Abstract
Mutations in the internal ribosome entry site (IRES) of hepatitis A virus (HAV) have been associated with enhanced in vitro replication and viral attenuation in animal models. To address the possible role of IRES variability in clinical presentation, IRES sequences were obtained from HAV isolates associated with benign (n = 8) or severe (n = 4) hepatitis. IRES activity was assessed using a bicistronic dual-luciferase expression system in adenocarcinoma (HeLa) and hepatoma (HuH7) cell lines. Activity was higher in HuH7 than in HeLa cells, except for an infrequently isolated genotype IIA strain. Though globally low, significant variation in IRES-dependent translation efficiency was observed between field isolates, reflecting the low but significant genetic variability of this region (94.2% ± 0.5% nucleotide identity). No mutation was exclusive of benign or severe hepatitis, and variations in IRES activity were not associated with a clinical phenotype, indirectly supporting the preponderance of host factors in determining the clinical presentation.
Hepatitis A virus (HAV) is a nonenveloped RNA virus of the Picornaviridae family. The viral genome consists of an approximately 7,500-nucleotide (nt)-long, positive-stranded RNA divided in three parts: a 5′ untranslated region (5′ UTR), a single open reading frame that encodes both structural and nonstructural proteins, and a 3′ UTR with a short poly(A) tail. By sequencing of the VP1-2A junction and the VP1 gene, 3 genotypes (I, II, and III) divided into A and B subtypes have been described in humans (7, 27). HAV is the main cause of acute viral hepatitis worldwide. The majority of cases follow a benign course, but some may be present with fulminant forms, characterized by acute liver failure (factor V levels of <50% and encephalopathy). HAV-induced liver disease appears to result primarily from immunologic mechanisms, chiefly on the basis of in vitro studies. Most HAV strains have no detectable cytopathic effect in cell culture and no apparent effect on cell growth or metabolism (16), and HAV-infected cells are lysed by cytotoxic T cells isolated from the liver of acutely infected patients (30, 31). Clinical studies have suggested that host factors such as age and underlying liver disease were involved in the severity of liver diseases (32, 33) and that the host immune response also played a role in the fulminant forms of hepatitis A, as evidenced by markedly low viral loads (26).
Nevertheless, the existence of viral determinants of hepatitis A severity is suggested by both experimental and clinical studies. Indeed, mutations within the VP1-2A and 2C genes have been shown to enhance virulence in tamarins (9). It has also been suggested that 5′ UTR mutations associated with viral adaptation to cell culture were also responsible for viral attenuation in vivo (15). The 5′ UTR of HAV is about 735 nucleotides long and is considered the most conserved region of the genome. The 5′ UTR is involved in genome replication and translation initiation. Folding predictions and biochemical probing showed that this region forms a highly ordered secondary structure containing a pyrimidine-rich tract (PRT) and an internal ribosomal entry site (IRES) with 10 to 12 AUG triplets upstream of the initiator codon (18). The IRES allows the initiation of the cap-independent translation of the viral genome. Most knowledge of HAV IRES activity is derived from studies of the HM-175 reference strain and its cell culture-adapted variants (4, 5, 36). These experiments have shown that HAV presents the lowest IRES-dependent translation initiation activity among picornaviruses both in reticulocyte lysates and in a variety of cell lines, including the human hepatoma cell line HepG2 (type III IRES) (3, 6). These features have been attributed to a lower affinity of the HAV 5′ UTR for translation factors (6). The hypothesis that the slow growth of HAV in cell culture could be related to this inefficient translation is supported by the emergence of 5′ UTR mutations in cell culture-adapted variants with enhanced viral replication (8). The finding that these mutations were associated with viral attenuation in vivo supports the hypothesis of viral determinants of virulence in the 5′ UTR (15). Among the few clinical studies which have addressed this question, Fujiwara et al., by comparing full-length HAV genomes obtained from Japanese patients with benign or fulminant hepatitis, found less nucleotide variation in the 5′ UTRs from patients with fulminant hepatitis (12, 13) and suggested that two IRES mutations (G324A and C372G/T) might influence the course of HAV infection (14).
The aim of the present study was to further examine the genetic variability of 5′ UTR sequences from field isolates, to assess the potential impact of nucleotide variations on IRES activity by using validated techniques, and to search for a relationship with disease severity by comparing isolates obtained from patients with benign or fulminant forms of hepatitis A.
MATERIALS AND METHODS
Viral isolates.
Twelve HAV isolates were obtained from patients with acute hepatitis A. Sera were stored at −80°C after anti-HAV IgM detection: 8 sera were obtained from patients hospitalized in the hepatology unit of the Paul Brousse Hospital, Villejuif, France, between January 1987 and April 2000 (26) and 4 were obtained from the collection of the HAV National Reference Centre (CNR HAV, Hôpital Paul Brousse, Villejuif, France). As shown in Table 1, 8 isolates were associated with a benign form and 4 with a severe form of hepatitis. Of these, one presented with a factor V level of <50% without encephalopathy and three presented with a factor V level of <50% with encephalopathy, defining the fulminant form. None had serological markers of acute viral infection by hepatitis B virus, hepatitis E virus, Epstein-Barr virus, cytomegalovirus, or herpes simplex virus. All were hepatitis C virus RNA negative.
TABLE 1.
Clinical isolates and patients' characteristics
| Isolate/patient codea | HAV genotypeb | Hepatitis severity | Age (yr) | ALTc (IU/ml) | Prothrombin time (%) | Factor V (%) | Encephalopathy | Outcome |
|---|---|---|---|---|---|---|---|---|
| G1B1 | IA | Benign | 37 | 1,455 | 90 | NAd | None | Recovery |
| G1B2 | IA | Benign | 43 | NA | 90 | NA | None | Recovery |
| G1B3 | IA | Benign | 32 | 924 | 83 | NA | None | Recovery |
| G1B4 | IA | Benign | 19 | 3,006 | 79 | NA | None | Recovery |
| G1B5 | IB | Benign | 28 | NA | 83 | NA | None | Recovery |
| G1B6 | IB | Benign | 17 | NA | 58 | 100 | None | Recovery |
| G1S1 | IB | Severe | 22 | 6,456 | 18 | 42 | None | Recovery |
| G1F1 | IA | Fulminant | 52 | 4,180 | 23 | 55 | Grade 1 | Recovery |
| G1F2 | IB | Fulminant | 30 | 2,100 | 17 | 38 | Grade 4 | Death |
| G2B1 | IIA | Benign | 46 | NA | NA | NA | None | Recovery |
| G3B1 | IIIA | Benign | 12 | 2,815 | 81 | NA | None | Recovery |
| G3F1 | IIIA | Fulminant | 43 | 3,696 | 8 | 26 | Grade 1 | Recovery |
Each isolate was identified by a 4-letter code which includes the strain genotype (G1, G2, or G3 for genotype I, II, or III, respectively), the hepatitis A clinical presentation (B, S, or F for benign, severe, or fulminant, respectively), and the number of the strain.
HAV genotype was determined by the phylogenetic analysis of the VP1/2A junction.
ALT, alanine aminotransferase.
NA, not available.
The wild type HM-175 reference strain (code G1RW) of genotype IB was obtained from the National Institute for Biological Standards and Control (NIBSC). The HM-175/18f strain (code G1RC) was kindly provided by Sylvie Perelle (Agence Française de Sécurité Sanitaire des Aliments [AFSSA]). HM-175/18f was isolated from persistently infected BS-C1 cells and replicates rapidly with a cytopathic effect on these cells (22). Both strains were used as controls in IRES activity measures.
HAV RNA extraction, amplification, and sequencing.
RNA was extracted from 140 μl of serum by using the QIAamp viral RNA minikit (Qiagen, Les Ulis, France). Amplification of the VP1-2A region was performed by using the OneStep reverse transcription-PCR (RT-PCR) kit (Qiagen), in a final volume of 50 μl containing 10 μl of RNA extract, according to the manufacturer's instructions.
Strain genotyping was performed by analysis of a fragment of 508 nucleotides encompassing the VP1/2A junction with primers +2870 and −3381 (Table 2 ).
TABLE 2.
Primers used for amplification, cloning, sequencing, and mutagenesis protocols
| Purpose and code | Tmc (°C) | Regiona | Positionb | Sequence | Reference |
|---|---|---|---|---|---|
| Amplification, cloning, and sequencing | |||||
| IRES1 | 62 | 5′ UTR (GI) | 28 | GAGYCCCTCTTGGAAGTC | This study |
| IRES2 | 58 | 5′ UTR (GI) | 869 | AAAATAAGAAGCACCAGTCAC | This study |
| IRES3 | 67 | 5′ UTR (GII-III) | 63 | ATACCTCACCGCCGTTTGC | This study |
| IRES4 | 65 | 5′ UTR (GII-III) | 785 | GTCAAGRCCACTCCCAACAG | This study |
| +697 | 64 | 5′ UTR | 697 | TCAGGGGCATTTAGGTTTTTC | This study |
| 5′ Bgl2 | 75 | 5′ UTR | 63 | CCCAGATCTATACCTCACCGCCGTTTGC | This study |
| 3′ NcoG1RW | 72 | 5′ UTR | 738 | CCCCCATGGTCATTATTATTTAAGAATGAGGAAAAAC | This study |
| 3′ NcoG1F1 | 77 | 5′ UTR | 738 | CCCCCATGGTCATTGTTATTTGAGAATGAGGAAAAAC | This study |
| 3′ NcoG1F2 | 75 | 5′ UTR | 738 | CCCCCATGGTCATTATTGTTTAAGAATGAGGAAAAAC | This study |
| 3′ NcoG1B1 | 77 | 5′ UTR | 738 | CCCCCATGGTCATTATTGTTTGACAATGAGGAAAAAC | This study |
| 3′ NcoG1BF | 72 | 5′ UTR | 738 | CCCCCATGGTCATTATTATTTAAGAATGAGGAAAAAC | This study |
| 3′ NcoG2B1 | 75 | 5′ UTR | 738 | CCCCCATGGTCATAATTATTTGAGAGTGAGGAAAAAC | This study |
| 3′ NcoG3B1 | 75 | 5′ UTR | 738 | CCCCCATGGTCATAATCATTTATTGATGAGGAAAAAC | This study |
| 3′ NcoG3F1 | 71 | 5′ UTR | 738 | CCCCCATGGTCATAATTATTTATAGATGAGGAAAAAC | This study |
| Strain genotyping | |||||
| +2870 | 62 | VP1-2B | 2870 | GACAGATTCTACATTTGGATTGG | 4 |
| −3381 | 64 | VP1-2B | 3381 | CCATTTCAAGAGTCCACACACT | 4 |
| +2870G3 | 62 | VP1-2B | 2870 | AACAGATTCAACTTTTGGACTTG | This study |
| −3368G3 | 64 | VP1-2B | 3368 | CCACACACTCCTACCAGCAG | This study |
| 5′ UTR mutagenesis | |||||
| a324g | 5′ UTR | GAGTCTAAATTGGGGACGCAGATGTTTGGAACGTC | This study | ||
| a324g-R | 5′ UTR | GACGTTCCAAACATCTGCGTCCCCAATTTAGACTC | This study | ||
| t372c | 5′ UTR | AGTGTTAACTTGGCTTTCATGAACCTCTTTGATCTTCCACAAG | This study | ||
| t372c-R | 5′ UTR | CTTGTGGAAGATCAAAGAGGTTCATGAAAGCCAAGTTAACACT | This study | ||
| t255c | 5′ UTR | CTTGCCCTAGGCCCTGGCCGTTGCG | This study | ||
| t255c-R | 5′ UTR | CGCAACGGCCAGGGCCTAGGGCAAG | This study | ||
| t278c | 5′ UTR | CGCCCGGCGGGGCCAACTCCATGAT | This study | ||
| t278c-R | 5′ UTR | ATCATGGAGTTGGCCCCGCCGGGCG | This study |
Genotype specificity is indicated in parentheses (GI, GII, and GIII, genotype I, II, or III, respectively).
Position of the first 5′ nucleotide on the reference sequence HM-175 (GenBank accession number M14707).
Tm, melting temperature.
Amplification of the 5′ UTR was performed with the same reagents using IRES1 and IRES2 primers for genotype I isolates and IRES3 and IRES4 primers for genotype II and III isolates (Table 2). A fragment of 675 nucleotides (nt 63 to 738 according to HM-175 sequence numbering) was obtained and includes the end of the replication domain II (nt 63 to 98), the pyrimidine-rich tract (PRT; nt 99 to 154), and IRES domains III, IV, V, and VI (nt 155 to 738) (5).
VP1-2A and 5′ UTR amplification products and 5′ UTR cloned fragments were sequenced bidirectionally by using the BigDye Terminator cycle sequencing kit on an ABI 377 DNA sequencer (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions.
Cloning of 5′ UTR amplification products.
Before sequencing and use in functional studies, 5′ UTR products were TA cloned into the pDRIVE vector by using the PCR Cloning Plus kit (Qiagen), according to the manufacturer's instructions. After screening, insert-containing clones were grown overnight in selective liquid medium, and then the plasmid was extracted with the QIAprep Spin Miniprep kit (Qiagen) and stored at −20°C.
Sequence analysis.
Nucleotide sequences were aligned with the clustalW software. Phylogenetic trees were constructed with the MEGA software (http://www.megasoftware.net) using the neighbor-joining algorithm and the Kimura 2-parameter model (21). Statistical evaluation of the topology was based on 1,000 replications of bootstrap sampling. Only bootstrap values above 70% were shown.
Mean percentage of nucleotide identity (PNI), within a genotype and between genotypes, was computed with the MEGA software by using the p-distance model.
Sequences of a given genotype were compared to a consensus sequence to identify polymorphic positions. Substitutions absent from GenBank sequences were reported on the IRES structural model established by Brown et al. to investigate their potential impact on the IRES secondary structures (5). Consensus sequences were constructed with consensus maker software (http://www.hiv.lanl.gov/content/sequence/CONSENSUS). For genotype I, we used the 9 clinical isolates of this study (5 of genotype IA, 4 of genotype IB) and 15 full-length sequences from GenBank (12 of IA and 3 of IB). For genotype III, we used our 2 clinical isolates (both IIIA) and 6 full-length sequences from GenBank (4 of IIIA and 2 of IIIB). The genotype IIA isolate was compared to the unique IIA strain present in GenBank (reference sequences listed in Table 3).
TABLE 3.
HAV reference sequences retrieved from GenBank for bioinformatics analysis
| GenBank accession no. | Strain code | Subgenotype | Sequenced length (bp) | Yr, place of isolation, place of infection if different |
|---|---|---|---|---|
| K02990 | LA | IA | 7,478 | 1975, United States |
| X75215 | GBM | IA | 7,431 | 1976, Germany |
| X83302 | FG | IA | 7,421 | 1985, Italy |
| AF357222 | LU38 | IA | 7,477 | 1988, China |
| AF485328 | LY6 | IA | 7,477 | 2002, China |
| AF512536 | DL3 | IA | 7,476 | 2002, China |
| AB020564 | AH1 | IA | 7,477 | 1992, Japan |
| AB020565 | AH2 | IA | 7,477 | 1991, Japan |
| AB020566 | AH3 | IA | 7,445 | 1993, Japan |
| AB020567 | FH1 | IA | 7,477 | 1992, Japan |
| AB020568 | FH2 | IA | 7,478 | 1993, Japan |
| AB020569 | FH3 | IA | 7,477 | 1994, Japan |
| M14707 | HM-175 | IB | 7,478 | 1976, Australia |
| M20273 | MBB | IB | 7,470 | 1978, Germany, North Africa |
| AF268396 | HAF-203 | IB | 7,468 | 1992, Brazil |
| AY644676 | CF53-Berne | IIA | 7,426 | 1979, France |
| AY644670 | SLF88 | IIB | 7,414 | 1988, United States, Sierra Leone |
| AY644337 | HMH IIIA | IIIA | 7,166 | 2003, Germany |
| AB279732 | JNG04-90F | IIIA | 7,478 | 1990, Japan |
| AB279733 | JNG08-92F | IIIA | 7,476 | 1992, Japan, Madagascar |
| AB279734 | J95-8F | IIIA | 7,478 | 1995, Japan, Philippines |
| AB279735 | J85-1F | IIIB | 7,478 | 1985, Japan |
| AB258387 | JNG06-90F | IIIB | 7,462 | 1990, Japan |
| D00924 | AGM27 | V (SHAVa) | 7,400 | 1985, Kenya |
SHAV, simian hepatitis A virus.
Preparation of bicistronic plasmids.
IRES sequences from field isolates were subcloned into the bicistronic pCREL vector (2) containing an upstream Renilla luciferase gene (RLuc), a downstream firefly luciferase gene (FLuc), and the studied IRES in the intercistronic space. Briefly, 5′ UTR sequences were amplified from their corresponding TA cloning vectors with primers containing the BglII and NcoI restriction sites (Table 2) and inserted between RLuc and FLuc cistrons. Ligation products were transformed into DH5α competent cells. Positive clones (pCRHAVL) were identified by restriction fragment analysis.
Cell culture and transient transfection of cloned IRES.
HeLa cells (human cervical carcinoma cells) were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), and the human hepatoma cell line HuH7 was cultured in RPMI medium supplemented with 10% FBS. All cultures were incubated at 37°C in an atmosphere of 5% CO2. The day prior to transfection, 1.5 × 105 cells were seeded per well into 6-well plates. Cells were transfected using FuGene 6 (Roche Applied Science) following the manufacturer's instructions, with IRES pCRHAVL plasmids (2 μg/well), and cultured for 24 h at 37°C prior to lysing of the cells in passive lysis buffer (Promega, Charbonnières, France).
Measurement of bioluminescence activities.
The RLuc and FLuc bioluminescent activities were measured using the Dual Glo luciferase kit (Promega) and a microplate luminometer (EG and G Berthold, Thoiry, France). Ten microliters of posttransfection cell lysate was incubated sequentially with firefly and then Renilla luciferase-specific substrates according to the protocol supplied by the manufacturer. Light emission was measured 1 min after addition of each substrate and integrated over a 10-s interval. IRES activity was calculated as the mean of at least three independent experiments. A pCREL vector containing the human rhinovirus (HRV) IRES (pCRHRVL) was used as positive control.
The signal ratio FLuc/RLuc allows the balancing of the IRES activity with transfection efficacy. This ratio is then divided by the FLuc/RLuc ratio measured from a pCREL plasmid vector which contains a hairpin structure, pCRHL, between Renilla and luciferase cistrons which prevents the translation of the FLuc cistron. This calculation standardizes the translation initiation activities, which are expressed in arbitrary units (AU).
Site-directed mutagenesis.
Mutations thought to modify IRES efficiency were introduced into pCREL vector containing the HM-175 IRES sequence by using the QuikChange multisite-directed mutagenesis kit from Agilent Technologies (La Jolla, CA), according to the manufacturer's instructions, and the primers listed in Table 2. Mutagenesis products were then cloned into the provided XL10-Gold ultracompetent cells. Mutagenesis products were checked by sequencing. pCREL plasmids containing the mutated HM-175 IRES were then assessed for IRES efficiency as described above.
Statistical analysis.
Results are expressed as means ± standard deviations (SD) or medians (ranges). Nonparametric statistics were performed using Statistica version 6 software (StatSoft, Maisons-Alfort, France). The Mann-Whitney U test was used to compare continuous variables. A P value of <0.05 was considered significant.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the 12 5′ UTR sequences are FJ829473, FJ829474, FJ829475, FJ829476, FJ829477, FJ829478, FJ829479, FJ829480, FJ829481, FJ829482, FJ829483, and FJ829484. The GenBank accession numbers of the 12 VP1-2A sequences are FJ829485, FJ829486, FJ829487, FJ829488, FJ829489, FJ829490, FJ829491, FJ829492, FJ829493, FJ829494, FJ829495, and FJ829496.
RESULTS
Analysis of HAV 5′ UTR genetic diversity.
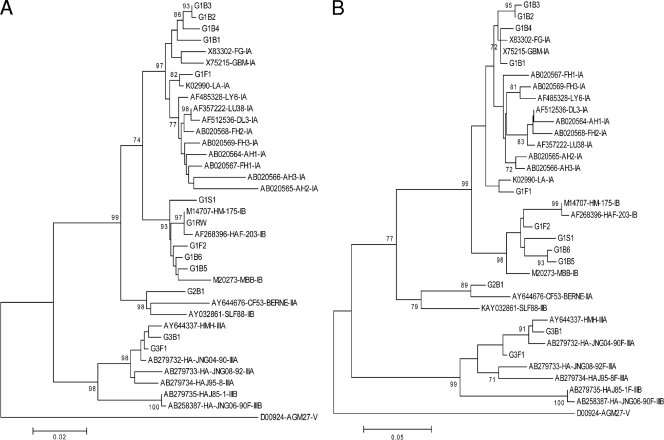
The 5′ UTR sequence of G1RW was totally identical to the HM-175 GenBank deposit, which validates the sequencing strategy of the 5′ UTR. The alignment of 35 sequences (12 clinical sequences and 23 references) showed 94.2% ± 0.5% nucleotide identity over the 675 studied nucleotides. Intragenotype homology was 96.2% ± 0.4% for genotype I, 97.2% ± 0.4% for genotype III, and 96.8% ± 0.5% for genotype II sequences. The alignment showed much higher variability in the nonstructured PRT (nt 99 to 154) than in the four structured domains of the IRES (nt 155 to 738). Due to deletions in the PRT, genotype II and III sequences were 17 to 20 nt shorter than genotype I sequences. The variability of 24 genotype I sequences (9 clinical plus 15 reference sequences) was higher than that of the 8 genotype III sequences (2 clinical plus 6 reference sequences), both in PRT (35 polymorphisms/60 positions for genotype I [58%] versus 7 polymorphisms/38 positions [18%] for genotype III) and in IRES (90/589 [15%] versus 40/583 [7%]). As shown in Fig. 1, phylogenetic clustering results of 5′ UTR and VP1/2A sequences were similar and sequences from benign and severe hepatitis isolates did not cluster together.
FIG. 1.
Phylogenetic analysis of studied and reference sequences (n = 35); the tree was constructed from a Kimura two-parameter genetic distance matrix using the neighbor-joining method followed by a bootstrap test (1,000 replicates). (A) 5′ UTR sequences (nucleotides 63 to 738, according to HM-175 strain numbering). (B) VP1-2A sequences (210 nucleotides).
Single-mutation analysis.
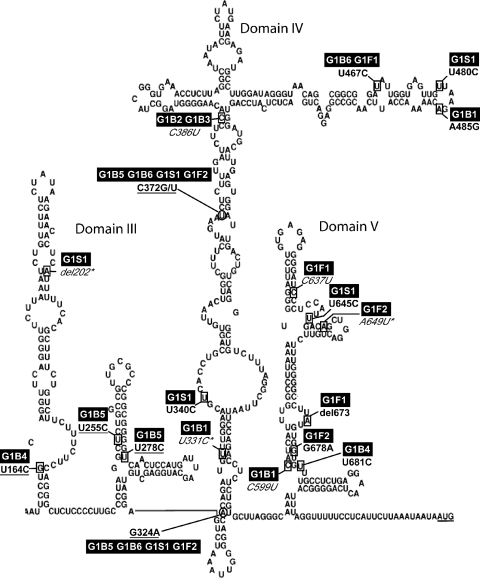
The alignment of the 24 clinical and reference sequences with the genotype I consensus showed 137 variable positions over the 675 studied nucleotides. Among those present in both reference and clinical sequences, we found G324A and C372G/T reported by Fujiwara et al. as potentially associated with benign hepatitis (14). These mutations were observed in clinical sequences from both benign (G1B5 and G1B6) and severe (G1S1 and G1F2) hepatitis, and also in the three genotype IB reference sequences, including HM-175. Among substitutions found only in clinical sequences, 17/32 were located in structured IRES domains and were reported in the model of Brown. As shown in Fig. 2, seven substitutions were located in single-strand RNA loops and 10 in double-helix RNA stems. Among stem mutations, three substitutions located in domain III resulted in an increase in the number of hydrogen bonds (U164C, U255C, and U278C) and thus in a potential stabilization of IRES structure. Figure 2 also shows the locations of G324A and C372G/T, in stems of domain IV: A324 and T372 increased the number of hydrogen bonds. The number of substitutions per isolate ranged from 1 to 4, but only the G1B5 sequence presented 2 mutations in the same putative RNA stem (U255C and U278C), both resulting in an increased number of hydrogen bonds.
FIG. 2.
Localization of variable positions of genotype I isolates in IRES secondary structures as described by Brown et al. (5). Each variable position is framed on the IRES structure. Isolate codes are highlighted. The substitution is depicted according to HM-175 numbering. “del” indicates a nucleotide deletion; an underlined substitution indicates an increase of bonds in a base pair, and an italicized substitution indicates a decrease; an asterisk indicates a mismatch.
The median number of substitutions within the IRES was not different between the 6 sequences from benign hepatitis and the 3 sequences from severe hepatitis (16 versus 8, P = 0.4). The difference was not significant either when the 675-nt sequences (PRT plus IRES) were analyzed (21 versus 9, P = 0.3). No mutations were exclusive of benign or severe hepatitis.
Our field G2B1 isolate was compared to the single genotype IIA sequence available in GenBank, CF53-Berne, obtained after 12 passages on cell culture and likely harboring adaptation mutations. Within the 583 nucleotides of the IRES domain, G2B1 differed from CF53-Berne by 18 substitutions (of which 10 were located in RNA loops and 6 in RNA stems) and from the reference HM-175 by 36 substitutions.
The alignment of genotype III sequences showed only 3 substitutions specific to clinical sequences and located in the IRES structured elements of the benign isolate. The field isolates, G3B1 and G3F1, differed from HM-175 IRES by 53 and 51 nucleotides, respectively, but differed from each other by only 5 nucleotides (genetic difference of <1%).
IRES activity measures.
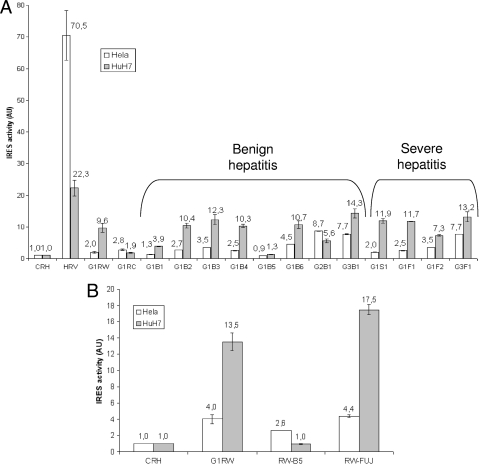
The FLuc/RLuc ratio represents the IRES activity normalized to the amount of RNA present in the cell. As shown in Fig. 3 A, the FLuc/RLuc ratio of the HRV IRES DNA construct was ∼70- and ∼20-fold the ratios obtained with the no-IRES construct pCRHL in HuH7 cells and HeLa cells, respectively. As expected, HAV IRES activity was much lower than HRV IRES activity, particularly in HeLa cells: the IRES of the HM-175 strain (G1RW vector) was 35 times less active than HRV IRES in HeLa cells (2.0 versus 70.5 AU) and 2 times less active in HuH7 cells (9.6 versus 22.3 AU). The cell culture-adapted strain G1RC presented a weak IRES activity in both HuH7 and HeLa cells, with, however, higher activity on HeLa cells. The median IRES activity of the 12 clinical isolates was 3.1 AU (range, 0.9 to 8.7) in HeLa cells and 10.6 AU (range, 1.3 to 14.3) in HuH7 cells. These IRES elements were globally 23 times and 2 times less active than HVR IRES in HeLa and HuH7 cells, respectively. Clinical IRES elements had higher activity in hepatoma cells than in adenocarcinoma cells, except for the IRES of the genotype IIA isolate, which has higher activity in HeLa cells (8.7 versus 5.6 AU, respectively).
FIG. 3.
IRES activity measured after transitory transfection of pCREL vectors in HuH7 and HeLa cells, expressed in arbitrary units (AU). Vertical bars represent standard deviations of the means from triplicate measures. (A) CRH (negative control), HRV (human rhinovirus), HM-175 strain (G1RW), HM-175/18f (G1RC), and studied clinical isolates (G1, G2, or G3 for genotype I, II, or III and B, S, or F for benign, severe, or fulminant hepatitis, respectively). (B) CRH (negative control); HM-175 (G1RW); HM-175 with T255C and T278C mutations, present in the G1B5 strain (RW-B5); HM-175 with G324 and C372 mutations (RW-FUJ).
The highest IRES activity on HuH7 cells was observed for genotype IIIA isolates (13.2 and 14.3 AU). Genotype I isolates presented up to 10-fold variation of IRES activity. The highest was measured for the G1B3 isolate (12.3 AU) and the lowest for the G1B5 isolate (1.3 AU), both associated with benign hepatitis. The IRES activity of the 8 benign hepatitis isolates was not different from that of the 4 severe hepatitis isolates in HuH7 cells (median, 10.4 AU [range, 1.3 to 14.3] versus 11.8 AU [range, 7.3 to 13.2], P = 0.4).
The impact of specific mutations on IRES activity was further investigated. G324A and C372G/T mutations reported as potentially associated with benign hepatitis (14) were found in genotype IB sequences, two from benign cases (G1B5 and G1B6, with IRES activities of 1.3 and 10.7 AU, respectively) and two from severe cases (G1S1 and G1F2, with IRES activities of 11.9 and 7.3 AU, respectively), and also in the genotype IB reference sequence HM-175. Nucleotides G324 and C372 were introduced by site-directed mutagenesis into the G1RW vector containing the HM-175 IRES. As shown in Fig. 3B, these mutations resulted in a slightly higher IRES activity (13.5 versus 17.5 AU; P, not significant). (Of note, IRES activity measurements of mutated vectors were performed on HuH7 cells used at a passage different from that of the initial experiments, explaining the higher activity values of the HM-175 reference IRES.)
In addition to G324A and C372T, two significant polymorphisms, U255C and U278C, were noted in the G1B5 isolate presenting the lowest IRES activity. These two mutations were introduced in the G1RW vector. As shown in Fig. 3B, they induced a dramatic reduction in IRES activity (13.5 versus 1.0 AU). We recall that these mutations were located in the same stem of domain III. The potential stabilization of the IRES structure has thus resulted in a decreased activity.
DISCUSSION
By studying field isolates, the present work explores the impact of 5′ UTR mutations on HAV IRES activity and disease severity. The low inter- and intragenotype variability of this region is confirmed, and nucleotide identity around 95% among our 12 field isolates is similar to that previously reported (18). As also reported (34) and at least for genotype I isolates, nucleotide conservation was particularly marked in structured IRES domains compared to the PRT region. This feature seems to be shared by genotype III isolates but needs to be confirmed from a broader set of sequences.
In contrast to enterovirus, which presents discordant discrimination patterns according to the genome region (25), the phylogenetic analysis of HAV 5′ UTR allowed a genotype discrimination similar to that obtained from the VP1-2A region and was supported by bootstrap values above 90%. These concordant discriminations may be interpreted as convergence of the negative selective pressures which preserve the IRES secondary structures and the amino acid sequence of capsid proteins, respectively (1, 5).
The role of IRES mutations in virulence or tissue tropism of picornavirus has been reported but is still a source of debate (10, 19). In the absence of an animal model for fulminant hepatitis A, several studies have searched for viral determinants of disease severity by comparing the sequences of field isolates associated with different clinical presentations (13, 26), and the potential role of 5′ UTR in disease severity has hence been suggested (12, 13). The existence of viral determinants of virulence is also suggested because cell culture adaptation is accompanied by mutations that often result in HAV attenuation in animal models. The sequence analysis of our field isolates did not support the existence of viral determinants of virulence in HAV 5′ UTR. First, as we have previously shown for VP1/2A (26), the phylogenetic analysis of 5′ UTR did not show any clustering of isolates according to clinical presentation. Second, no IRES mutations previously reported to be associated with cell culture adaptation and attenuation (A161G, U214del, A598G, G653A, C676U, and A694G) (8, 15, 28) were identified in our clinical sequences. Of note, we did not show any genetic variation of the PRT associated with hepatitis severity, which concords with the absence of influence of the PRT deletion on HAV replication or virulence in an animal model (29). And third, no obvious polymorphisms common to isolates responsible for benign hepatitis on the one hand or severe hepatitis on the other hand were identified.
Our study is the first to combine variability analysis with translation initiation activity measurements in relation to disease severity. These measurements were performed after transient transfection of a bicistronic vector, as previously described for other picornaviruses (17, 24) and also for HAV (23, 35). The HuH7 cell line was used, as a relevant model of liver cells, which are the final target cells of HAV (20, 28). Our results confirm from field isolates the low IRES activity of HAV and, as expected from previous work (3), also confirm that most wild-type isolates had 5′ UTR sequences that translated better in HuH7 cells, suggesting that hepatic factors are required for proper HAV translation in the liver. In contrast, translation driven by the 5′ UTR of the cell culture-adapted HM-175/18f strain was higher in HeLa than in HuH7 cells. HM-175/18f is thought to be attenuated in vivo (22) and presents important modifications of the 5′ UTR sequence, of which a 14-nt insertion is immediately upstream from the IRES. Though mutations responsible for attenuation may be located throughout the genome, this result suggest that some attenuation mutations may indeed affect translation efficiency in hepatoma cells.
The G2B1 isolate, associated with benign hepatitis, was the only one to present a higher activity in HeLa cells. Its IRES sequence differed by more than 6% from the HM-175 IRES (6.2%). A genetic background so different makes difficult the identification of mutations responsible for this paradoxical IRES activity. This phenotype has to be confirmed by the analysis of other clinical genotype II strains. As the French Reference Centre for HAV, we have identified recently additional IIA strains; the genetic characterization of their IRES sequences is in progress, and up to now we have shown up to 2% of genetic heterogeneity in this region (unpublished data).
Except for G2B1, field isolates had higher activity in hepatoma cells. Though globally low, IRES activity revealed significant variation between isolates. However, this variation was not linked to the clinical phenotype, indicating that IRES activity is not a major determinant of disease presentation. The localization of substitutions in the IRES model did not indicate a straightforward modification of its structure stability that could explain differences in IRES activity, except for 2 mutations present in the same RNA stem of the G1B5 sequence that increased the number of hydrogen bonds. We showed by introducing these mutations into the wild-type G1RW vector a dramatic reduction of G1RW IRES activity in HuH7 cells, thus confirming the hypothesis based on sequence analysis. In addition to these deleterious mutations, the G1B5 IRES also presented the G324A and C372T polymorphisms, both located in stems of the IRES domain III and thought to limit the severity of the disease (14). In our study, these polymorphisms were detected in both severe and mild hepatitis due to genotype IB strains and were also present in the HM-175 reference strain. The introduction of G324 and C372 (supposedly wild-type nucleotides) into the HM-175 IRES sequence slightly enhanced its activity. However, the important difference in translation activity of clinical sequences harboring these polymorphisms suggests that mutations at positions 324 and 372 do not have a major influence on translation activity or disease severity.
Among clinical isolates, the highest activity was observed for genotype III IRES sequences that differed from each other by less than 1%. A broader set of clinical strains, representative of the genetic diversity of this genotype, has to be studied to investigate its possible higher translation activity.
Finally, significant variation in IRES activity was observed between field isolates, but these variations were not associated with a clinical phenotype. Though genetic variations of different regions may cooperate to influence the level of replication and possibly influence HAV virulence, as recently suggested (11), our present data indirectly support the preponderance of host factors in determining clinical presentation. The variation observed in IRES activity appears to be the result of a low but significant genetic variability.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Aragones, L., S. Guix, E. Ribes, A. Bosch, and R. M. Pinto. 2010. Fine-tuning translation kinetics selection as the driving force of codon usage bias in the hepatitis A virus capsid. PLoS Pathog. 6:e1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnal, S., F. Pileur, C. Orsini, F. Parker, F. Pujol, A. C. Prats, and S. Vagner. 2005. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 280:4144-4153. [DOI] [PubMed] [Google Scholar]
- 3.Borman, A. M., P. Le Mercier, M. Girard, and K. M. Kean. 1997. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 25:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman, A. M., Y. M. Michel, and K. M. Kean. 2001. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J. Virol. 75:7864-7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, E. A., S. P. Day, R. W. Jansen, and S. M. Lemon. 1991. The 5′ nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J. Virol. 65:5828-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, E. A., A. J. Zajac, and S. M. Lemon. 1994. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa-Mattioli, M., J. Cristina, H. Romero, R. Perez-Bercof, D. Casane, R. Colina, L. Garcia, I. Vega, G. Glikman, V. Romanowsky, A. Castello, E. Nicand, M. Gassin, S. Billaudel, and V. Ferre. 2002. Molecular evolution of hepatitis A virus: a new classification based on the complete VP1 protein. J. Virol. 76:9516-9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day, S. P., P. Murphy, E. A. Brown, and S. M. Lemon. 1992. Mutations within the 5′ nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J. Virol. 66:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerson, S. U., Y. K. Huang, H. Nguyen, A. Brockington, S. Govindarajan, M. St. Claire, M. Shapiro, and R. H. Purcell. 2002. Identification of VP1/2A and 2C as virulence genes of hepatitis A virus and demonstration of genetic instability of 2C. J. Virol. 76:8551-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, D. M., G. Dunn, P. D. Minor, G. C. Schild, A. J. Cann, G. Stanway, J. W. Almond, K. Currey, and J. V. Maizel, Jr. 1985. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature 314:548-550. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara, K., H. Kojima, Y. Yonemitsu, S. Yasui, F. Imazeki, M. Miki, K. Suzuki, I. Sakaida, K. Okita, E. Tanaka, M. Omata, and O. Yokosuka. 2009. Phylogenetic analysis of hepatitis A virus in sera from patients with hepatitis A of various severities. Liver Int. 29:838-845. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara, K., O. Yokosuka, T. Ehata, F. Imazeki, and H. Saisho. 2000. PCR-SSCP analysis of 5′-nontranslated region of hepatitis A viral RNA: comparison with clinicopathological features of hepatitis A. Dig. Dis. Sci. 45:2422-2427. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara, K., O. Yokosuka, T. Ehata, H. Saisho, N. Saotome, K. Suzuki, K. Okita, K. Kiyosawa, and M. Omata. 2002. Association between severity of type A hepatitis and nucleotide variations in the 5′ non-translated region of hepatitis A virus RNA: strains from fulminant hepatitis have fewer nucleotide substitutions. Gut 51:82-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara, K., O. Yokosuka, K. Fukai, F. Imazeki, H. Saisho, and M. Omata. 2001. Analysis of full-length hepatitis A virus genome in sera from patients with fulminant and self-limited acute type A hepatitis. J. Hepatol. 35:112-119. [DOI] [PubMed] [Google Scholar]
- 15.Funkhouser, A. W., G. Raychaudhuri, R. H. Purcell, S. Govindarajan, R. Elkins, and S. U. Emerson. 1996. Progress toward the development of a genetically engineered attenuated hepatitis A virus vaccine. J. Virol. 70:7948-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauss-Muller, V., and F. Deinhardt. 1984. Effect of hepatitis A virus infection on cell metabolism in vitro. Proc. Soc. Exp. Biol. Med. 175:10-15. [DOI] [PubMed] [Google Scholar]
- 17.Jang, S. K., M. V. Davies, R. J. Kaufman, and E. Wimmer. 1989. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J. Virol. 63:1651-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi, M. S., A. M. Walimbe, and S. D. Chitambar. 2008. Evaluation of genomic regions of hepatitis A virus for phylogenetic analysis: suitability of the 2C region for genotyping. J. Virol. Methods 153:36-42. [DOI] [PubMed] [Google Scholar]
- 19.Kauder, S. E., and V. R. Racaniello. 2004. Poliovirus tropism and attenuation are determined after internal ribosome entry. J. Clin. Invest. 113:1743-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konduru, K., and G. G. Kaplan. 2006. Stable growth of wild-type hepatitis A virus in cell culture. J. Virol. 80:1352-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 22.Lemon, S. M., P. C. Murphy, P. A. Shields, L. H. Ping, S. M. Feinstone, T. Cromeans, and R. W. Jansen. 1991. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J. Virol. 65:2056-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, J. H., G. Dveksler, and G. G. Kaplan. 2004. Rescue of hepatitis A virus from cDNA-transfected but not virion RNA-transfected mouse Ltk- cells. Arch. Virol. 149:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 25.Poyry, T., L. Kinnunen, T. Hyypia, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 26.Rezende, G., A. M. Roque-Afonso, D. Samuel, M. Gigou, E. Nicand, V. Ferre, E. Dussaix, H. Bismuth, and C. Feray. 2003. Viral and clinical factors associated with the fulminant course of hepatitis A infection. Hepatology 38:613-618. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, B. H., R. W. Jansen, B. Khanna, A. Totsuka, O. V. Nainan, G. Siegl, A. Widell, H. S. Margolis, S. Isomura, K. Ito, et al. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J. Gen. Virol. 73:1365-1377. [DOI] [PubMed] [Google Scholar]
- 28.Schultz, D. E., M. Honda, L. E. Whetter, K. L. McKnight, and S. M. Lemon. 1996. Mutations within the 5′ nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J. Virol. 70:1041-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaffer, D. R., S. U. Emerson, P. C. Murphy, S. Govindarajan, and S. M. Lemon. 1995. A hepatitis A virus deletion mutant which lacks the first pyrimidine-rich tract of the 5′ nontranslated RNA remains virulent in primates after direct intrahepatic nucleic acid transfection. J. Virol. 69:6600-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallbracht, A., P. Gabriel, K. Maier, F. Hartmann, H. J. Steinhardt, C. Muller, A. Wolf, K. H. Manncke, and B. Flehmig. 1986. Cell-mediated cytotoxicity in hepatitis A virus infection. Hepatology 6:1308-1314. [DOI] [PubMed] [Google Scholar]
- 31.Vallbracht, A., K. Maier, Y. D. Stierhof, K. H. Wiedmann, B. Flehmig, and B. Fleischer. 1989. Liver-derived cytotoxic T cells in hepatitis A virus infection. J. Infect. Dis. 160:209-217. [DOI] [PubMed] [Google Scholar]
- 32.Vento, S., T. Garofano, C. Renzini, F. Cainelli, F. Casali, G. Ghironzi, T. Ferraro, and E. Concia. 1998. Fulminant hepatitis associated with hepatitis A virus superinfection in patients with chronic hepatitis C. N. Engl. J. Med. 338:286-290. [DOI] [PubMed] [Google Scholar]
- 33.Willner, I. R., M. D. Uhl, S. C. Howard, E. Q. Williams, C. A. Riely, and B. Waters. 1998. Serious hepatitis A: an analysis of patients hospitalized during an urban epidemic in the United States. Ann. Intern. Med. 128:111-114. [DOI] [PubMed] [Google Scholar]
- 34.Witwer, C., S. Rauscher, I. L. Hofacker, and P. F. Stadler. 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 29:5079-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi, M., and S. M. Lemon. 2002. Replication of subgenomic hepatitis A virus RNAs expressing firefly luciferase is enhanced by mutations associated with adaptation of virus to growth in cultured cells. J. Virol. 76:1171-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi, M., D. E. Schultz, and S. M. Lemon. 2000. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J. Virol. 74:6459-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]