lack of detectable TMPRSS4 expression in Caco-2 cells (Fig. 3) precluded evaluation of the efficiency of siRNA-mediated TMPRSS4 knockdown. However, analysis of 293T cells transiently cotransfected with a TMPRSS4 expression plasmid and siRNAs demonstrated that the TMPRSS4-specific siRNA was active (data not shown). When PR8 (H1N1) infection of siRNA-treated Caco-2 cells was analyzed, a substantial reduction in the level of viral spread was observed upon transfection of either TMPRSS2- or TMPRSS4-specific siRNAs, and this effect was enhanced upon cotransfection of both TMPRSS2- and TMPRSS4-specific siRNAs. Notably, the blockade of viral spread by siRNAs could be fully rescued by trypsin treatment (Fig. 5B, left), demonstrating that a lack of proteolytic activation of influenza virus was the underlying inhibitory mechanism. In stark contrast, TMPRSS2- and TMPRSS4-specific siRNAs had no significant effect on SC35M (H7N7) spread (Fig. 5B, right). These observations suggest that TMPRSS2 and TMPRSS4 expression in Caco-2 cells is responsible, at least in part, for the ability of these cells to support the trypsin-independent spread of influenza viruses.
FIG. 5.
Knockdown of TMPRSS2 and TMPRSS4 inhibits the trypsin-independent spread of influenza virus in Caco-2 cells. (A) Analysis of TMPRSS2 knockdown. Caco-2 cells either were transfected, by use of Lipofectamine, with a nonsense control siRNA or a TMPRSS2- or TMPRSS4-specific siRNA in the presence or absence of the control siRNA or were cotransfected with a 1:1 mixture of TMPRSS2- and TMPRSS4-specific siRNAs. TMPRSS2 expression in transfected cells was analyzed by Western blotting. Detection of β-actin served as a loading control. All siRNA transfection mixtures contained 200 pmol siRNA. Thus, if the transfection mixture contained a single siRNA species, 200 pmol of the siRNA was added. If two different siRNA species were cotransfected, 100 pmol of each was added. (B) Caco-2 cells were first transfected with siRNAs as described for panel A and then infected with PR8 (H1N1) or SC35M (H7N7) at an MOI of 0.01 in the presence or absence of trypsin. The release of infectious particles into the supernatant was determined by a focus formation assay. The results of a representative experiment ± standard deviations are shown and were confirmed by three separate experiments for PR8 (H1N1) and by one separate experiment for SC35M (H7N7). The statistical significance (asterisks) of the inhibitory effects of TMPRSS2- and TMPRSS4-specific siRNAs relative to that of the nonsense control siRNA was calculated by using the two-tailed Student t test for correlated samples.
TMPRSS2 is coexpressed with 2,6-linked sialic acids on type II pneumocytes and alveolar macrophages.
The important contribution of TMPRSS2 expression to the trypsin-independent spread of influenza virus in Caco-2 cells raised the question of whether this protease is expressed in influenza virus target cells in the human lung. To address this question, we immunostained human lung tissue with labeled Sambucus nigra lectin, which recognizes 2,6-linked sialic acids, and a monoclonal antibody specific for TMPRSS2 (29). Microscopic examination of stained tissues revealed coexpression of TMPRSS2 and 2,6-linked sialic acids in type II pneumocytes and alveolar macrophages (Fig. 6). The former have been shown to bind to human influenza viruses, albeit less frequently than type I pneumocytes (44), and to constitute viral target cells (35), indicating that TMPRSS2 might also support influenza virus spread in this cell type.
FIG. 6.
Expression of TMPRSS2 in human alveolar epithelium. (A) Hematoxylin-and-eosin-stained section of a normal lung showing several alveolar spaces, in which alveolar macrophages (M), type I pneumocytes (P1), and type II pneumocytes (P2) are labeled. Bar, 20 μm. (B) A serial section of panel A immunostained for TMPRSS2 using the peroxidase technique (brown) shows strong positive staining in type II pneumocytes and alveolar macrophages. (C) A serial section of panel B stained with Sambucus nigra lectin shows strong positive membrane staining of all cell types, including type II pneumocytes and alveolar macrophages. (D) A serial section of panel C immunostained with an irrelevant mouse primary antibody (Melan-A), as a negative control for panel B, shows no immunostaining. Alveolar macrophages show a faint brown tint, due to the presence of carbon, but not the strong brown staining of macrophages seen in panel B. A serial section of panel D immunostained using a goat polyclonal serum as a primary antibody, as a negative control for panel C, appeared very similar to panel D (data not shown).
DISCUSSION
We investigated the contributions of TMPRSS2 and TMPRSS4 to the proteolytic activation of influenza viruses in cell lines and in primary target cells. Transient expression of these proteases facilitated HA cleavage and allowed influenza virus to spread in 293T cells in the absence of exogenously added trypsin. The related protein hepsin was also able to cleave HA but failed to render HA-bearing viruses infectious, for reasons that remain unclear. Caco-2 cells, which allowed trypsin-independent viral spread, were found to express endogenous TMPRSS2 (mRNA and protein) and TMPRSS4 (mRNA), and expression of these proteases was shown to be required for efficient influenza virus activation. Finally, analysis of human lung tissue revealed that type II pneumocytes and alveolar macrophages coexpressed TMPRSS2 and 2,6-linked sialic acids, indicating that TMPRSS2 could support viral spread in the infected host.
Proteolytic activation of influenza virus HA primes the protein for low-pH-induced membrane fusion and is indispensable for viral infectivity (23, 24, 26, 27). Our previous study showed that TMPRSS2 and TMPRSS4 can activate influenza virus HA (5, 6), in agreement with other published work (2, 3, 45), and raised the possibility that other members of the TTSP family might also cleave and activate HA. Indeed, the TTSP hepsin mediated HA cleavage (Fig. 1A and B), and this cleavage resulted in the production of HA1 and HA2 fragments of the expected sizes (Fig. 1 and data not shown), indicating that it occurred at the junction between HA1 and HA2. However, hepsin-dependent HA cleavage did not confer appreciable infectivity on influenza virus PR8 (H1N1) or on lentiviral pseudotypes bearing HA, despite detectable incorporation of cleaved HA into virions (Fig. 2). The reasons for this apparent contradiction are at present unclear and might involve quantitative differences between HA cleavage by hepsin and that by TMPRSS2 and TMPRSS4. In this context, it is noteworthy that hepsin and TMPRSS2 interfered with HA1 glycosylation; the underlying mechanism and its potential relevance for HA function remain to be clarified.
Transient expression of TMPRSS2 and TMPRSS4 increased influenza virus spread at least 10-fold above background in 293T cells (the readily measurable background was due to the infection of cells with activated viruses generated in hens' eggs) (Fig. 2B). This observation prompted us to investigate whether endogenous expression of these proteases also facilitates the spread of influenza virus in cell lines in the absence of exogenously added proteases. Many cell lines, including 293T, Huh-7, and Vero E6, support the spread of human influenza virus in the presence of exogenous trypsin (5, 9, 13, 26). However, only the Caco-2 cell line, which was derived from intestinal epithelium, has been shown to allow the spread of different influenza viruses in the absence of trypsin (48). Our findings that Caco-2 cells released fully infectious HA-bearing pseudotypes and allowed the replication of PR8 (H1N1) virus in the absence of trypsin confirmed these results (Fig. 4A to C), although we did not obtain evidence that HA was activated upon viral uptake, as previously suggested (48). This discrepancy might be due to differences in the experimental systems used; we examined HA pseudotypes, while Zhirnov and colleagues employed replication-competent influenza viruses (48). Alternatively, different cell culture conditions might account for the conflicting results, since Zhirnov and coworkers observed HA activation upon viral uptake only in proliferating Caco-2 cells (48). Nevertheless, both studies demonstrated efficient HA activation in Caco-2 cells in the absence of trypsin and raised questions concerning the nature of the protease(s) responsible.
We detected robust expression of TMPRSS2 (mRNA and protein) and TMPRSS4 (mRNA) in Caco-2 cells (trypsin independent) but not in Huh-7, Vero E6, or 293T cells (trypsin dependent) (Fig. 3), suggesting a correlation between TMPRSS2 and TMPRSS4 expression and trypsin-independent replication of influenza virus. Furthermore, siRNA knockdown demonstrated that expression of both TMPRSS2 and TMPRSS4 was essential for the efficient trypsin-independent spread of influenza virus in Caco-2 cells (Fig. 5). Why TMPRSS2 knockdown was not fully rescued by the presence of TMPRSS4 and vice versa is at present unclear, and a definite answer might have to await the availability of efficient TMPRSS4 detection reagents. However, the enhanced inhibitory effect of a cocktail of TMPRSS2- and TMPRSS4-specific siRNAs relative to each siRNA alone clearly indicates that both proteins facilitate HA activation and can, at least in part, functionally complement each other. Collectively, these results provide a molecular mechanism to explain the ability of Caco-2 cells to support the spread of influenza virus in the absence of exogenous HA-activating protease, and they demonstrate for the first time that endogenous TTSPs can cleave and activate influenza viruses.
A contribution of TMPRSS2 and TMPRSS4 to the spread of influenza virus in the host requires that these proteases be expressed on permissive cells. A major determinant for permissiveness to infection by human influenza viruses is the expression of surface structures modified with 2,6-linked sialic acids, which are bound by HA (38). Seasonal and, in particular, pandemic influenza viruses, including the 2009 H1N1 virus (46), can spread to the alveolar epithelium, and infection of these target cells parallels the development of primary viral pneumonia (25, 42). Immunohistochemistry of human alveoli revealed that type II pneumocytes and alveolar macrophages coexpress TMPRSS2 and 2,6-linked sialic acids (Fig. 6), suggesting that TMPRSS2 might promote the spread of influenza virus in these cells. Indeed, attachment of human influenza viruses to type II pneumocytes and, rarely, to alveolar macrophages has been demonstrated previously (44), and recent evidence indicates that type II pneumocytes and, occasionally, macrophages are viral targets (35). Notably, type I pneumocytes have been reported to bind influenza virus more efficiently than type II pneumocytes (44), and the protease responsible for viral activation in these target cells remains to be defined—with TMPRSS4 being an attractive candidate. In contrast, it remains unknown whether TMPRSS2 (and TMPRSS4) is expressed and promotes viral spread in target cells in the upper airways. In addition, it is noteworthy that TMPRSS2 expression in the colon, stomach, and kidney has been demonstrated previously (29), and it is possible that this protease might contribute to the rare extrapulmonary spread (25) of influenza viruses in humans.
In summary, we provide evidence that TMPRSS2 and TMPRSS4 activate human influenza viruses in cell culture and potentially promote the spread of influenza viruses in and between humans. The finding that knockdown of a single protease, either TMPRSS2 or TMPRSS4, was sufficient to reduce viral spread markedly in Caco-2 cells lends further weight to the concept of targeting host cell proteases for influenza therapy—an approach that has already been shown to be feasible and effective in animals (50) and humans (49). Partial inhibition of viral spread by the blockade of a single protease might well be sufficient to produce a clinical benefit. However, our findings that two proteases can activate influenza virus in Caco-2 cells also indicate that high antiviral activity might be achieved only by targeting several proteases simultaneously. In any case, the characterization of the exact roles of TMPRSS2 and TMPRSS4 in influenza virus spread and pathogenesis is an important task, and Tmprss2 knockout mice, which do not show an obvious phenotype in the absence of infection (20), might be valuable tools for these endeavors.
Acknowledgments
We thank T. F. Schulz for support, M. Guipponi for the TMPRSS3 expression plasmid, J. Lucas and P. Nelson for antibody P5H9, and O. Dittrich-Breiholz for supervision of quantitative RT-PCR experiments.
S.P. was supported by the BMBF (grant 01KI 0703), I.S. by a fellowship from the Center of Infection Biology, K.S. by the BMBF (grant 01KI07137), and P.B. by the Georg-Christoph-Lichtenberg Foundation. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: p24 hybridoma 183-H12-5C (catalog no. 1547), contributed by N. Pedersen, and plasmid p96ZM651gag-opt (catalog no. 8675), contributed by Y. Li, F. Gao, and B. Hahn.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Bosch, F. X., M. Orlich, H. D. Klenk, and R. Rott. 1979. The structure of the hemagglutinin, a determinant for the pathogenicity of influenza viruses. Virology 95:197-207. [DOI] [PubMed] [Google Scholar]
- 2.Böttcher, E., C. Freuer, T. Steinmetzer, H. D. Klenk, and W. Garten. 2009. MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine 27:6324-6329. [DOI] [PubMed] [Google Scholar]
- 3.Böttcher, E., T. Matrosovich, M. Beyerle, H. D. Klenk, W. Garten, and M. Matrosovich. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böttcher-Friebertshäuser, E., C. Freuer, F. Sielaff, S. Schmidt, M. Eickmann, J. Uhlendorff, T. Steinmetzer, H. D. Klenk, and W. Garten. 2010. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J. Virol. 84:5605-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaipan, C., D. Kobasa, S. Bertram, I. Glowacka, I. Steffen, T. S. Tsegaye, M. Takeda, T. H. Bugge, S. Kim, Y. Park, A. Marzi, and S. Pohlmann. 2009. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 83:3200-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, S. Y., S. Bertram, I. Glowacka, Y. W. Park, and S. Pohlmann. 2009. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol. Med. 15:303-312. [DOI] [PubMed] [Google Scholar]
- 7.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 8.Cox, N. J., and K. Subbarao. 2000. Global epidemiology of influenza: past and present. Annu. Rev. Med. 51:407-421. [DOI] [PubMed] [Google Scholar]
- 9.de Wit, E., M. I. Spronken, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2004. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res. 103:155-161. [DOI] [PubMed] [Google Scholar]
- 10.Dushoff, J., J. B. Plotkin, C. Viboud, D. J. Earn, and L. Simonsen. 2006. Mortality due to influenza in the United States—an annualized regression approach using multiple-cause mortality data. Am. J. Epidemiol. 163:181-187. [DOI] [PubMed] [Google Scholar]
- 11.Garten, W., S. Hallenberger, D. Ortmann, W. Schafer, M. Vey, H. Angliker, E. Shaw, and H. D. Klenk. 1994. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie 76:217-225. [DOI] [PubMed] [Google Scholar]
- 12.Glaser, L., J. Stevens, D. Zamarin, I. A. Wilson, A. Garcia-Sastre, T. M. Tumpey, C. F. Basler, J. K. Taubenberger, and P. Palese. 2005. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 79:11533-11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govorkova, E. A., N. V. Kaverin, L. V. Gubareva, B. Meignier, and R. G. Webster. 1995. Replication of influenza A viruses in a green monkey kidney continuous cell line (Vero). J. Infect. Dis. 172:250-253. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung, H., K. P. Lee, S. J. Park, J. H. Park, Y. S. Jang, S. Y. Choi, J. G. Jung, K. Jo, D. Y. Park, J. H. Yoon, J. H. Park, D. S. Lim, G. R. Hong, C. Choi, Y. K. Park, J. W. Lee, H. J. Hong, S. Kim, and Y. W. Park. 2008. TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial-mesenchymal transition. Oncogene 27:2635-2647. [DOI] [PubMed] [Google Scholar]
- 16.Kaverin, N. V., and R. G. Webster. 1995. Impairment of multicycle influenza virus growth in Vero (WHO) cells by loss of trypsin activity. J. Virol. 69:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaoka, Y., and R. G. Webster. 1988. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 85:324-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kido, H., Y. Okumura, H. Yamada, T. Q. Le, and M. Yano. 2007. Proteases essential for human influenza virus entry into cells and their inhibitors as potential therapeutic agents. Curr. Pharm. Des. 13:405-414. [DOI] [PubMed] [Google Scholar]
- 19.Kido, H., Y. Yokogoshi, K. Sakai, M. Tashiro, Y. Kishino, A. Fukutomi, and N. Katunuma. 1992. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J. Biol. Chem. 267:13573-13579. [PubMed] [Google Scholar]
- 20.Kim, T. S., C. Heinlein, R. C. Hackman, and P. S. Nelson. 2006. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol. Cell. Biol. 26:965-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klenk, H. D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 22.Klenk, H. D., and R. Rott. 1988. The molecular biology of influenza virus pathogenicity. Adv. Virus Res. 34:247-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klenk, H. D., R. Rott, and M. Orlich. 1977. Further studies on the activation of influenza virus by proteolytic cleavage of the haemagglutinin. J. Gen. Virol. 36:151-161. [DOI] [PubMed] [Google Scholar]
- 24.Klenk, H. D., R. Rott, M. Orlich, and J. Blodorn. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68:426-439. [DOI] [PubMed] [Google Scholar]
- 25.Kuiken, T., and J. K. Taubenberger. 2008. Pathology of human influenza revisited. Vaccine 26(Suppl. 4):D59-D66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazarowitz, S. G., and P. W. Choppin. 1975. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68:440-454. [DOI] [PubMed] [Google Scholar]
- 27.Lazarowitz, S. G., A. R. Goldberg, and P. W. Choppin. 1973. Proteolytic cleavage by plasmin of the HA polypeptide of influenza virus: host cell activation of serum plasminogen. Virology 56:172-180. [DOI] [PubMed] [Google Scholar]
- 28.Li, S. Q., M. Orlich, and R. Rott. 1990. Generation of seal influenza virus variants pathogenic for chickens, because of hemagglutinin cleavage site changes. J. Virol. 64:3297-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucas, J. M., L. True, S. Hawley, M. Matsumura, C. Morrissey, R. Vessella, and P. S. Nelson. 2008. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J. Pathol. 215:118-125. [DOI] [PubMed] [Google Scholar]
- 30.Murakami, M., T. Towatari, M. Ohuchi, M. Shiota, M. Akao, Y. Okumura, M. A. Parry, and H. Kido. 2001. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur. J. Biochem. 268:2847-2855. [DOI] [PubMed] [Google Scholar]
- 31.Parrish, C. R., and Y. Kawaoka. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 59:553-586. [DOI] [PubMed] [Google Scholar]
- 32.Poland, G. A., R. M. Jacobson, and I. G. Ovsyannikova. 2009. Influenza virus resistance to antiviral agents: a plea for rational use. Clin. Infect. Dis. 48:1254-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheiblauer, H., A. P. Kendal, and R. Rott. 1995. Pathogenicity of influenza A/Seal/Mass/1/80 virus mutants for mammalian species. Arch. Virol. 140:341-348. [DOI] [PubMed] [Google Scholar]
- 34.Scott, H. S., J. Kudoh, M. Wattenhofer, K. Shibuya, A. Berry, R. Chrast, M. Guipponi, J. Wang, K. Kawasaki, S. Asakawa, S. Minoshima, F. Younus, S. Q. Mehdi, U. Radhakrishna, M. P. Papasavvas, C. Gehrig, C. Rossier, M. Korostishevsky, A. Gal, N. Shimizu, B. Bonne-Tamir, and S. E. Antonarakis. 2001. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet. 27:59-63. [DOI] [PubMed] [Google Scholar]
- 35.Shieh, W. J., D. M. Blau, A. M. Denison, M. Deleon-Carnes, P. Adem, J. Bhatnagar, J. Sumner, L. Liu, M. Patel, B. Batten, P. Greer, T. Jones, C. Smith, J. Bartlett, J. Montague, E. White, D. Rollin, R. Gao, C. Seales, H. Jost, M. Metcalfe, C. S. Goldsmith, C. Humphrey, A. Schmitz, C. Drew, C. Paddock, T. M. Uyeki, and S. R. Zaki. 2010. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am. J. Pathol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirogane, Y., M. Takeda, M. Iwasaki, N. Ishiguro, H. Takeuchi, Y. Nakatsu, M. Tahara, H. Kikuta, and Y. Yanagi. 2008. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 82:8942-8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons, G., J. D. Reeves, C. C. Grogan, L. H. Vandenberghe, F. Baribaud, J. C. Whitbeck, E. Burke, M. J. Buchmeier, E. J. Soilleux, J. L. Riley, R. W. Doms, P. Bates, and S. Pohlmann. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115-123. [DOI] [PubMed] [Google Scholar]
- 38.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 39.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1-20. [DOI] [PubMed] [Google Scholar]
- 40.Stieneke-Gröber, A., M. Vey, H. Angliker, E. Shaw, G. Thomas, C. Roberts, H. D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11:2407-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szabo, R., and T. H. Bugge. 2008. Type II transmembrane serine proteases in development and disease. Int. J. Biochem. Cell Biol. 40:1297-1316. [DOI] [PubMed] [Google Scholar]
- 42.Taubenberger, J. K., and D. M. Morens. 2008. The pathology of influenza virus infections. Annu. Rev. Pathol. 3:499-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Towatari, T., M. Ide, K. Ohba, Y. Chiba, M. Murakami, M. Shiota, M. Kawachi, H. Yamada, and H. Kido. 2002. Identification of ectopic anionic trypsin I in rat lungs potentiating pneumotropic virus infectivity and increased enzyme level after virus infection. Eur. J. Biochem. 269:2613-2621. [DOI] [PubMed] [Google Scholar]
- 44.van Riel, D., V. J. Munster, E. de Wit, G. F. Rimmelzwaan, R. A. Fouchier, A. D. Osterhaus, and T. Kuiken. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 171:1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, W., E. N. Butler, V. Veguilla, R. Vassell, J. T. Thomas, M. Moos, Jr., Z. Ye, K. Hancock, and C. D. Weiss. 2008. Establishment of retroviral pseudotypes with influenza hemagglutinins from H1, H3, and H5 subtypes for sensitive and specific detection of neutralizing antibodies. J. Virol. Methods 153:111-119. [DOI] [PubMed] [Google Scholar]
- 46.Yeh, E., R. F. Luo, L. Dyner, D. K. Hong, N. Banaei, E. J. Baron, and B. A. Pinsky. 2010. Preferential lower respiratory tract infection in swine-origin 2009 A(H1N1) influenza. Clin. Infect. Dis. 50:391-394. [DOI] [PubMed] [Google Scholar]
- 47.Zhirnov, O. P., M. R. Ikizler, and P. F. Wright. 2002. Cleavage of influenza A virus hemagglutinin in human respiratory epithelium is cell associated and sensitive to exogenous antiproteases. J. Virol. 76:8682-8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhirnov, O. and H. D. Klenk. 2003. Human influenza A viruses are proteolytically activated and do not induce apoptosis in CACO-2 cells. Virology 313:198-212. [DOI] [PubMed] [Google Scholar]
- 49.Zhirnov, O. P., A. V. Ovcharenko, A. G. Bukrinskaia, L. P. Ursaki, and L. A. Ivanova. 1984. Antiviral and therapeutic action of protease inhibitors in viral infections: experimental and clinical observations. Vopr. Virusol. 29:491-497. (In Russian.) [PubMed] [Google Scholar]
- 50.Zhirnov, O. P., A. V. Ovcharenko, and A. G. Bukrinskaya. 1982. Protective effect of protease inhibitors in influenza virus infected animals. Arch. Virol. 73:263-272. [DOI] [PubMed] [Google Scholar]