Abstract
We report an RNA-negative, temperature-sensitive (ts) mutant of Murine hepatitis virus, Bristol ts31 (MHV-Brts31), that defines a new complementation group within the MHV replicase gene locus. MHV-Brts31 has near-normal levels of RNA synthesis at the permissive temperature of 33°C but is unable to synthesize viral RNA when the infection is initiated and maintained at the nonpermissive temperature of 39.5°C. Sequence analysis of MHV-Brts31 RNA indicated that a single G-to-A transition at codon 1307 in open reading frame 1a, which results in a replacement of methionine-475 with isoleucine in nonstructural protein 3 (nsp3), was responsible for the ts phenotype. This conclusion was confirmed using a vaccinia virus-based reverse genetics system to produce a recombinant virus, Bristol tsc31 (MHV-Brtsc31), which has the same RNA-negative ts phenotype and complementation profile as those of MHV-Brts31. The analysis of protein synthesis in virus-infected cells showed that, at the nonpermissive temperature, MHV-Brtsc31 was not able to proteolytically process either p150, the precursor polypeptide of the replicase nonstructural proteins nsp4 to nsp10, or the replicase polyprotein pp1ab to produce nsp12. The processing of replicase polyprotein pp1a in the region of nsp1 to nsp3 was not affected. Transmission electron microscopy showed that, compared to revertant virus, the number of double-membrane vesicles in MHV-Brts31-infected cells is reduced at the nonpermissive temperature. These results identify a new cistron in the MHV replicase gene locus and show that nsp3 has an essential role in the assembly of a functional MHV replication-transcription complex.
Coronaviruses are positive-stranded RNA viruses with genomes ranging in size from 27 to 32 kb. Approximately two-thirds of the genome encodes proteins that are involved in viral RNA synthesis. The majority of these proteins are encoded in two 5′-proximal, overlapping open reading frames (ORFs), ORF1a and ORF1b, and are translated as polyproteins, pp1a and pp1ab, which are then processed by virus-encoded proteinases into 16 nonstructural proteins (nsp's): nsp1 to nsp11, encoded in ORF1a, and nsp12 to nsp16, encoded in ORF1b. The translation products of ORF1a and ORF1ab, together with other viral proteins (41, 60) and, possibly, cellular proteins, assemble into replication-transcription complexes (RTCs) responsible for the synthesis of genome-sized RNA and a set of 3′-coterminal subgenome-sized mRNAs. The subgenome-sized mRNAs are produced by an unusual mechanism that involves discontinuous transcription during negative-strand RNA synthesis (for recent reviews, see references 39 and 43).
The coronavirus RTCs accumulate at perinuclear regions and are associated with a network of endoplasmic reticulum-derived, modified membranes (16, 20, 28). The interconnected structures within this network are known as convoluted membranes (CMs) and double-membrane vesicles (DMVs). Both structures are associated with viral replicase proteins and double-stranded RNA (16, 28, 53), but it is not yet entirely clear if both are the sites of active RNA synthesis. Hydrophobic transmembrane domains are present in nsp3, nsp4, and nsp6 and likely serve to anchor the nascent pp1a/pp1ab polyproteins to membranes during the first steps of RTC formation (1, 6, 16, 21, 26, 32-34, 54). A variety of studies suggest that most, if not all, coronavirus nsp's are recruited to the viral RTCs (16, 21, 35, 45), and a complex and dynamic pattern of interactions between intermediate and mature RTC proteins is thought to play a role in the regulation of viral RNA synthesis. Crystallographic analysis of individual and oligomeric complexes of coronavirus nsp's, as well as studies involving Saccharomyces cerevisiae two-hybrid, coimmunoprecipitation, cosedimentation, and cross-linking analysis (3, 47, 48, 55, 56), provides insights into some of the structural aspects of these RTC interactions.
Many of the coronavirus nsp's have been shown, or are predicted, to have enzymatic functions, including papain-like cysteine proteinases (nsp3), ADP-ribose 1"-phosphatase (nsp3), 3C-like cysteine proteinase (nsp5), RNA-dependent RNA polymerase primase (RdRP-primase) (nsp8), RdRp (nsp12), 5′-to-3′ helicase (nsp13), 3′-to-5′ exonuclease (nsp14), N7-methyltransferase (nsp14), endoribonuclease (nsp15), and S-adenosylmethionine-dependent 2′-O-methyltransferase (nsp16) (5, 22, 39, 44, 49). Clearly, some of these activities are common to many RNA viruses and are directly related to the biogenesis of the RTC and viral RNA synthesis. However, others (e.g., the endoribonuclease of nsp15) are unique to coronaviruses or viruses closely related to them (23) and may be involved in unusual activities that are related to the size of the coronavirus genome or virus-host interactions (15). Also, the finding that 2′-O-ribose-methylated RNA substrates are resistant to cleavage by the severe acute respiratory syndrome coronavirus (SARS-CoV) endoribonuclease indicates that there may be functional links between different nsp's: for example, the endonuclease activity of nsp15 and the S-adenosylmethionine-dependent 2′-O-methyltransferase activity of nsp16 (23).
The nsp3 protein is the largest and, possibly, most complex of the proteins encoded in the murine hepatitis virus (MHV) replicase gene. It is comprised of 2,005 amino acids (222 kDa) and is thought to contain as many as 15 domains that can be defined by structure, function, and phylogeny (31). These domains include amino-terminal ubiquitin-related (UB1) and hypervariable acidic (AC) domains, a papain-like cysteine proteinase domain (PLP1), an X domain with poly(ADP-ribose) binding activity (X or ADRP domain), a second ubiquitin-related domain (UB2), a second papain-like cysteine proteinase domain (PLP2), group 2 specific nucleic acid binding (NAB) and marker (G2M) domains, a transmembrane domain (TMD) that includes at least 2 membrane-spanning helices, a putative metal-binding domain (ZF), and a carboxyl-terminal Y domain that contains three subdomains (Y1, Y2, and Y3) with characteristic patterns of conserved Cys/His residues. The nsp3 protein is excised from its precursors, pp1a and pp1ab, by the concerted activity of the PLP1 and PLP2 domains, with PLP1 being responsible for the amino-terminal cleavage (cleavage site 2 [CS2]) and PLP2 being responsible for the carboxyl-terminal cleavage (cleavage site 3 [CS3]) (2, 12, 26, 59).
The genetic analysis of functions associated with the coronavirus RTC has employed both classical and reverse approaches. Reverse genetics has, for example, been used to introduce mutations at the active site of the coronavirus nsp14 exonuclease domain, which results in a failure to rescue recombinant virus (30) or the rescue of mutant viruses with significant growth and RNA synthesis defects (11). Similarly, mutant MHVs that are unable to liberate nsp1 from the nascent polyprotein (i.e., nsp1/nsp2 cleavage mutants) and mutant MHVs that lack extensive regions of the nsp1 or, indeed, the entire nsp2 coding region could be rescued but exhibited decreased or delayed replication (18). In contrast to these attenuated phenotypes, mutation of the active site of the Human coronavirus strain 229E (HCoV-229E) nsp3 ADRP domain has no significant effect on virus RNA synthesis or virus titer (36).
The analysis of classical temperature-sensitive (ts) mutants is also a powerful tool for investigating the complexities of coronavirus RNA synthesis. We have recently reported a genetic and functional analysis of a panel of MHV ts mutants that are unable to synthesize viral RNA when the infection is initiated and maintained at the nonpermissive temperature, i.e., mutants with an RNA-negative phenotype. One of the main conclusions of this analysis is that mutations in the nsp12, nsp14, and nsp16 proteins define distinct cistrons (cistrons II, IV, and VI) while mutations in the nsp4, nsp5, and nsp10 proteins define a single cistron (cistron I). On the basis of these results, we have predicted that most of the ORF1a gene products (more specifically, nsp4 to nsp10) function as a polyprotein before cleavage into individual polypeptides, or they are assembled into the RTC and are then cleaved, with a gain of function expressed in individual polypeptides. In contrast, the ORF1b-encoded nsp's (nsp12 to nsp16) are diffusible and assemble and function in viral RNA synthesis after cleavage from pp1ab. We have also suggested that replicase polyprotein processing may have an important role in regulating the different functions associated with the coronavirus RTC: for example, minus- and plus-strand RNA synthesis and the aging of the active complex (40).
The model that we have presented is consistent with our understanding of the organization and expression of the coronavirus replicase gene. However, there are still many unanswered questions. For example, it has not been shown that, as we predict, the nsp6, nsp7, nsp8, and nsp9 proteins function in cis, together with nsp4, nsp5, and nsp10. To do this, it will be necessary to find MHV RNA-negative ts mutants with causal mutations in the relevant proteins and to complement them with existing mutants. Similarly, our model leaves open the question of whether or not other ORF1a-encoded proteins will be able to complement defects in cistron I mutants. As mentioned above, it is known that particular domains of nsp1 and nsp3 are not essential for virus replication, but it is, of course, still possible that these proteins do have functions in RNA synthesis. Finally, the classical genetic approach can be further exploited to provide mechanistic insights into the functional inactivation of nsp's at the nonpermissive temperature. For example, a recent biochemical analysis of the MHV RNA-negative mutant LA ts6 has suggested that nsp10 acts as a regulatory cofactor of nsp5 proteinase activity (9). It would be interesting to investigate the polyprotein processing pattern of additional mutants in order to establish whether similar interactions exist between other proteins encoded in the MHV replicase gene locus.
MATERIALS AND METHODS
Cells and viruses.
17Cl-1 (17 clone 1) mouse fibroblast cells (46) and HeLa-D98OR (27) cells were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 5% tryptose phosphate broth (TPB). Monkey kidney (CV-1) cells and baby hamster kidney (BHK-21) cells were obtained from the European Collection of Cell Cultures (ECACC) and cultured in minimal essential medium (MEM) supplemented with HEPES (25 mM), 5% FCS, and antibiotics. MHV strain A59 (MHV-A59) was originally obtained from the laboratory of L. Sturman, Wadsworth Center for Laboratories and Research, Albany, NY. The characterized ts mutant viruses used in this study have been described previously (40) and were derived from the original mutant isolates after plaque purification and propagation in 17Cl-1 cells cultured at 33°C in low-pH MEM (pH 6.6 to 6.8) containing 5% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 5% TPB (38). Revertant viruses were picked from plaques of mutants whose titers were determined at 39.5°C and were plaque purified three times at 39.5°C. Virus stocks were obtained by using virus from a single plaque (∼107 PFU) to infect ∼4 × 106 cells to yield 6 ml of stock virus, which was then used to infect ∼1 × 107 cells at 33°C for 16 h to yield passage 2 virus stocks with a titer of ∼5 × 108 PFU/ml. Vaccinia virus (WR strain) and vaccinia virus recombinants were propagated, titrated, and purified as described previously (50).
Isolation of Bristol ts mutants.
17Cl-1 cells were infected with MHV-A59 at a multiplicity of infection (MOI) of 5 PFU/cell in Hanks' balanced salt solution (HBSS) containing 50 μg/ml DEAE-dextran and 0.2% bovine serum albumin (BSA). After 30 min at 33°C, the inoculum was removed and the cells were incubated in DMEM containing 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 150 μg/ml of 5-fluorouracil at 33°C for 16 h. This concentration of pyrimidine analogue was determined to inhibit virus replication by 80%. The mutagenized virus stock (∼2 × 106 PFU/ml) was stored at minus 80°C. The mutagenized virus was diluted to 1.5 PFU/ml, and 200-μl aliquots were incubated with ∼104 17Cl-1 cells at 33°C for 48 h. Supernatants from cultures displaying cytopathic effects were taken and used to infect duplicate cultures of 17Cl-1 cells that were incubated at 33°C and 39.5°C for 24 h. Supernatants from cultures showing cytopathic effect at 33°C but not 39.5°C were taken as potential ts mutants and isolated by two rounds of plaque purification.
Characterization of ts mutant stocks for titer and efficiency of plating.
17Cl-1 cells in six-well plates were infected in duplicate at room temperature with 0.5 ml of 10-fold dilutions of virus in HBSS containing 50 μg/ml DEAE-dextran and 0.2% BSA. The inoculum was removed after 30 min, and the cells were washed with 2 ml of DMEM. Monolayers were overlaid with MEM containing 5% FCS, 100 U/ml of penicillin, 100 μg/ml streptomycin, and 0.1% Gelrite gellan gum (Sigma-Aldrich, Poole, Dorset, United Kingdom) and incubated at the appropriate temperature with 7.5% CO2. Infected cells were incubated at 33°C for 3 days or 37°C/39.5°C for 2 days. Cells were fixed with 5% formaldehyde in phosphate-buffered saline (PBS) and stained with a solution of 0.2% toluidine blue, 0.2% azure blue, and 1% boric acid. The efficiency of plating (EOP) was determined by dividing the titer at 39.5°C by the titer at 33°C.
Viral RNA synthesis.
Viral RNA synthesis was measured by [3H]uridine incorporation into acid-precipitable material in the presence of 20 μg/ml actinomycin D. [5-3H]uridine (>1.0 TBq/mmol; Amersham Radiochemicals, Chalfont, United Kingdom) was added to the medium at 1.85 MBq/ml or 7.4 MBq/ml. After incubation, the radioactive medium was removed and the cells were lysed with 5% lithium dodecyl sulfate and 200 μg/ml proteinase K in LEH buffer (0.1 M LiCl, 0.001 M EDTA, 0.01 M HEPES [pH 6.6]) at 2 × 105 to 5 × 105 cells per ml. DNA was sheared by repeated passage of the lysate through a 27-gauge needle attached to a 1-ml tuberculin syringe. Duplicate or triplicate samples of 5 × 104 cells were precipitated with trichloroacetic acid, the precipitates were collected on glass fiber filters (Fisher Scientific, Leicestershire, United Kingdom) and dried under a heat lamp, and the radioactivity was determined by liquid scintillation spectroscopy.
Northern blotting.
RNA for Northern blotting was obtained by infecting ∼107 17Cl-1 cells with virus at an MOI of 10, incubating the cells for 8 h at 33°C or 6 h at 39.5°C, and isolating total RNA using the Trizol reagent (Invitrogen, Paisley, United Kingdom) as described by the manufacturer. The poly(A)-containing RNA was isolated using oligo(dT)25 Dynabeads (Dynal, Oslo, Norway) as previously described (52). The RNA was then incubated with formamide (50%) and formaldehyde (2.2 M) at 70°C for 10 min and electrophoresed in a 1% agarose gel containing 20 mM MOPS (3-N-morpholinopropanesulfonic acid) and 600 mM formaldehyde. After electrophoresis, the gel was soaked in 0.05 N NaOH, neutralized, and equilibrated with 20× SSC (3 M NaCl, 0.3 M sodium citrate, 1 mM EDTA) before being vacuum blotted onto a nylon membrane (Optitran BA-S 83; Schleicher and Schuell, Sigma-Aldrich, Poole, United Kingdom). The RNA was cross-linked to the membrane using UV light, and MHV RNAs were detected by hybridization with a 466-bp, α-32P-random-prime-labeled PCR product corresponding to sequences in the nucleocapsid protein ORF of the MHV-A59 genome (10).
Complementation analysis.
Complementation analysis was done by infecting 17Cl-1 cells in 35-mm dishes either singly with one ts mutant or doubly with two ts mutants to give a total MOI of 20 PFU/cell. After incubation at room temperature for 30 min, the inoculum was removed and the monolayers were washed three times with HBSS. The cells were then incubated with low-pH MEM containing 5% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 5% TPB prewarmed to 39.5°C. Infected cells were incubated at 39.5°C for 8 h. Supernatants were harvested and clarified by centrifugation at 10,000 rpm for 5 min. The virus titer was determined by plaque assay at 33°C. The complementation indices (CI) were calculated as described previously (40), and a CI of >2 between mutant pairs was consistent with complementation, i.e., ±4-fold difference above background, while a CI of <2 was negative for complementation.
RT-PCR and sequencing.
Viral RNA for reverse transcription-PCR (RT-PCR) and sequencing was obtained by infecting ∼107 17Cl-1 cells with virus at an MOI of 10, incubating the cells for 8 h at 33°C, and isolating total RNA using the Trizol reagent as described by the manufacturer. The replicase gene coding region (ORF1a and ORF1b) of MHV-Brts31 and MHV-Br31R1 virus (nucleotides [nt] 211 to 21751) was amplified and sequenced using a set of 121 oligonucleotides (P1 to P121) that are complementary to sequences spaced at approximately 350-nucleotide intervals along the positive- and negative-strand copies of the viral RNA (sequences available on request). A part of the nsp3 protein coding region of MHV-Brtsc31 and MHV-Br31cR1 (nt 3704 to 4517) was amplified and sequenced using six oligonucleotides of the oligonucleotide set. RT-PCR of viral RNA and sequencing were done using standard protocols. Sequencing analysis was done by MWG Eurofins (Ebersberg, Germany). Computer-assisted analysis of sequence data was done using the Lasergene biocomputing software (DNAStar).
Recombinant viruses.
Recombinant MHV-inf-1 and MHV-Brtsc31 were derived from vaccinia virus vMHV-inf-1, which contains a cloned, full-length MHV-A59 cDNA (GenBank accession number AY700211). Mutagenesis was done using the reverse genetics system described previously (7). Briefly, two rounds of vaccinia virus-mediated homologous recombination were done using the Escherichia coli guanine-phosphoribosyltransferase (GPT) gene as a selection marker. First, part of the nsp3 protein coding region within the vMHV-inf-1 cDNA was replaced by the GPT gene using homologous recombination with plasmid pGPT-nsp3(N). pGPT-nsp3(N) encodes the GPT gene flanked on its left by MHV-A59 nt 3751 to 4161 and on its right by MHV-A59 nt 5044 to 5460. Second, the GPT gene within the recombinant vaccinia virus vMHV-nsp3GPT cDNA was replaced by a mutated nsp3 protein coding region using homologous recombination with plasmid pnsp3-M475I (MHV-A59 nt 4129 to 4131; ATG→ATA). The plasmid pnsp3-M475I was produced by overlap PCR using mutagenic primers (details are available from the authors upon request) and vMHV-inf-1 DNA as template. The resulting PCR product, which encompasses MHV-A59 nt 3751 to 5460, was then cloned into pCR-Blunt II-TOPO (Invitrogen) to produce pnsp3-M475I. The identities of plasmids and recombinant vaccinia viruses were confirmed by sequence analysis of the mutated regions. Further cloning details, plasmid maps, and sequences are available from the authors on request.
Recombinant coronaviruses, MHV-inf-1 and MHV-Brtsc31, were rescued from cloned cDNA using purified, EagI-cleaved vaccinia virus DNA as a template for the transcription of recombinant, full-length MHV genomic RNA, which was electroporated into BHK-MHV-N cells as described previously (7). Following electroporation, the transfected BHK-MHV-N cells were mixed with a 4-fold excess of 17Cl-1 cells and cultured at 33°C. At days 1 and 2 postelectroporation, tissue culture supernatants were taken and recombinant coronaviruses were plaque purified three times. Virus stocks were obtained by using virus from a single plaque to infect 17Cl-1 cells to yield passage 1 virus stocks with a titer greater than 1 × 108 PFU/ml. The identities of recombinant MHV-A59 and recombinant mutant viruses were confirmed by sequence analysis of the mutated regions.
Viral protein synthesis and immunoprecipitation of MHV replicase products.
17Cl-1 cells (∼106 cells) in 60-mm-diameter cell culture dishes were either mock infected or infected with viruses at an MOI of 20.0 and incubated at 33°C in serum-free medium. After 1 h, the medium was replaced with DMEM supplemented with 5% FCS and containing 5 μg/ml of actinomycin D. At 4.0 h postinfection, the medium was replaced with medium lacking methionine and cysteine and supplemented with 2% dialyzed fetal calf serum. At 4.5 h postinfection, 35S-trans label (100 μCi/ml) was added and one dish was transferred to 39.5°C. At 6.5 h postinfection, cells were washed with PBS and then lysed in 250 μl of lysis buffer A (4% SDS, 3% dithiothreitol [DTT], 40% glycerol, and 0.065 M Tris-HCl, pH 6.8). Cell lysates were immunoprecipitated as described previously (42). Briefly, radiolabeled cell lysates (1 × 105 cell equivalents) were diluted in 1 ml RIPA buffer (0.5% Triton X-100, 0.1% SDS, 300 mM NaCl, 4 mM EDTA, and 50 mM Tris-HCl, pH 7.4) and immunoprecipitated with 2 μl of nsp-specific antibodies and protein A Sepharose beads (GE Healthcare). Protein-bead conjugates were washed four times in RIPA buffer, and the proteins were eluted from beads by incubation with 2× sample buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromphenol blue, 0.125 M Tris HCl, pH 6.8) at 37°C for 30 min. Protein products were separated by SDS-PAGE in 5 to 12.5% gradient polyacrylamide gels and were analyzed by autoradiography. The 14C molecular mass marker (GE Healthcare) was used as a molecular mass standard.
TEM analysis.
17Cl-1 cells were infected with virus at an MOI of 20.0 and incubated at 33°C. At 3.5 h postinfection, one set of infected cells was shifted to 39.5°C. At 5.5 h postinfection, the cells were washed two times with PBS, scraped from the dish, and pelleted in a microcentrifuge tube at 1,000 × g for 5 min at room temperature. PBS was replaced with 0.1 M cacodylate-buffered 4% glutaraldehyde before processing for transmission electron microscopy (TEM) analysis as previously described (16). Briefly, cells fixed in glutaraldehyde were rinsed two times for 10 min each in 0.1 M cacodylate buffer and postfixed in 0.1 M cacodylate-buffered 1% osmium tetroxide for 1 h. After two rinses in buffer, the cells were serially dehydrated in increasing concentrations of ethanol. After the dehydration step, the ethanol was replaced with 100% propylene oxide (transition fluid) by 3 changes of 10 min each. The cells were then immersed in a 1:1 solution of resin-propylene oxide and left on a rotator overnight at room temperature. Next day, the solution was replaced with a 3:1 solution of resin-propylene oxide for 7 to 8 h, after which the solution was changed to 100% resin and left on a rotator overnight at room temperature. The samples were then embedded in a freshly prepared 100% Embed 812 resin (Electron Microscopy Sciences) for 7 to 8 h. The molds were then placed in a 56°C oven overnight. Sections 0.5 μm thick were stained with 1% toluidine blue, and selected areas were thin sectioned (80 nm), mounted on copper grids, and stained with 5% uranyl acetate and Reynolds' lead citrate (37). The grids were examined and photographed on a Hitachi H600 at 75 kV. Negatives were scanned into digital files using a Microtek i800 scanner.
RESULTS
Isolation and phenotype of MHV-Brts31.
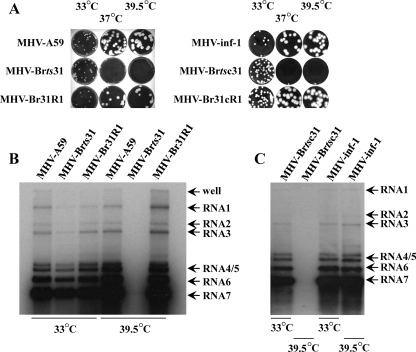
MHV-Brts31 was isolated as described in Materials and Methods and identified as a putative RNA-negative, ts mutant due to its inability to cause cytopathic effects in 17Cl-1 cells that were infected and maintained at the nonpermissive temperature. To analyze the phenotype of MHV-Brts31 in more detail, we determined first the EOP of a high-titer (passage 2) virus stock. At 33°C, the stock had a titer of 1 × 108 PFU/ml, and at 39.5°C, the stock had a titer of 1.5 × 103 PFU/ml, which is an EOP of 1.5 × 10−5. This value is indicative of a single point mutation. Of the plaques that formed at the nonpermissive temperature, all that we tested were no longer ts and are, therefore, revertants. One of these revertants was designated MHV-Br31R1. The plaque size and morphology of MHV-Brts31 and MHV-Br31R1 at 33°C and of MHV-Br31R1 at 39.5°C were identical to those of the parental MHV-A59 (Fig. 1A, left panel). MHV-Brts31 also failed to form plaques at 37°C.
FIG. 1.
(A) Plaque morphology of MHV-A59, MHV-Brts31, MHV-Br31R1, MHV-inf-1, MHV-Brtsc31, and MHV-Br31cR1. 17Cl-1 cells were infected with 10 to 20 PFU of virus, overlaid with solid medium, and maintained at 33°C for 3 days or 37°C and 39.5°C for 2 days. The overlay was removed, and the cells were fixed and stained as described in Materials and Methods. (B) Virus RNA accumulation in MHV-Brts31 and MHV-Br31R1 virus-infected cells at permissive and nonpermissive temperatures. RNA was extracted from cells that had been infected with MHV-A59, MHV-Brts31, or MHV-Br31R1 and maintained at either 33°C or 39.5°C. After electrophoresis and transfer to membrane, the virus RNA was detected with an α-32P-random-prime-labeled PCR probe derived from the MHV-A59 nucleocapsid protein gene. (C) Virus RNA accumulation in MHV-inf-1 and MHV-Brtsc31 virus-infected cells at permissive and nonpermissive temperatures. RNA was extracted from cells that had been infected with MHV-inf-1 or MHV-Brtsc31 and maintained at either 33°C or 39.5°C. After electrophoresis and transfer to membrane, the virus RNA was detected with an α-32P-random-prime-labeled PCR probe derived from the MHV-A59 nucleocapsid protein gene.
Next, we next asked whether or not MHV-Brts31 is an RNA-negative mutant. First, we determined the amounts of viral RNA synthesized in cells infected with MHV-A59, MHV-Brts31, or MHV-Br31R1 and maintained at the permissive and nonpermissive temperatures of 33°C and 39.5°C, respectively. Figure 1B shows that at 33°C, all three viruses produce genome-sized and subgenome-sized virus RNA in the expected ratios, although the Northern blot suggests that MHV-Brts31 may synthesize slightly less viral RNA than does MHV-A59 or MHV-Br31R at the permissive temperature. At the nonpermissive temperature of 39.5°C, MHV-Brts31 did not synthesize a detectable level of RNA. In contrast, MHV-Br31R1 synthesizes amounts of virus RNA that are comparable to those of parental MHV-A59. These results confirm that MHV-Brts31 is an RNA-negative mutant.
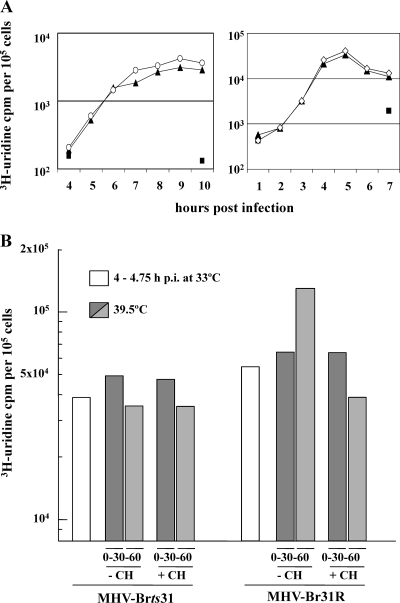
In order to look more closely at the RNA synthesis phenotype of MHV-Brts31 and MHV-Br31R1, we next analyzed the kinetics of viral RNA synthesis in cells infected with MHV-A59, MHV-Brts31, or MHV-Br31R1 and maintained at the permissive and nonpermissive temperatures. Figure 2A (left panel) shows that, at the permissive temperature, the kinetics and levels of virus RNA synthesis in 17Cl-1 cells infected with MHV-Brts31 and MHV-A59 are very similar. Figure 2A (right panel) shows that the kinetics and levels of RNA synthesis are also similar when cells infected with MHV-Br31R1 and MHV-A59 are maintained at the nonpermissive temperature, although, as expected, the peak of RNA synthesis is earlier at the higher temperature. We have also confirmed that the kinetics and levels of virus RNA synthesis in MHV-Brts31-infected cells maintained at the nonpermissive temperature cannot be distinguished from those of mock-infected cells and that cells infected with MHV-Br31R1 and MHV-A59 are indistinguishable in terms of virus RNA synthesis at the permissive temperature (data not shown). These results show that MHV-Brts31 has near-normal levels of total RNA synthesis at the permissive temperature. They also suggest that MHV-Br31R1 is most likely to be a true revertant. To show definitively whether or not the kinetics and levels of viral RNA synthesis in MHV-Brts31-infected cells at the permissive temperature are identical to those of MHV-A59-infected cells will require a more detailed analysis, possibly using different multiplicities of infection and a more accurate measure of RNA levels, e.g., quantitative RT-PCR (qRT-PCR).
FIG. 2.
(A) RNA synthesis phenotype of the MHV-Brts31 mutant. RNA synthesis was determined using 1-h pulse-labels with [3H]uridine in the presence of actinomycin D, given to mock (▪)-, MHV-A59 (▴)-, and MHV-Brts31 (○)-infected cells 4 to 10 h postinfection at 33°C (left panel) or mock (▪)-, MHV-A59 (▴)-, and MHV-Br31R1 (⋄)-infected cells 1 to 7 h postinfection at 39.5°C (right panel). Duplicate samples of 5 × 104 cells were precipitated with trichloroacetic acid, and the incorporated radioactivity was determined. The duplicates were reproducible to within 2% of each other. (B) Temperature sensitivity of RNA synthesis in MHV-Brts31 and MHV-Br31R1 virus-infected cells. RNA synthesis was determined using 30-min pulse-labels with [3H]uridine in the presence of actinomycin D, with or without the addition of cycloheximide (CH), after shifting the incubation temperature of MHV-Brts31- or MHV-Br31R1-infected cells from 33°C to 39.5°C at 4.75 h postinfection (p.i.). Dark gray bars, 0- to 30-min pulse; light gray bars, 30- to 60-min pulse. Triplicate samples of 5 × 104 cells were precipitated with trichloroacetic acid, and the incorporated radioactivity was determined. The triplicates were reproducible to within 2% of each other.
A major advantage of using ts mutants is that they can be analyzed by temperature shift protocols, i.e., by allowing them to gain function at the permissive temperature, shifting them to the nonpermissive temperature, and then determining if the function is lost. Thus, we determined the effect of temperature shift on the levels of total viral RNA synthesis in MHV-Brts31- and MHV-Br31R1-infected 17Cl-1 cells. To do this, infections were initiated at 33°C and infected cells were incubated until RNA synthesis reached about 20% of the maximum rate, which is approximately 4 to 5 h postinfection. At this time, one set of cells were pulse-labeled with [3H]uridine in the presence of actinomycin D. Another set of virus-infected cells were shifted to 39.5°C and pulse-labeled for 30 min at 0 and 30 min after shifting to the nonpermissive temperature. The cells were pulse-labeled with [3H]uridine in the presence of actinomycin D and in the presence or absence of 100 μg/ml of cycloheximide. It should be noted that at 5 h postinfection the virus RTC produces mainly (>90%) plus-strand RNA (38).
The results are shown in Fig. 2B. The pulse-labeling prior to temperature shift shows again that MHV-Brts31- and MHV-BR31R1-infected cells have comparable levels of RNA synthesis. After temperature shift, total RNA synthesis increased in MHV-Br31R1-infected cells in the absence of cycloheximide. This reflects the amplification of virus RTCs and the increased metabolic rate at higher temperatures. In the presence of cycloheximide, the activity fails to increase, but after shift, significant activity remains. We conclude that the MHV-Br31R1 replicase-transcriptase activity formed before temperature shift is relatively resistant to higher temperature. In MHV-Brts31-infected cells, there is also significant RNA synthesis activity after temperature shift, irrespective of the presence or absence of cycloheximide. We interpret this to mean that for MHV-Brts31, preexisting complexes are also relatively resistant and continue to synthesize plus-strand RNA at the nonpermissive temperature. To decide whether or not RTCs in MHV-Brts31-infected cells are able to synthesize minus-strand RNA at the nonpermissive temperature will require further experiments.
Complementation analysis.
Previous work by Sawicki et al. (40) has divided a large panel of RNA-negative, ts mutants of MHV into four complementation groups. We crossed MHV-Brts31 with at least one virus from each of these known complementation groups using a classical complementation assay. The results are shown in Table 1. Basically, MHV-Brts31 was able to complement all of the mutant viruses tested from each complementation group, with complementation indices (CI) of between 28 and 638. This contrasts with a CI of 1.8 obtained for a cross involving mutants in the same complementation group (MHV-Albts6, CG1, versus MHV-Albts16, CG1) and a CI of 240 for a cross involving mutants from different complementation groups (MHV-Albts16, CG1, versus MHV-Albts22, CG2). These results indicate that MHV-Brts31 forms the first member of a new complementation group and defines a new MHV replicase gene cistron, which we wish to provisionally designate cistron 0.
TABLE 1.
Complementation analysis of MHV-Brts31 and MHV-Brtsc31a
| Cross | CI |
|
|---|---|---|
| Expt 1 | Expt 2 | |
| MHV-Albts6 (C1) × MHV-Albts16 (C1) | 1.83 | 0.61 |
| MHV-Albts6 (C1) × MHV-Albts22 (C2) | ND | 705 |
| MHV-Albts16 (C1) × MHV-Albts22 (C2) | 240 | ND |
| MHV-Brts31 × MHV-Albts6 (C1) | 500 | ND |
| MHV-Brts31 × MHV-Albts16 (C1) | 28 | ND |
| MHV-Brts31 × MHV-Albts22 (C2) | 130 | ND |
| MHV-Brts31 × MHV-Albts17 (C4) | 267 | ND |
| MHV-Brts31 × MHV-Wüts36 (C6) | 638 | ND |
| MHV-Brtsc31 × MHV-Albts6 (C1) | ND | 27 |
| MHV-Brtsc31 × MHV-Albts16 (C1) | ND | 33 |
| MHV-Brtsc31 × MHV-Albts22 (C2) | ND | 717 |
| MHV-Brtsc31 × MHV-Albts17 (C4) | ND | 488 |
| MHV-Brtsc31 × MHV-Wüts36 (C6) | ND | 710 |
MHV-Brts31 and MHV-Brtsc31 were crossed with viruses from each of the four known complementation groups. The complementation indices (CI) for two independent experiments are shown. ND, cross not done. An index greater than 2 is taken as indicative of complementation (40).
Identification of the mutation responsible for the ts phenotype of MHV-Brts31.
Thiel et al. have shown that MHV replicase gene products suffice for subgenome-length mRNA synthesis (51). We reasoned, therefore, that MHV-Brts31 would have a mutation somewhere in the replicase gene, and to identify this mutation, we sequenced the entire replicase genes of both MHV-Brts31 and MHV-Br31R1 (data not shown; sequence available on request). This comparative analysis identified a single coding mutation, G to A at nucleotide 4131 (codon 1307 in ORF1a, amino acid 1307 in pp1a), which resulted in a conservative replacement of methionine-475 with isoleucine in nsp3. The methionine residue at position 475 in the MHV nsp3 is conserved among group 2a coronaviruses (e.g., MHV, bovine coronavirus [BCoV], and human coronavirus OC43) but is not conserved in other coronaviruses. Both MHV-A59 and the revertant MHV-Br31R1 have a methionine at this position, showing that MHV-Br31R1 is a true revertant. The numbering system used was that assigned to our infectious clone of the MHV-A59 genome (GenBank accession number AY700211).
In order to confirm that G4131 to A is the mutation responsible for the ts phenotype of MHV-Brts31, we used a vaccinia virus-based reverse genetics system to produce a recombinant virus, MHV-Brtsc31, that has a single G-to-A nucleotide exchange at position 4131 compared to the parental recombinant MHV-A59, which we call MHV-inf-1. MHV-Brtsc31 was rescued and plaque purified three times at 33°C, and a passage 2 virus stock was obtained. Sequence analysis of RNA from this stock of virus confirmed the presence of the G4131-to-A mutation (data not shown). At 33°C, the MHV-Brtsc31 stock had a titer of 6 × 108 PFU/ml, and at 39.5°C, the stock had a titer of 1.0 × 104 PFU/ml, which is an EOP of 1.7 × 10−5. MHV-Brtsc31 also failed to form plaques at 37°C (Fig. 1A, right panel). One of the plaques that formed in cells infected with MHV-Brtsc31 and maintained at the nonpermissive temperature of 39.5°C was picked and designated MHV-Br31cR1. The plaque size and morphology of MHV-Brtsc31 and MHV-Br31cR1 at 33°C and those of MHV-Br31cR1 at 39.5°C were identical to those of the parental MHV-inf-1 (Fig. 1A, right panel). Sequence analysis of RNA isolated from 17Cl-1 cells infected with a passage 2 stock of MHV-Br31cR1 showed that it was, indeed, a true revertant (data not shown). Next, we determined the amounts of viral RNA synthesized in cells infected with MHV-inf-1 and MHV-Brtsc31 and maintained at the permissive and nonpermissive temperatures of 33°C and 39.5°C, respectively. Figure 1C shows that at 33°C both viruses produce genome-sized and subgenome-sized virus RNA in the expected ratios. At the nonpermissive temperature of 39.5°C, MHV-Brtsc31 did not synthesize a detectable level of RNA. Finally, we repeated the same complementation analysis described above using MHV-Brtsc31 (Table 1). Again, the ts mutant virus was able to complement all of the mutant viruses tested from each complementation group, with complementation indices (CI) of between 27 and 717. Taken together, these results confirm that a single G4131-to-A mutation is responsible for the ts phenotype of MHV-Brts31, conclusively locate cistron 0 in ORF1a, and show that nsp3 has, at least, one essential role in MHV RNA synthesis.
Analysis of replicase polyprotein processing in MHV-Brtsc31- and MHV-31cR1-infected cells.
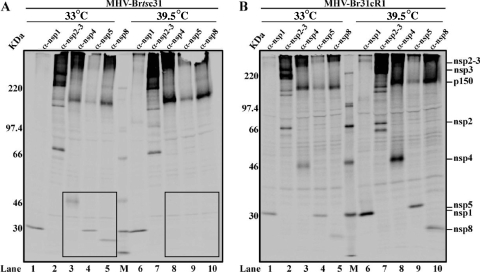
To determine if proteolytic processing of the replicase polyprotein(s) is affected in MHV-Brtsc31- and MHV-Br31cR1-infected cells at the nonpermissive temperature, we did temperature shift experiments with metabolic labeling and immunoprecipitation using nsp-specific antibodies (Fig. 3). As described in Materials and Methods, 17Cl-1 cells were infected with either MHV-Brtsc31 or MHV-Br31cR1 at 33°C and incubated for 4 h. At this time, one set of virus-infected cells was shifted to 39.5°C and a duplicate set was left at 33°C. All the virus-infected cells were then radiolabeled with 35S-trans label from 4.5 to 6.5 h postinfection. At the end of the radiolabeling, whole-cell lysates were prepared, the viral nsp's were immunoprecipitated with nsp-specific antisera, and the products were analyzed by SDS-PAGE.
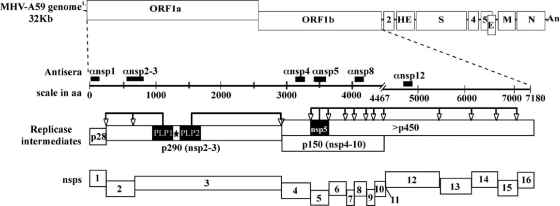
FIG. 3.
Regions recognized by MHV nsp-specific antisera and location of the MHV-Brts31 M475I mutation. The MHV replicase gene comprises two ORFs, ORF1a and ORF1b, connected by a (−1) ribosomal frameshift, which encode the viral replicase polyproteins. The replicase polyproteins are processed by papain-like cysteine protease activities 1 and 2 (PLP1 and PLP2, respectively, in nsp3) and the 3C-like cysteine protease activity (3CLpro in nsp5) to generate intermediates and mature nonstructural proteins (nsp's), which assemble to generate functional replication-transcription complexes. The site of the ts lesion in the MHV-Brts31 nsp3 open reading frame is indicated by the star. Domains recognized by the polyclonal antibodies used in the immunoprecipitation assays are indicated by black bars.
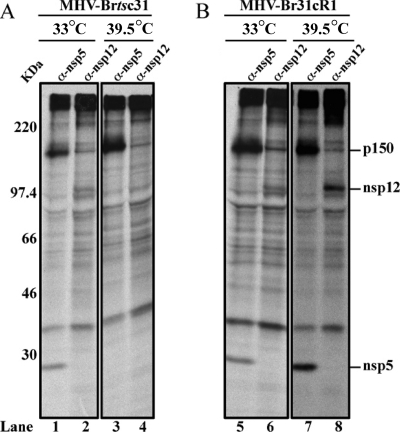
Our first experiment (Fig. 4) showed that, in MHV-Brtsc31-infected cells incubated at the nonpermissive temperature, compared to the permissive temperature, there is a major disruption of replicase polyprotein processing with respect to the processing of the pp1a p150 intermediate to its products, nsp4, nsp5 and nsp8 (compare boxed areas in Fig. 4A, lanes 3 to 5, with those in lanes 8 to 10). This disruption of p150 processing was not seen in cells infected with MHV-Br31cR1 (Fig. 4B, lanes 3 to 5 and lanes 8 to 10). In contrast, there was no obvious impairment of the processing of nsp1, nsp2, and nsp3 in the MHV-Brtsc31-infected cells at the nonpermissive temperature (Fig. 4A, lanes 1 and 2 and lanes 6 and 7). Also, there was no temperature-sensitive effect on nsp1, nsp2, and nsp3 processing in MHV-Br31cR1-infected cells at either permissive or nonpermissive temperatures (Fig. 4B, lanes 1 and 2 and lanes 6 and 7). These results indicate that, at the nonpermissive temperature, the replacement of methionine-475 with isoleucine in nsp3 of MHV-Brtsc31 is affecting nsp5-mediated processing of pp1a and, more specifically, the processing of p150 to generate nsp4, nsp5, and nsp8. These results were unexpected and the first demonstration of a relationship between nsp3 function and pp1a processing by the 3CLpro cysteine protease activity of nsp5.
FIG. 4.
Analysis of proteolytic products generated in MHV-Brtsc31 and MHV-Br31cR1 virus-infected cells at permissive and nonpermissive temperatures. Newly synthesized proteins were radiolabeled with 35S-trans label in MHV-Brtsc31- (A) or MHV-Br31cR1-infected cells (B) at either the permissive (33°C) or nonpermissive (39.5°C) temperature and subjected to immunoprecipitation with polyclonal antisera specific to nsp1 (lanes 1 and 6), nsp2 and nsp3 (lanes 2 and 7), nsp4 (lanes 3 and 8), nsp5 (lanes 4 and 9), and nsp8 (lanes 5 and 10). The products were analyzed by 5 to 12.5% SDS-PAGE and visualized by autoradiography. 14C molecular mass markers (GE Healthcare) are shown in lanes M.
In addition to mediating the proteolytic processing of the pp1a polypeptide, nsp5 also mediates the processing of the ORF1b-encoded region of pp1ab, i.e., the production of nsp12 to nsp16. To see if the replacement of methionine-475 with isoleucine in nsp3 of MHV-Brtsc31 has any effect on pp1ab processing, we repeated the experiment described above using an antiserum specific for nsp12, the viral RdRp. The results shown in Fig. 5 lead us to conclude that the processing of nsp12 is also impaired at the nonpermissive temperature compared to the permissive temperature in cells infected with MHV-Brtsc31 but not in MHV-Br31cR1-infected cells (Fig. 5, lanes 2 and 4 and lanes 6 and 8). As a control, we once again demonstrated the impaired processing of nsp5 in MHV-Brtsc31-infected cells at the nonpermissive temperature (Fig. 5, lanes 1 and 3 and lanes 5 and 7).
FIG. 5.
Analysis of proteolytic products generated in MHV-Brtsc31 and MHV-Br31cR1 virus-infected cells at permissive and nonpermissive temperatures. Newly synthesized proteins were radiolabeled with 35S-trans label in MHV-Brtsc31- (A) or MHV-Br31cR1-infected cells (B) at either the permissive (33°C) or nonpermissive (39.5°C) temperature and subjected to immunoprecipitation with polyclonal antisera specific to nsp5 (odd-numbered lanes) and nsp12 (even-numbered lanes). The products were analyzed by 5 to 12.5% SDS-PAGE and visualized by autoradiography.
Analysis of double-membrane vesicles (DMVs) in MHV-Brts31 and MHV-Br31R1 virus-infected cells.
Since coronavirus RNA synthesis occurs upon modified membranes (14, 16, 45), we next investigated if the ts lesion in nsp3 of MHV-Brts31 had any effect on the number of DMVs in virus-infected cells. Two sets of 17Cl-1 cells were initially infected with MHV-Brts31and MHV-Br31R1 at 33°C. At 3.5 h postinfection, one set of infected cells was shifted to 39.5°C. At 5.5 h postinfection, the cells were harvested and processed for transmission electron microscopy (TEM) analysis as described in Materials and Methods.
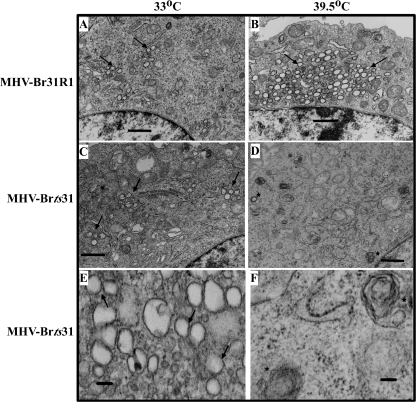
TEM analysis of MHV-infected cells reveals that DMVs can be visualized as dark-ringed vesicles in the cytoplasm adjacent to the nucleus (16). In cells infected with MHV-Br31R1, the typical accumulation of DMV structures (indicated by arrows) was evident after incubation at the permissive (Fig. 6A) and nonpermissive (Fig. 6B) temperatures, with an increased number of DMVs in the cells incubated at 39.5°C. In contrast, we found that although the number of DMVs in MHV-Brts31-infected cells incubated at the permissive temperature (Fig. 6C) was similar to the number found in revertant virus-infected cells, the number of DMVs in MHV-Brts31-infected cells that had been shifted to the nonpermissive temperature was reduced (Fig. 6D and F). We also noted a small number of aberrant vesicles at the nonpermissive temperature (indicated by asterisks in Fig. 6D and F) that stained with uranyl acetate and Reynolds' lead citrate but do not have the typical double-membrane morphology. Further work will be necessary to determine if these are possibly residual DMVs.
FIG. 6.
Analysis of double-membrane vesicles in MHV-Brts31 and MHV-Br31R1 virus-infected cells at permissive and nonpermissive temperatures. 17Cl-1 cells were infected with either MHV-Brts31 or MHV-Br31R1 at an MOI of 20.0 and incubated at 33°C for 5.5 h (A, C, and E) or initially incubated at 33°C and then shifted to 39.5°C at 3.5 to 5.5 h postinfection (B, D, and F). At 5.5 h postinfection, cells were harvested and processed for TEM analysis. DMVs can be visualized by TEM as dark-ringed vesicles in the cytoplasm of MHV-infected cells (arrows). Asterisks denote rare aberrant vesicles detected at the nonpermissive temperature in MHV-Brts31-infected cells. Bars, 1 μm (A to D) or 0.1 μm (E and F).
Taken together, these results indicate that the replacement of methionine-475 with isoleucine in nsp3 of MHV-Brts31 impairs both 3CLpro-mediated processing of replicase polyproteins and the assembly or stability of DMVs at the nonpermissive temperature.
DISCUSSION
Previously, we have identified four complementation groups of MHV RNA-negative ts mutants. Three of these groups, groups II, IV, and VI, were defined by ts viruses with mutations in the ORF1b-encoded proteins nsp12, nsp14, and nsp16, respectively, while the fourth group, group I, was defined by ts viruses with mutations in three of the ORF1a-encoded proteins, nsp4, nsp5, and nsp10. This led us to the hypothesis that each of the five MHV ORF1b cleavage products (nsp12 to nsp16) may represent distinct cistrons but that the majority of the ORF1a cleavage products will function as a single cistron. More specifically, it was proposed that cistron I would encompass the regions of ORF1a that encode nsp4 to nsp10.
The complementation analysis of MHV-Brts31 described here identifies a new cistron (cistron 0) in the MHV replicase gene. Furthermore, the identification of the mutation responsible for the ts phenotype of MHV-Brts31 (G4131 to A) shows that cistron 0 encompasses, at least, the region encoding nsp3. For a number of reasons, we suggest that cistron 0 will not extend beyond the coding region of nsp3. First, as mentioned above, extensive deletion of sequences encoding the carboxyl region of nsp1 or, indeed, the entire coding region of nsp2 has an attenuating effect but is not lethal for MHV replication (13, 18). Second, although it has been shown that an RNA stem-loop in the bovine coronavirus (BCoV) 5′ proximal coding region of nsp1 is essential for RNA replication (4) (which would explain the lethality of deleting the entire nsp1 coding region), the majority of evidence suggests that the coronavirus nsp1 protein primarily has a role in the virus-host interaction (8, 24, 25, 29) or regulatory, rather than essential, functions in RNA synthesis (19). Although the possibility that other ORF1a- or ORF1b-encoded nsp's (i.e., nsp6, nsp7, nsp8, nsp9, nsp11, nsp13, or nsp15) could fall into the same complementation group as MHV-Brts31 cannot be ruled out, it would, in our opinion, be difficult to reconcile this with our current view of MHV replicase gene expression and function.
Our analysis of replicase polyprotein processing and DMV structures in MHV-Brtsc31-infected cells at the nonpermissive temperature allows us to reach two important conclusions. First, the ts lesion of MHV-Brtsc31 is affecting nsp5-mediated processing of pp1a and pp1ab, specifically, the processing of p150 to generate nsp4, nsp5, and nsp8 and the processing of pp1ab to generate nsp12. However, there is no effect on the processing of the replicase polyproteins to produce nsp1, nsp2, and nsp3. We interpret this to mean one of two things. Either there is an interaction between nsp3 and another component of the RTC, which is necessary to facilitate nsp5-mediated polyprotein processing, or the normal structure or function of nsp3 is needed to provide pp1a/pp1ab substrates upon which nsp5 can act. In this respect, the precedent for protein-protein interactions within the coronavirus RTC that result in a modification of nsp5 activity has been given by Donaldson et al. (9), who have suggested that nsp10 may act as a cofactor for 3CLpro-mediated processing.
Second, the decrease in the number of DMV structures in MHV-Brts31-infected cells at the nonpermissive temperature indicates that the assembly or stability of the RTC is impaired. A quantitative analysis of DMV numbers in cells infected with MHV-Brts31, together with experiments to determine the turnover rate of DMV structures in MHV-A59-infected cells, will be needed to determine the precise defect in DMV biogenesis. However, our data suggest that preformed RTCs are able to continue plus-strand RNA synthesis at the nonpermissive temperature, and it seems more likely that the failure to correctly process the replicase polyproteins would affect assembly rather than stability of the RTC. On the other hand, an earlier analysis of cells infected with the RNA-negative, ts mutant MHV-Albts6 (6) has shown that DMV formation and replicase polyprotein processing are not always linked. In this case, DMV formation was severely impaired even though replicase polyprotein processing appeared to be normal. The finding that both MHV-Brts31 and MHV-Albts6 fail to accumulate DMVs at the nonpermissive temperature is consistent with the idea that DMVs either are required for coronavirus RNA synthesis or are generated as a result of viral RNA synthesis.
In the light of these conclusions, the obvious challenge is to identify the specific function or functions of nsp3 that are impaired at the nonpermissive temperature in MHV-Brts31/MHV-Brtsc31-infected cells and to relate this to the phenotype of the mutant virus. In this context, it is important to note that the data do not allow us to discern whether the phenotypic changes that we observe are directly or indirectly related to the causal mutation. The M475I mutation, which is located, essentially, at the junction of the PLP1 and X domains of nsp3, could lead to multiple effects that may then be separately related to the different phenotypic manifestations. Indeed, Graham and Denison (17) have shown that recombinant MHV mutants with changes in the nsp1-to-nsp3 protein coding region may have a variety of different phenotypes with respect to replicase polyprotein processing, replication, and RNA synthesis.
In general, we can offer three possible ways in which the M475I mutation may exert its effect. First, it is possible that one or more of the enzymatic activities associated with nsp3 are rendered inactive at the nonpermissive temperature. For a number of reasons, we consider this the least likely explanation. For example, we consider it unlikely that inactivation of PLP1 activity is responsible for the phenotype of MHV-Brts31 (or MHV-Brtsc31). Our data do not provide any evidence for a defect in nsp1, nsp2, and nsp3 processing, and previous studies have shown that MHV PLP1 activity is not essential for virus replication in cell culture (17). Nevertheless, mutants of this sort are highly impaired on recovery and we cannot rule out that the inactivation of PLP1 may somehow be involved. Similarly, although it has been shown that the HCoV-229E PLP2 activity is essential for replication (58), our results do not indicate any defect in this activity at the nonpermissive temperature in MHV-Brtsc31-infected cells. A second type of function associated with nsp3 is the ADP-ribose 1"-phosphatase and poly(ADP-ribose) binding activities of the X domain. Again, we consider it unlikely that inactivation of one or both of these activities can account for the phenotype of MHV-Brts31 because it has been shown previously that mutation of the active site of the HCoV-229E nsp3 X domain has no significant effect on virus RNA synthesis (36). The inactivation of nsp3-associated deubiquitination activity (57) and other, as-yet-unidentified enzymatic activities remains a possibility.
A second and perhaps more likely explanation is that, at the nonpermissive temperature, interactions between nsp3 and cellular membranes are compromised. Although there is still some uncertainty, the current model of MHV nsp3 topology predicts at least 2 transmembrane helices (26, 31, 32). This would place the bulk of nsp3 on the same membrane face occupied by nsp5 and the nsp12 to nsp16 proteins processed from pp1ab. Kanjanahaluethai et al. (26) recently showed that removing the nsp3 transmembrane domain abolished PLP2 activity, presumably by preventing the interaction of PLP2 with its substrate. If the MHV-Brts31/MHV-Brtsc31 nsp3 protein is no longer stably inserted into the cellular membrane at the nonpermissive temperature, then this could compromise the integrity of the whole replicase-transcriptase complex. The same might be true if the replacement of methionine-475 with isoleucine in nsp3 had a global effect upon the structure of nsp3, or even the pp1a and pp1ab polyproteins as a whole. Our current view of nsp3 as a large protein with a functional and structural domain architecture might argue against a global effect (31), and the position of this substitution, which is essentially predicted to be in a flexible region separating conserved functional domains, would be more consistent with the idea that intramolecular nsp3 domain interactions are affected at the nonpermissive temperature.
The third possibility is that, at the nonpermissive temperature, nsp3 protein-protein, or perhaps nsp3 protein-RNA, interactions within the RTC are compromised. This could be manifested as an inability to assemble new complexes or as the assembly of a nonfunctional complex. Putative metal-binding domains are located in the PLP, ZF, and Y domains of the MHV nsp3, and they may represent zinc-finger motifs that are involved in the recognition of other protein components of the RTC. The MHV nsp3 protein also contains a putative nucleic acid-binding domain. Neuman et al. (31) have shown that the corresponding SARS-coronavirus NAB domain of nsp3 has both single-stranded nucleic acid-binding activity and double-stranded unwinding activity, and they suggest that these activities are consistent with a nucleic acid chaperone function. It is easy to see how the disruption of MHV nsp3 protein-protein or nsp3 protein-RNA interactions could prevent the assembly of a functional RTC and result in the ts phenotype that we observe for MHV-Brts31 and MHV-Brtsc31.
Work is under way to investigate some of these possibilities, and we are confident that RNA-negative ts mutants such as MHV-Brts31 will prove to be valuable tools for unraveling the complexities of coronavirus RNA synthesis.
Acknowledgments
This work was supported by the Wellcome Trust Programme grant 06548 to S.G.S. and NIH Public Health Service research grants AI 060915 and HHSN2662040035C to S.C.B.
We thank Volker Thiel for the pGPT-nsp3(N) plasmid.
Footnotes
Published ahead of print on 28 July 2010.
REFERENCES
- 1.Baliji, S., S. A. Cammer, B. Sobral, and S. C. Baker. 2009. Detection of nonstructural protein 6 in murine coronavirus-infected cells and analysis of the transmembrane topology by using bioinformatics and molecular approaches. J. Virol. 83:6957-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brockway, S. M., X. T. Lu, T. R. Peters, T. S. Dermody, and M. R. Denison. 2004. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J. Virol. 78:11551-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, C. G., K. S. Nixon, S. D. Senanayake, and D. A. Brian. 2007. An RNA stem-loop within the bovine coronavirus nsp 1 coding region is a cis-acting element in DI RNA replication. J. Virol. 81:7716-7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., H. Cai, J. Pan, N. Xiang, P. Tien, T. Ahola, and D. Guo. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 106:3484-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clementz, M. A., A. Kanjanahaluethai, T. E. O'Brien, and S. C. Baker. 2008. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375:118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coley, S. E., E. Lavi, S. G. Sawicki, L. Fu, B. Schelle, N. Karl, S. G. Siddell, and V. Thiel. 2005. Recombinant mouse hepatitis virus strain A59 from cloned, full-length cDNA replicates to high titers in vitro and is fully pathogenic in vivo. J. Virol. 79:3097-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornillez-Ty, C. T., L. Liao, J. R. Yates III, P. Kuhn, and M. J. Buchmeier. 2009. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 83:10314-10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson, E. F., R. L. Graham, A. C. Sims, M. R. Denison, and R. S. Baric. 2007. Analysis of murine hepatitis virus strain A59 temperature sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J. Virol. 81:7086-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dye, C., and S. G. Siddell. 2005. Genomic RNA sequence of feline coronavirus strain FIPV WSU-79/1146. J. Gen. Virol. 86:2249-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckerle, L. D., X. Lu, S. M. Sperry, L. Choi, and M. R. Denison. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 81:12135-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egloff, M. P., H. Malet, A. Putics, M. Heinonen, H. Dutartre, A. Frangeul, A. Gruez, V. Campanacci, C. Cambillau, J. Ziebuhr, T. Ahola, and B. Canard. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 80:8493-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadlage, M. J., R. L. Graham, and M. R. Denison. 2008. Murine coronaviruses encoding nsp2 at different genomic loci have altered replication, protein expression, and localization. J. Virol. 82:11964-11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, R. L., and M. R. Denison. 2006. Replication of murine hepatitis virus is regulated by papain-like proteinase 1 processing of nonstructural proteins 1, 2, and 3. J. Virol. 80:11610-11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham, R. L., J. S. Sparks, L. D. Eckerle, A. C. Sims, and M. R. Denison. 2008. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 133:88-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustin, K. M., B. J. Guan, A. Dziduszko, and D. A. Brian. 2009. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J. Virol. 83:6087-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagemeijer, M. C., M. H. Verheije, M. Ulasli, I. A. Shaltiel, L. A. de Vries, F. Reggiori, P. J. Rottier, and C. A. de Haan. 2010. Dynamics of coronavirus replication-transcription complexes. J. Virol. 84:2134-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imbert, I., J. C. Guillemot, J. M. Bourhis, C. Bussetta, B. Coutard, M. P. Egloff, F. Ferron, A. E. Gorbalenya, and B. Canard. 2006. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25:4933-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U. S. A. 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamitani, W., C. Huang, K. Narayanan, K. G. Lokugamage, and S. Makino. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 16:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamitani, W., K. Narayanan, C. Huang, K. Lokugamage, T. Ikegami, N. Ito, H. Kubo, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U. S. A. 103:12885-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanjanahaluethai, A., Z. Chen, D. Jukneliene, and S. C. Baker. 2007. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 361:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerr, S. M., and G. L. Smith. 1991. Vaccinia virus DNA ligase is nonessential for virus replication: recovery of plasmids from virus-infected cells. Virology 180:625-632. [DOI] [PubMed] [Google Scholar]
- 28.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law, A. H., D. C. Lee, B. K. Cheung, H. C. Yim, and A. S. Lau. 2007. Role for nonstructural protein 1 of severe acute respiratory syndrome coronavirus in chemokine dysregulation. J. Virol. 81:416-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 103:5108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neuman, B. W., J. S. Joseph, K. S. Saikatendu, P. Serrano, A. Chatterjee, M. A. Johnson, L. Liao, J. P. Klaus, J. R. Yates III, K. Wuthrich, R. C. Stevens, M. J. Buchmeier, and P. Kuhn. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 82:5279-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oostra, M., M. C. Hagemeijer, M. van Gent, C. P. Bekker, E. G. te Lintelo, P. J. Rottier, and C. A. de Haan. 2008. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 82:12392-12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oostra, M., E. G. te Lintelo, M. Deijs, M. H. Verheije, P. J. Rottier, and C. A. de Haan. 2007. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 81:12323-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78:9977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putics, A., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 79:12721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawicki, S. G., and D. L. Sawicki. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schelle, B., N. Karl, B. Ludewig, S. G. Siddell, and V. Thiel. 2006. Nucleocapsid protein expression facilitates coronavirus replication. Adv. Exp. Med. Biol. 581:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siddell, S., J. Ziebuhr, and E. J. Snijder. 2005. Coronaviruses, toroviruses, and arteriviruses, p. 823-856. In B. W. J. Mahy and V. Ter Meulen (ed.), Topley & Wilson's microbiology and microbial infections. Virology. Hodder Arnold, London, United Kingdom.
- 44.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sturman, L. S., and K. K. Takemoto. 1972. Enhanced growth of a murine coronavirus in transformed mouse cells. Infect. Immun. 6:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su, D., Z. Lou, F. Sun, Y. Zhai, H. Yang, R. Zhang, A. Joachimiak, X. C. Zhang, M. Bartlam, and Z. Rao. 2006. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J. Virol. 80:7902-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.te Velthuis, A. J., J. J. Arnold, C. E. Cameron, S. H. van den Worm, and E. J. Snijder. 2010. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 38:203-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 51.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 75:6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thiel, V., A. Rashtchian, J. Herold, D. M. Schuster, N. Guan, and S. G. Siddell. 1997. Effective amplification of 20-kb DNA by reverse transcription PCR. Anal. Biochem. 252:62-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ulasli, M., M. H. Verheije, C. A. de Haan, and F. Reggiori. 2010. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 12:844-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Brunn, A., C. Teepe, J. C. Simpson, R. Pepperkok, C. C. Friedel, R. Zimmer, R. Roberts, R. Baric, and J. Haas. 2007. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One 2:e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhai, Y., F. Sun, X. Li, H. Pang, X. Xu, M. Bartlam, and Z. Rao. 2005. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng, D., G. Chen, B. Guo, G. Cheng, and H. Tang. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 18:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziebuhr, J., B. Schelle, N. Karl, E. Minskaia, S. Bayer, S. G. Siddell, A. E. Gorbalenya, and V. Thiel. 2007. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 81:3922-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuniga, S., J. L. Cruz, I. Sola, P. A. Mateos-Gomez, L. Palacio, and L. Enjuanes. 2010. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 84:2169-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]